Abstract
Adenosine diphosphate (ADP)-ribosylation factor-like tumour suppressor gene 1(ARLTS1) might be associated with an increased risk of several types of familial cancers. However, previous studies have shown that cancer susceptibility is not completely consistent with ARLTS1 polymorphisms, and the precise mechanism remains unknown. Therefore, we conducted a meta-analysis of case-control studies by searching the PubMed, Embase, OVID, Science Direct and Chinese National Knowledge Infrastructure (CNKI) databases. In total, 12 studies met the inclusion criteria and were included in this meta-analysis. Statistical analyses were performed using STATA 11.0 software. Overall, the Cys148Arg T > C variant significantly increased cancer risk (CC vs. TT: OR = 1.27, 95% CI = 1.15–1.41, P < 0.05). The stratification indicated that the Cys148Arg variant is significantly associated with sporadic cancer (CC vs. TT: OR = 1.36, 95% CI = 1.18–1.55) and familial cancer (CC vs. TT: OR = 1.26, 95% CI = 1.12–1.43). Trp149Stop, Pro131Leu, Ser99Ser and Leu132Leu were not correlated with cancer susceptibility. Based on these results, we demonstrated that the ARLTS1 Cys148Arg polymorphism is associated with an increased risk of sporadic cancer and familial cancer, and there were no associations between the other four SNPs (i.e., Trp149Stop, Pro131Leu, Ser99Ser and Leu132Leu) and cancer risk.
Similar content being viewed by others
Background
Adenosine diphosphate (ADP)-ribosylation factor-like tumour suppressor gene 1 (ARLTS1), which is also known as ADP-ribosylation factor-like protein 11 (ARL11), islocated in chromosomal region 13q14.3 and is a member of the Alternative Reading Frame (ARF) family of the RAS super family of small GTPases, which is involved in apoptotic signalling [1, 2]. ARLTS1 comprises two exons, its second exon encompasses the entire open reading frame andencodes 196 amino acid guanine-nucleotide-binding proteins that are critical components of several different eukaryotic vesicle trafficking pathways [3].
In 2005, Calin et al. found that chronic lymphocytic leukaemia was associated with a polymorphism in ARLTS1 [2]. Subsequently, ARLTS1 variants were shown to be associated with an increased risk of several types of sporadic and familial cancers, such as prostate cancer [4], ovarian cancer [5], breast cancer [6] and other cancers [7]. However, ARLTS1 variants did not exhibit a significantly increased risk of familial colorectal cancer [8]. Consequently, the results of these studies remain inconsistent. Hence, in the present study, a meta-analysis of all relevant case-control studies published before 1 November 2016 was performed to more precisely estimate the relationship between the ARLTS1 variants listed in Table 1 and the susceptibility to cancer.
Materials and methods
Literature search
Searches of the Medline, PubMed, Embase, Web of Science and China National Knowledge Infrastructure (CNKI) databases were performed (from January 2005 to November 2016). The following search words and their combinations were used: (“genetic polymorphism” or “polymorphism” or “SNP” or “gene mutation” or “genetic variants”) and (“cancer” or “melanoma” or “lymphocytic leukaemia” or “carcinoma” or “malignancy”) and “ARLTS1”. All identified studies were retrieved, and their bibliographies were reviewed to identify other relevant publications. Review articles were manually searched for additional studies. In the case of overlapping and republished studies, only the most recent study or the study with the largest sample size was selected for this meta-analysis.
Inclusion and exclusion criteria
The studies included in this meta-analysis met the following criteria: (a) the studies used a case-control design; (b) the studies evaluated the association between ARLTS1 variants (Cys148Arg, Trp149Stop, Pro131Leu, Ser99Ser and Leu132Leu) and cancer susceptibility; and (c) the studies had sufficient published data to calculate the odds ratio (OR) with a 95% confidence interval (CI). The primary reasons for the exclusion of studies were as follows: (a) no control population was included, (b) the studies contained overlapping data, and (c) the genotype frequency datawere not available.
Data extraction
Two investigators independently collected the data from all eligible publications according to the inclusion and exclusion criteria. Whenever disagreements occurred, a third investigator was consulted to resolve the dispute, and a final decision was made by the majority of the votes. The following data were extracted from each study: the first author’s last name, publication year, ethnicity of the study population, study design, cancer type, total number of cases and controls and the Hardy-Weinberg equilibrium (HWE) p-value. The different cancer types were classified as sporadic tumours or familial tumours. The source of the controls was stratified into hospital-based studies and population-based studies.
Quality assessment
The quality of the included publications was assessed using a quality assessment scale (Table 2), which was modified from previously published meta-analyses [9,10,11]. Two investigators assessed the quality of the studies according to the scale for quality assessment, and disagreements were resolved by discussion and consensus. The total scores ranged from 0 to 10. Scores of 0–4,5–8 and 9–10 were defined as low, moderate and high quality, respectively.
Statistical analysis
In our meta-analysis,we investigated the association between ARLTS1 polymorphisms and cancer risk according to a pooled OR with the corresponding 95% CIs. The stratification analysis was performed by type of cancer, control source and study quality. The significance of the pooled ORs was determined by a Z test, and P < 0.05 was considered statistically significant.
In our study, the HWE was tested using the χ 2 test (significance was set as P < 0.05). The heterogeneity of the studies was examined using the Q-test and the I2 test [12, 13]. If the between-study heterogeneity was significant (P < 0.05 for the Q-test and I2 > 50%), we used a random-effects model (DerSimonian-Laird method) [14]; otherwise, the fixed-effects model (Mantel-Haenszel method) was used [15]. The sensitivity was assessed by the omission of individual studies to determine the stability of the results in this meta-analysis. To assess the potential publication bias, a Begg’s funnel plot was generated to visually detect bias [16]. In addition, Egger’s test was also conducted to analyse the publication bias statistically [17]. All p-values were two-sided. All analyses were calculated using STATA Version 11.0 software.
Results
Characteristics of the included studies
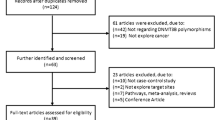
By searching the databases, 31 abstracts were collected according to the search criteria. Of these 31 studies, 11 studies were excluded after reviewing the titles and abstracts. Another 10 studies were excluded after reviewing the full texts. The study selection process is shown in Fig. 1. Finally, 10 studies met the inclusion criteria and were included in the meta-analysis. Of the 10 studies, one study [18] involved three cohorts, and, thus, each cohort was considered a single study. In total, 12 case-control studies [4, 6, 8, 18,19,20,21,22,23,24], evaluating the association between ARLTS1 polymorphisms and cancer susceptibility, were included in this meta-analysis. The characteristics of all studies are summarized in Table 3. Not all studies genotyped the SNPs included in this meta-analysis. Therefore, the number of included studies differs across the SNPs. All results are presented as the ORs (95% CI), p-value.
Quantitative analysis
Cys148Arg
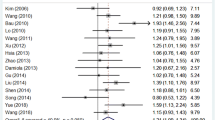
Among the 12 eligible studies, 11 studies included in this meta-analysis involved SNP C148R with 7152 cases and 6698 controls. Overall, when the 11 studies were pooled into the meta-analysis, the significant main effects on cancer risk were associated with ARLTS1 Cys148Arg (CC vs. TT: 1.27 (1.15–1.41), p = 0.000 Fig. 2; CC + TC vs. TT: 1.13 (1.02–1.27), p = 0.026 Fig. 3; CC vs. TC + TT: 1.16 (1.00–1.35), p = 0.045). The meta-analysis results are listed in Table 4. Furthermore, several stratified analyses were performed by control source, study quality and type of cancer (Table 4). Regarding the control source, the ARLTS1 Cys148Arg CC genotype increased the cancer risk in a population-based (CC vs. TT: 1.38 (1.22–1.55), p = 0.000; CC + TC vs. TT:1.16(1.03,1.30), p = 0.003; CC vs. TC + TT:1.26(1.07–1.49), p = 0.000), but not in a hospital-based setting. Regarding the study quality, the results were only significant in the high-quality studies (CC vs. TT: 1.35(1.16–1.57), p = 0.001; CC + TC vs. TT: 1.14 (1.00–1.31), p = 0.008). In addition, positive results were observed in both familial cancer (CC vs. TT: 1.26 (1.12–1.43), p = 0.000; CC + TC vs. TT: 1.15 (1.02–1.29), p = 0.021) and sporadic cancer (CC vs. TT: 1.36 (1.18–1.55), p = 0.000; CC + TC vs. TT: 1.13 (1.02–1.25), p = 0.016; CC vs. TC + TT: 1.22 (1.02–1.46), p = 0.030).
Trp149Stop, Pro131Leu, Ser99Ser and Leu132Leu
All studies were pooled in the meta-analysis. No significant results were observed for any of these SNPs (Trp149Stop GA vs. AA: 1.11 (0.66–1.86), p = 0.700; Pro131Leu CT vs. CC: 0.94 (0.81–1.08), p = 0.374; Ser99Ser AG vs. AA: 1.02 (0.86–1.20) p = 0.847; Leu132Leu GC vs. GG: 0.90 (0.58–1.39) p = 0.845), which are referred to in Table 5. Furthermore, the stratified analysis by familial and sporadic cancer revealed no association between these variants and cancer.
Sensitivity analysis and publication bias
Each study was evaluated in this meta-analysis individually to detect the influence of the individual data set on the pooled ORs. The results were not substantially altered, indicating that our results were stable and robust. Begg’s funnel plots were generated to assess the publication bias, and the shapes revealed no evidence of obvious asymmetry (Fig. 4). Egger’s test was performed based on the linear regression of the standard normal deviation against its precision, which was performed to test for the existence of publication bias. The results did not show any statistical evidence of publication bias for ARLTS1 Cys148Arg (p = 0.595 for CC vs. TT) (Fig. 5).
Discussion
ARLTS1 is a cancer-associated gene with notable tumour suppressor properties. ARLTS1 has been shown to induce apoptosis [25]. However, the precise functional properties of the ARLTS1 gene remain unclear, such as its access, interactions in the 13q14 region and the structure of the protein. The proposed mechanism is that the ARLTS1 gene produces proteins that affect acceptors of the immune system via transcription and translation, and then, endogenous inflammatory cells become activated to suppress tumour formation. The ARLTS1 gene may have a restraining effect on cancer development in lung [26] and ovarian cancer [5]. The expression of ARLTS1 is frequently down-regulated in prostate cancer [25] and chronic lymphocytic leukaemia samples [27]. The potential mechanisms leading to the down-regulated expression of ARLTS1 include promoter hypermethylation or loss of heterozygosity (LOH) in the gene region. The reduction or absence of ARLTS1 expression has been reported to contribute to DNA mutations with an LOH in breast cancer and a mutation with methylation in thyroid cancer [2]. This down-regulated expression immediately impacts immune responses, resulting in diminished apoptosis, reduced defence mechanisms and cancer progression. Additionally, the ARLTS1 polymorphism was not only associated with sporadic cancer but also with familial cancer [28]. Consequently, the association between ARLTS1 polymorphisms and cancer risk, particularly familial cancer, has recently become a high priority research topic.
To the best of our knowledge, the present study is the first systematic review to investigate the relationship between the ARLTS1 polymorphisms and cancer risk based on 12 case-control informative studies. Regarding Cys148Arg, we found that the SNP C148R polymorphism was significantly associated with cancer in all subjects. When a subgroup analysis was carried out based on the size of the total sample, we observed different associations with sporadic cancer and familial cancer, which was consistent with the result obtained by Calin et al [2]. Regarding Trp149Stop, Pro131Leu, Ser99Ser and Leu132Leu, however, we found no correlations with cancer risk. Therefore, we speculate that the ARLTS1 Cys148Arg CC genotype may significantly contribute to decreased apoptosis, thereby promoting the overall cancer incidence. However, the other variants may still have associations with cancer. These variants may have minor effects individually, but they are likely to contribute adequately in combination to lead to the failure of theimmune response. Therefore, gene-gene interactions should be considered in future studies to elucidate the precise role of ARLTS1.
Some limitations of this meta-analysis should be addressed. First, this study did not analyse the potential and gene-environment interactions due to a lack of original data, such as data involving environmental risk factors and genotypes. Second, the controls were not uniformly defined. Although most of the controls were selected mainly from healthy populations, some controls had a benign disease. In addition, our results were based on unadjusted estimates without adjusting for other risk factors, such as age, smoking status, drinking status, obesity and environmental factors. Therefore, a more precise analysis should be conducted if individual data were available. Although this study has several limitations, as previously stated, this study is the first to reveal the association between Cys148Arg and cancer risk and exclude the association between the other four SNPs (i.e., Trp149Stop, Pro131Leu, Ser99Ser and Leu132Leu) and cancer risk. Additionally, our study provides powerful evidence for future large-scale population-based cohort and case-control studies.
Conclusion
In summary, the ARLTS1 Cys148Arg polymorphism was associated with sporadic cancer risk and familial cancer risk. However, well-designed studies with larger sample sizes should be performed to confirm this association. Moreover, further studies estimating the effect of gene-gene and gene-environment interactions may eventually provide a better and more comprehensive understanding to clarify the role of ARLTS1 in the pathogenesis of cancer.
References
Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–6.
Calin GA, Trapasso F, Shimizu M, Dumitru CD, Yendamuri S, Godwin AK, Ferracin M, Bernardi G, Chatterjee D, Baldassarre G, Rattan S, Alder H, Mabuchi H, Shiraishi T, Hansen LL, Overgaard J, Herlea V, Mauro FR, Dighiero G, Movsas B, Rassenti L, Kipps T, Baffa R, Fusco A, Mori M, Russo G, Liu CG, Neuberg D, Bullrich F, Negrini M, Croce CM. Familial cancer associated with a polymorphism in ARLTS1. N Engl J Med. 2005;352:1667–76.
Yendamuri S, Trapasso F, Calin GA. ARLTS1 - a novel tumor suppressor gene. Cancer Lett. 2008;264:11–20.
Siltanen S, Wahlfors T, Schindler M, Saramaki OR, Mpindi JP, Latonen L, Vessella RL, Tammela TL, Kallioniemi O, Visakorpi T, Schleutker J. Contribution of ARLTS1 Cys148Arg (T442C) variant with prostate cancer risk and ARLTS1 function in prostate cancer cells. PLoS One. 2011;6:e26595.
Petrocca F, Iliopoulos D, Qin HR, Nicoloso MS, Yendamuri S, Wojcik SE, Shimizu M, Di Leva G, Vecchione A, Trapasso F, Godwin AK, Negrini M, Calin GA, Croce CM. Alterations of the tumor suppressor gene ARLTS1 in ovarian cancer. Cancer Res. 2006;66:10287–91.
Akisik E, Yazici H, Dalay N. ARLTS1, MDM2 and RAD51 gene variations are associated with familial breast cancer. Mol Biol Rep. 2011;38:343–8.
Masojc B, Mierzejewski M, Cybulski C, van de Wetering T, Debniak T, Gorski B, Jaworowska E, Tarnowska C, Lenner M, Scott RJ, Lubinski J. Cancer familial aggregation (CFA) and G446A polymorphism in ARLTS1 gene. Breast Cancer Res Treat. 2006;99:59–62.
Frank B, Hemminki K, Brenner H, Hoffmeister M, Chang-Claude J, Burwinkel B. ARLTS1 variants and risk of colorectal cancer. Cancer Lett. 2006;244:172–5.
Xiao XY, Wang XD, Zang DY. MMP1-1607 1G/2G polymorphism and lung cancer risk: a meta-analysis. Tumour Biol. 2012;33:2385–92.
Huang SX, Wu FX, Luo M, Ma L, Gao KF, Li J, Wu WJ, Huang S, Yang Q, Liu K, Zhao YN, Li LQ. The glutathione S-transferase P1 341C > T polymorphism and cancer risk: a meta-analysis of 28 case-control studies. PLoS One. 2013;8:e56722.
Salanti G, Amountza G, Ntzani EE, Ioannidis JP. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet. 2005;13:840–8.
Chen C, Li S, Lu X, Tan B, Huang C, Qin L. High resolution melting method to detect single nucleotide polymorphism of VKORC1 and CYP2C9. Int J Clin Exp Pathol. 2014;7:2558–64.
Czogalla KJ, Biswas A, Rost S, Watzka M, Oldenburg J. The Arg98Trp mutation in human VKORC1 causing VKCFD2 disrupts a di-arginine-based ER retention motif. Blood. 2014;124:1354–62.
Matagrin B, Montagut-Romans A, Damin M, Lemaire M, Popowycz F, Benoit E, Lattard V. Identification of VKORC1 genotype leading to resistance to tecarfarin. J Clin Pharmacol. 2014;54:896–900.
Kamal El-Din MA, Farhan MS, El Shiha RI, El-Kaffas RM, Mousa SM. Frequency of CYP2C9 and VKORC1 gene polymorphisms and their influence on warfarin dose in Egyptian pediatric patients. Paediatr Drugs. 2014;16:337–41.
Tomek A, Matoska V, Kolarova T, Neumann J, Sramek M, Sarbochova I, Taborsky L, Bojar M, Goetz P, Serebruany VL. The bleeding risk during warfarin therapy is associated with the number of variant alleles of CYP2C9 and VKORC1 genes. Cardiology. 2013;125:182–91.
Shrif NE, Won HH, Lee ST, Park JH, Kim KK, Kim MJ, Kim S, Lee SY, Ki CS, Osman IM, Rhman EA, Ali IA, Idris MN, Kim JW. Evaluation of the effects of VKORC1 polymorphisms and haplotypes, CYP2C9 genotypes, and clinical factors on warfarin response in Sudanese patients. Eur J Clin Pharmacol. 2011;67:1119–30.
Siltanen S, Syrjakoski K, Fagerholm R, Ikonen T, Lipman P, Mallott J, Holli K, Tammela TL, Jarvinen HJ, Mecklin JP, Aittomaki K, Blomqvist C, Bailey-Wilson JE, Nevanlinna H, Aaltonen LA, Schleutker J, Vahteristo P. ARLTS1 germline variants and the risk for breast, prostate, and colorectal cancer. Eur J Hum Genet. 2008;16:983–91.
Yang XY, Yu H, Xi MR, Yang KX, Pan XL, Hu M, Peng ZL. Association of the ARLTS1 variants with familial ovarian cancer risk in China. Int J Gynecol Cancer. 2009;19:585–90.
Li X, Gast A, Rudnai P, Gurzau E, Koppova K, Hemminki K, Kumar R. ARLTS1 polymorphisms and basal cell carcinoma of the skin. Hereditary Cancer Clin Prac. 2007;5:25–9.
Castellvi-Bel S, Castells A, de Cid R, Munoz J, Balaguer F, Gonzalo V, Ruiz-Ponte C, Andreu M, Llor X, Jover R, Bessa X, Xicola RM, Pons E, Alenda C, Paya A, Carracedo A, Pique JM, Gastrointestinal Oncology Group of the Spanish Gastroenterological, A. Association of the ARLTS1 Cys148Arg variant with sporadic and familial colorectal cancer. Carcinogenesis. 2007;28:1687–91.
Sellick GS, Catovsky D, Houlston RS. Relationship between ARLTS1 polymorphisms and risk of chronic lymphocytic leukemia. Leuk Res. 2006;30:1573–6.
Frank B, Meyer P, Boettger MB, Hemminki K, Stapelmann H, Gast A, Schmitt C, Kumar R, Sergi C, Burwinkel B. ARLTS1 variants and melanoma risk. Int J Cancer. 2006;119:1736–7.
Frank B, Hemminki K, Meindl A, Wappenschmidt B, Klaes R, Schmutzler RK, Untch M, Bugert P, Bartram CR, Burwinkel B. Association of the ARLTS1 Cys148Arg variant with familial breast cancer risk. Int J Cancer. 2006;118:2505–8.
Siltanen S, Fischer D, Rantapero T, Laitinen V, Mpindi JP, Kallioniemi O, Wahlfors T, Schleutker J. ARLTS1 and prostate cancer risk--analysis of expression and regulation. PLoS One. 2013;8:e72040.
Zambon CF, Pengo V, Padrini R, Basso D, Schiavon S, Fogar P, Nisi A, Frigo AC, Moz S, Pelloso M, Plebani M. VKORC1, CYP2C9 and CYP4F2 genetic-based algorithm for warfarin dosing: an Italian retrospective study. Pharmacogenomics. 2011;12:15–25.
Ng D, Toure O, Fontaine L, McMaster ML, Goldin LR, Caporaso N, Toro JR. No association of ARLTS1 polymorphisms and risk for familial chronic lymphocytic leukaemia. Br J Haematol. 2007;137:173–5.
De Brakeleer S, Teugels E, De Greve J. Familial cancer and ARLTS1. N Engl J Med. 2005;353:313–4. author reply 313-4.
Acknowledgements
Not applicable.
Availability of data and materials
Not applicable.
Authors’ contributions
Conception and design: H-YL; drafting the manuscript: YJ; data extraction: C-YZ, L-CC and BX; quality assessment: YJ and C-YZ; quantitative analysis: YJ. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Jiang, Y., Zhao, CY., Cheng, LC. et al. ARLTS1 polymorphism is associated with an increased risk of familial cancer: evidence from a meta-analysis. Hered Cancer Clin Pract 15, 8 (2017). https://doi.org/10.1186/s13053-017-0068-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13053-017-0068-7