Abstract
Small RNAs (also referred to as small noncoding RNAs, sncRNA) are defined as polymeric ribonucleic acid molecules that are less than 200 nucleotides in length and serve a variety of essential functions within cells. Small RNA species include microRNA (miRNA), PIWI-interacting RNA (piRNA), small interfering RNA (siRNA), tRNA-derived small RNA (tsRNA), etc. Current evidence suggest that small RNAs can also have diverse modifications to their nucleotide composition that affect their stability as well as their capacity for nuclear export, and these modifications are relevant to their capacity to drive molecular signaling processes relevant to biogenesis, cell proliferation and differentiation. In this review, we highlight the molecular characteristics and cellular functions of small RNA and their modifications, as well as current techniques for their reliable detection. We also discuss how small RNA modifications may be relevant to the clinical applications for the diagnosis and treatment of human health conditions such as cancer.
Similar content being viewed by others
Introduction
RNA molecules play essential and diverse roles in numerous biological functions, as studied in organisms ranging from prokaryotes to eukaryotes [1,2,3]. From those studies, it emerged that post-transcriptional modifications are essential for the functions of RNA molecules to carry out their cellular functions. In the 1960s, scientists first discovered modifications in RNA bases through enzymatic digestion and electrophoresis [4]. Since then, over 170 different types of RNA post-transcriptional chemical modifications have been described across all currently known RNA species [5]. Over the course of these investigations, the enzymes responsible for writing (catalyzing and modifying nucleotides), reading (recognizing and binding modified nucleotides) and erasing (catalyzing the removal of specific modifications) RNA modifications have also been discovered [6,7,8,9]. Small RNAs, which are a class of noncoding RNAs that are less than 200 nucleotides in length, are widely present in various cell types and tissues [10,11,12]. Over the past 20 years, extensive research has led to their classification on the basis of their size and structural characteristics, as follows: traditional small RNAs, structural small RNAs and derived small RNAs (also called non-canonical small RNAs, Fig. 1 and Table 1) [13]. These small RNAs are involved in various biological processes through different mechanisms. For example, traditional small RNA species, including, microRNA (miRNA), PIWI-interacting RNA (piRNA) and small interfering RNA (siRNA), interact with Argonaute proteins to mediate RNA-silencing effects. Furthermore, structural small RNAs (including tRNA, rRNA, snoRNA, snRNA, yRNA and vtRNA) are essential components within cells that regulate physiological homeostasis. In contrast, non-canonical small RNAs represent structural RNAs of poorly characterized functions independent of Argonaute proteins, and these are generated following enzymatic cleavage by evolutionarily ancient RNases [14]. Further to these small RNAs, new evidence suggests that small RNAs can be modified in a variety of ways which significantly influence their functions across various biological processes [14, 15]. Here, we detail the roles for small RNA modifications in the biogenesis and functions of small RNAs, with a focus on the following modifications: N6-methyladenosine (m6A), 2′-O-methylation (Nm), 5-methylcytosine (m5C) and pseudouridine (Ψ). Also, we summarize the current methods of detecting these small RNAs, highlight the evidence for this molecular process in cell and tissue homeostasis and discuss the potential clinical application of small RNAs and its modifications in human disease.
The structure and classification of small RNAs. The left panel displays traditional small RNAs, including miRNA, piRNA and siRNA; the middle panel displays structural small RNAs, including tRNA, yRNA, vtRNA, rRNA (containing 5s rRNA and 5.8s rRNA, and 5s rRNA was showed here), snoRNA and snRNA; the right panel displays derived small RNAs, whose fragment sizes are less than 50nt, and predominantly includes tsRNA from tRNA, ysRNA from yRNA, vtsRNA from vtRNA, rsRNA from 5s rRNA, snosRNA from snoRNA and snsRNA from snRNA. This figure was developed using BioRender.com
Small RNA modifications
N6-methyladenosine (m6A)
N6-methyladenosine (m6A), first discovered in the 1970s [40, 41], is a methylation modification of the sixth nitrogen (N) atom of adenine (A). The m6A modification is one of the most abundant type of modification to messenger RNAs (mRNAs) in the biological world [42, 43], including in small RNAs of eukaryotic species [44]. This m6A modification can be catalyzed by S-adenosylmethionine (SAM) binding proteins methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14) [45, 46]. Notably, other cofactors, such as Wilms tumor-associating protein (WTAP) [47], methyltransferase-like 16 (METTL16) [48], RNA-binding motif protein 15 (RBM15) [49], KIAA1429 (also called VIRMA) and zinc finger CCCH domain-containing protein 13 (ZC3H13) [50], are also known to be essential for catalyzing function of m6A methyltransferases. The proteins known as fat mass and obesity-associated protein (FTO) and ALKB homolog 5 (ALKBH5) are both also identified as m6A demethylases [51, 52]. On the other hand, members of YT521-B homology domain family 1/2/3 (YTHDF1/2/3) [7, 53], YT521-B homology domain-containing proteins 1/2 (YTHDC1/2) [7, 54], members of the heterogeneous nuclear ribonucleoprotein protein families (including HNRNPC, HNRNPA2/B1) [7, 55], eukaryotic translation initiation factor 3 (eIF3) [49], as well as insulin-like growth factor-2 mRNA-binding proteins 1/2/3 (IGF2BP1/2/3) [56] have all been characterized as reader proteins that recognize m6A methylation (Fig. 2A).
The m6A modification in miRNA. A. The chemical structure of adenosine and the site of methylation on N6 are shown alongside the enzymes (writers, eraser and readers) known to be involved. B. The m6A modification of pri-miRNA mediated by METTL3 or METTL14 is recognized by HNRNPA2/B1, so as to promote the interaction between DGCR8 and pri-miRNA, leading to acceleration of miRNA biosynthesis. On the contrary, ALKBH15 demethylates pri-miRNA, preventing DGCR8 from interacting with pri-miRNA, and this results in blockade of mature miRNA synthesis. HNR-: HNRNPA2/B1. C. The m6A modification in pre-miRNA mediated by METTL3 promotes the binding of Dicer to pre-miRNA which, in turn, accelerates the biosynthesis of miRNA (note that the specific m6A modification sites remain unknown). D. While studies have found that m6A modifications can be detected on mature miRNAs, the origin and biological relevance of such modifications remains to be better characterized
In 2015, Alarcon and colleagues reported that alterations to levels of the methyltransferase METTL3 affected the expression of mature miRNA as well as unprocessed primary miRNA (pri-miRNA), in addition to its known effect on mRNAs [57]. This result suggests that m6A is associated with miRNA biosynthesis. As described [13], the first step in miRNA biosynthesis involves binding and recognition by the double-stranded RNA-binding protein (DGCR8) to the junction between the pri-miRNA hairpin stem and the flanking single-stranded RNA within the nucleus, followed by recruitment of RNase III endonuclease Drosha to form a microprocessor complex. This leads to cleavage of pri-miRNA to produce a precursor miRNA (pre-miRNA) species. The pre-miRNA then binds exportin 5 and is transported to the cytoplasm to be cleaved into mature miRNA by Dicer. Interestingly, Alarcon and colleagues found that this biological process is dependent on m6A modification of RNA [57]. Indeed, METTL3 can methylate pri-miRNAs for HNRNPA2/B1 recognition, following which HNRNPA2/B1 recruits and interacts with DGCR8 to bind to pri-miRNA, leading to acceleration of miRNA production. Thus, m6A is an important post-transcriptional modification of efficient miRNA biosynthesis within cells (Fig. 2B) [57]. This finding provides important insight into the role of m6A in various biological processes, as well as in the progression of human diseases. For example, a mechanism for aberrant cell proliferation in bladder cancer implicates a pathway in which high METTL3 expression enhances DGCR8 recognition and binding of m6A-modified pri-miR221/222 which, in turn, potentiates miR221/222 maturation and subsequent reduction in levels of phosphatase and tensin homolog (PTEN), a known target of miR221/222 [58]. Also, in the context of patients spinal tissue degeneration, it has been reported that METTL14 regulates m6A modification of pri-miR-34a-5p to accelerate DGCR8 recognition and, through this mechanism, increases miR-34a-5p to target silent information regulator sirtuin 1 (SIRT1) and, in the process, promotes Tumor necrosis factor-alpha (TNF-α)-induced cell senescence within the nucleus pulposus invertebral disc tissue [59]. In addition to the actions of methyltransferases, m6A demethylases and reader proteins also affect the biological processes of miRNAs. For example, In lung cancer, the m6A reader HNRNPA2B1 interacts with LINC01234 to recruit DGCR8, and this leads to potentiation and accumulation of miR-106b-5p which, in turn, exerts a downregulatory effect on cryptochrome circadian regulator 2 (CRY2) levels, leading to elevated c-Myc levels and lung cancer growth [60]. In the scenario lung fibroblast activation and silica-induced lung fibrosis, it has been reported that ALKBH5, a demethylase, can demethylate pri-miR-320a-3p to prevent its interaction with DGCR8 and this, in turn, blocks the generation of miR-320a-3p, leading to dysregulation in the expression of its target genes, including forkhead box M1 (FOXM1), which ultimately leads to lung tissue damage [61]. Table 2 provides a summary of the roles of small RNA modifications in various cellular contexts in homeostasis and disease.
In addition to its functions with methylation readers, writers and erasers, m6A modification may also facilitate miRNA maturation by promoting Dicer splicing of precursor miRNAs (Fig. 2C). In the context of non-small cell lung cancer, METTL3 has been shown to increase pre-miR-143-3p splicing in an m6A-dependent manner to promote miR-143-3p biogenesis, leading to lung cancer invasion and angiogenesis through a mechanism involving dysregulation of vasohibin 1 (VASH1) expression [62]. This finding provides a potential avenue of investigation through which to develop novel treatments for patients with non-small cell lung cancer, as well as brain tissue metastasis by cancer cells.
Interestingly, in the absence of changes to its primary transcript, it has been reported that levels of several mature miRNAs are decreased within cells after downregulation of the m6A demethylase known as FTO, and this suggests that m6A can negatively regulate miRNA biogenesis [63]. Consistent with this observation, NSUN2 methyltransferase inhibits processing of pri-miR-125b to miR-125b, and this results in a decrease in miR-125b expression levels [64]. This process of NSUN2-dependent miR-125b downregulation may be facilitated by the actions of the protease activation receptor 2 (PAR2) and can ultimately promote rectal cancer metastasis through dysregulation of the expression of GRB2-associated binding protein 2 (Gab2) gene [65]. Contrastingly, in studies of endocrine-resistant breast cancer cells, HNRNPA2/B1 appears to play a more complex role in miRNA biogenesis. HNRNPA2/B1 is found to be overexpressed in endocrine-resistant breast cancer cells, and this leads to upregulation of miR-1266-5p, miR-1268a and miR-671-3p, as well as reductions in levels of miR-29a-3p, miR29b-3p and miR-222 which, collectively, is linked to a reduction in the sensitivity of such cells to cancer drugs 4-hydroxytamoxifen and fulvestrant [66]. Thus, while m6A methylation is important to miRNA homeostasis and in the context of cancer, the underlying mechanisms remain to be better characterized. Since m6A methylation signatures can regulate mRNA degradation, inhibition of miRNA processing by m6A could be explained by a mechanism involving downregulation of the mRNA expression of protein factors that read, process and interact with m6A RNA species, such as DGCR8, Drosha and Dicer. Further to this issue, Chen and colleagues suggest that m6A modification on pri-miRNAs may be selectively recognized by specialized readers involved in regulating miRNA instability or degradation, or both. Evidence for such a mechanism has been demonstrated for several ncRNAs and mRNA [67, 68].
Furthermore, m6A can be found in mature miRNAs [69]. However, the origin of m6A on mature RNAs and its impact is poorly understood (Fig. 2D).
Taken together, m6A modification is essential to miRNA processing, and our understanding of the functional impact of m6A modification of miRNAs remains to be better clarified.
2′-O-methylation (Nm)
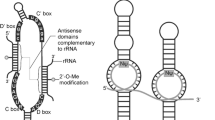
Another RNA modification is 2′-O-methylation (Nm), which is an abundant and highly conserved modification that replaces hydrogen (-H) atom on the ribose fraction 2′-hydroxyl (-OH) with a methyl group (-CH3) [107]. The Nm modification can be found at various sites within transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs) [108] and messenger RNAs (mRNAs) [109]. Also, Nm modifications are detected on the 3′-ends of small RNAs such as miRNAs and siRNAs in plants [110, 111], Argonaute 2 (AGO2)-loaded siRNAs and miRNAs in flies, as well as piRNAs in animals [112, 113]. During the maturation process of small RNAs in humans, small RNAs undergo Nm modification at their 3′ end nucleotides after their processing by Dicer or PIWI proteins [114], and this modification is important for them to form stable structures protected from 3′–5′ truncation and 3′-uridine-triggered degradation (Fig. 3) [115,116,117,118]. In addition, Nm is postulated to affect the stability of small RNAs by affecting thermodynamic properties such as base stacking and structural rigidity [119, 120].
The 2′-O-methylation (Nm) modification in piRNA. HEN1 catalyzes Nm modification of piRNA, replacing a hydrogen (–H) atom on the ribose fraction 2′-hydroxyl (–OH) with a methyl-group (–CH3). During piRNA maturation, the 3′-terminal nucleotide undergoes Nm modification, leading to formation of a stable piRNA structure that is protected from 3′-uridine degradation
The first identified 2′-O-methyltransferase responsible for Nm modification of small RNAs was the HUA ENHANCER 1 (HEN1) protein, discovered in Arabidopsis as a methylase for miRNAs and siRNAs [111, 121]. Subsequently, HEN1 homologs were found in other plants, as well as homologs that methylated piRNAs in animals, as well as AGO2-associated small RNAs in Drosophila [112, 122,123,124,125,126,127]. Studies have shown that HEN1 knockout or mutation in Arabidopsis leads to elevated levels of heterogeneous 3′-ends and poly-U, both features of which are known to disrupt RNA stability [110, 118], and which result in aberrant lengths as well as decreased levels of small RNAs, respectively. This phenotype is predicted to be associated with the role of enzymes responsible for 3′ uridylation of small RNAs, such as HEN1 SUPPRESSOR1 (HESO1) [116, 117] and UTP:RNA uridylyltransferase 1 (URT1) [128]. For example, in Drosophila, loss of Pimet (HEN1 homolog) activity leads to deletion of Nm in piRNA and siRNA [129]. In addition, mutations in the HEN1 gene accelerate neurodegeneration and shorten lifespan, suggesting that Nm of small RNAs may affect age-related signaling events within cells [130]. In zebrafish, the absence of Hen1 (HEN1 homolog) results in a decrease in piRNA content within oocytes and a shortening of exonuclease-mediated piRNAs, which ultimately leads to oocyte loss and infertility [124]. Collectively, these findings highlight the roles for HEN1 and its orthologs across a broad range of plant and animal model systems in stabilizing germline small RNAs, with species-specific consequences.
In mammals, the HEN1 homolog, HENMT1, plays an essential role in fertility. For example, in mice, loss of HENMT1 expression leads to piRNA instability, observed as reductions to piRNA volume and length, as well as developmental arrest of germ cells during the process of spermatogenesis. Particularly, loss of HENMT1 and the associated loss of piRNA collectively lead to defective meiosis and precocious and selective expression of haploid transcripts during meiosis [131]. This finding shows that HENMT1 is critical to piRNA homeostasis in maintaining TE inhibition and spermatogenesis in germ cells [131]. Of note, Nm modifications have also recently been detected in mature miRNAs in mammals. Different 3′-terminal Nm patterns of miRNAs, particularly miR-21-5p, have been reported in RNAs from non-small cell lung cancer cells from human subjects, as well as within cells of their paired normal tissue extracts. This methylation is reported to enhance the capacity for miR-21-5p to resist degradation by the polyribonucleotide nucleotidyltransferase enzyme PNPase 1 (PNPT1) which, in turn, leads to their prolonged loading onto AGO2 to form a complex that enhances the inhibition of expression of programmed cell death 4 (PDCD4) [132]. This suggests that Nm modification of miRNA can enhance miRNA stability and prevent their degradation by enzymes such as PNTP1 [132].
In addition to HEN1, other 2′-O-methyltransferases, such as FTSJ1 and FBL, are also crucial for Nm modification on small RNAs and their functions. For instance, studies have shown that the reduction of human FTSJ1 orthologs-mediated tRNA Nm modification in Drosophila leads to small RNA pathway dysfunction and increased susceptibility to RNA virus infection. This phenomenon is also associated with small RNA-induced gene silencing pathways [133]. FTSJ1-mediated Nm modification on tRNA also can effectively suppress DRAM1 expression, consequently inhibiting the progression of non-small cell lung cancer [134]. Yi et al. discovered that EZH2 has a direct interaction with FBL, a 2′-O-methyltransferase, leading to an enhancement in the 2′-O-methylation of rRNA, which promotes the assembly of box C/D small nucleolar ribonucleoproteins and facilitates tumor cell translation [135]. Overall, Nm modifications on tRNA play important roles in RNA silencing, translation and antiviral defense.
5-Methylcytidine (m5C)
Another RNA modification is 5-Methylcytidine (m5C) in which the fifth carbon atom (C) of cytosine is methylated within RNAs [108]. The highly conserved NSUN (NOL1/NOP2/Sun) domain family has been identified as specific enzymes responsible for m5C RNA modification [136, 137]. Also, the enzyme DNA methyltransferase 2 (DNMT2) has been found to catalyze the formation of m5C at position C38 of tRNAs (Fig. 4A) [136, 137].
The m5C modification in small noncoding RNAs. A. The chemical structure of cytosine and the site of methylation on C5 are both shown alongside the relevant enzymes (writers, eraser and readers). B. The m5C modification in vtRNA. The m5C modification of vtRNA catalyzed by NSUN2 affects cleavage by Dicer, resulting in persistence of svRNA4 products and a relative decrease in svRNA1, svRNA2 and svRNA3 (↓ denotes downregulation, while ↑ denotes upregulation). C. The m5C modification in miRNAs. Nsun2-mediated m5C modification in miRNA can interfere with the formation of miRNA;mRNA pairing, resulting in the loss of miRNA-mediated gene silencing activity. D. The m5C modification in tRNA. DNMT2-mediated m5C modification of tRNA can reduce the affinity of ANG to tRNA, leading to a reduction in tsRNA synthesis in cells
All these methyltransferases can also methylate small RNAs. For example, Shobbir Hussain and coworkers found that vault RNAs, a class of small RNAs of approximately 88 to 100 nucleotides in length and transcribed by RNA polymerase III, contain 6 methylated cytosines by NSUN and formed riboprotein granules called Vault with proteins [138]. The authors also used NSUN2-deficient patient cells to further demonstrate that loss of cytosine-5 methylation in vt-RNA leads to abnormal processing of argonaute-associated small RNA fragments that could function as miRNAs (Fig. 4B) [139]. The methylation of cytosine 69 in vtRNA occurs frequently in human cells and is jointly regulated by NSUN2 and serine/arginine splicing factor 2 (SRSF2), such that vtRNA processing produces different small-vault RNA (svRNA), which is implicated in the regulation of epidermal differentiation [140]. In addition, m5C modification is also present on miRNAs [141].m5C modifications on miRNAs have been reported to interfere with the formation of miRNA/mRNA pairing, resulting in loss of gene silencing activity of the miRNAs themselves (Fig. 4C). For example, m5C modification abolishes the capacity for miRNA-181a-5p to function as a tumor suppressor and correlates with poor prognosis in glioblastoma patients [142]. The m5C modification of miRNAs is also found to result in structural changes in the RNA-induced silencing complex (RISC). For example, m5C modification at position 9 of miR-200c-3p adjacent to its recognition with the RISC complex can disrupt the hydrogen bond formed between miRNA and AGO Ser220, resulting in guanine interaction at miRNA position 8 with Arg761 of AGO translocation [69].
In addition, m5C modifications on tRNAs affects the generation of tsRNA species. The formation of tsRNAs is reported to be induced in response to stress, since unconventional 5′ initiation sites can be found in the 5′ UTR of stress response transcripts to inhibit canonical translation and favor ribosome assembly [143]. Thus, m5C modification on tRNA protects them from angiogenin (ANG) processing into tsRNA (Fig. 4D) [144,145,146,147,148]. In contrast, DNMT2-mediated loss of cytosine 38 methylation of tRNA leads to accumulation of tsRNAs, leading to mis-translation of specific codons and disruptions to protein synthesis [146]. These tRNA and tsRNA alterations therefore inhibit protein synthesis and negatively affects cell function and development [144, 146, 147, 149]. Furthermore, in sperm, small RNAs can encode paternal information through m5C modifications with the capacity for intergenerational transmission of paternally acquired phenotypes. For example, Zhang and colleagues altered the expression profile of sperm small RNAs, including the levels of tsRNAs and rRNA-derived small RNAs, by knocking out mouse tRNA methyltransferase DNMT2. This intervention led to a prevention of the high-fat diet-induced elevation of RNA modifications (m5C, m2G) in the 30-40nt RNA fraction in sperm and subsequent abolishment of the transmission of small RNA-mediated metabolic disorders from high-fat diet-induced sperm. This finding suggests that DNMT2-mediated m5C modification of RNAs contributes to the secondary structure and biological characteristics of small RNAs that underlie their capacity for paternal epigenetic memory, programmed as "coding marks" within sperm RNAs [150].
Other modifications on small RNAs
In addition to the modifications mentioned above, various other small RNA modifications have also been reported, including pseudouridine (Ψ) modifications, m7G modifications and m1A modifications, and of which also seem to play important roles in cell biological processes, as described below. One caveat is that the underlying effects of such modifications on the structure and functions of RNAs remain to be better characterized.
Pseudouridine (Ψ)
Pseudouridine (Ψ), an isomeric form of uridine also known as 5-ribosyluracil, was discovered in the 1950s and is described as the most abundant type of post-transcriptional RNA modification discovered in all kingdoms of life [151,152,153]. Pseudouridine is catalyzed by an evolutionarily conserved family of pseudouridine synthases (PUS) or by RNA-dependent mechanisms that involve a significant number of H/ACA box small nucleolar RNA (snoRNA) [154]. Pseudouridine can be detected in tRNAs [155], rRNAs [156, 157], small nuclear RNAs (snRNAs) [158,159,160] and mRNAs [161, 162]. Its presence is known to affect the biogenesis, structure, function and coding potential of RNA, with attendant effects on downstream signaling and cell homeostasis.
Recent studies have found that pseudouridine catalyzed by PUS7 could activate tsRNAs that are involved in protein synthesis which are vital to the functions of mammalian stem cells. For example, inhibition of a pseudouridine-driven regulatory network can severely affect hematopoiesis and promote the incidence and pathogenesis of human myeloid malignancies [163]. With regard to the impact of pseudouridine synthesis on miRNAs, it has been reported that inhibition of PUS10 results in reduced mature miRNA and accumulation of primary miRNA, yet this process is independent of catalytic activity of ubiquitin-specific peptidase 10 (USP10) [164]. In another example, endogenous TruB1, a predominant mammalian pseudouridine synthase, is able to bind the stem-loop of pri-let-7 to enhance the interaction of this miRNA with the microRNA processor protein DGCR8, so as to enhance the maturation of let-7 microRNA family members that signal to inhibit cell proliferation [165]. These studies suggest that ability for cells to synthesize pseudouridine, as well as the presence of pseudouridine on multiple small RNA species are all critical to the regulation of critical biological functions.
N7-methylguanosine (m7G)
N7-methylguanosine (m7G) is a methylated modification of the seventh nitrogen atom of guanine in RNA, and the modification is documented in tRNAs, rRNAs, mRNAs and miRNAs [166,167,168,169,170]. The m7G modification within cells can be catalyzed by METTL1 and WD repeat domain 4 (WDR4) proteins [171]. For example, METTL1 binds directly to the miRNA precursor via m7G and accelerates miRNA maturation [168]. After pre-miRNA processing, m7G can persist on mature miRNAs and its presence can influence the function of mature miRNAs. For example, in lung cancer cells, a mature let-7e miRNA that encodes a m7G modification is capable of downregulating the stability and translation efficiency of the mRNA transcript encoding high mobility group protein 2 (HMGA2). This, in turn, leads to a reduction in levels of translated HMGA2 protein and subsequent arrest of lung cancer cell proliferation and migration [168].
N1-methyladenosine (m1A)
N1-methyladenosine (m1A) modification involves the methylation of the first nitrogen atom of adenine within RNA. This modification is mainly detected in tRNAs, rRNAs, mRNAs and small RNAs, and its presence has been found to influence the structure and functions of these RNA species [172,173,174]. Particularly, small RNAs can regulate gene expression through m1A. For example, Su and colleagues found that TRMT6/61A, an RNA methylase enzyme, was highly expressed in urothelial carcinoma cells derived from human bladder, compared with normal cells, and this was accompanied by higher levels of m1A modification across multiple RNAs including tsRNAs. This resulted in dysregulation of the tsRNA targetome and contributed to cellular functions related to malignant transformation, including a direct effect on the unfolded protein response within these cancer cells [175].
Clinical applications
Prevention
Current evidence in the literature has indicated that therapeutic manipulation of small RNA modification may be clinically beneficial in the treatment and prevention of human disease conditions. For example, Wang and colleagues investigated changes in methylation of 22 miRNAs in 57 cases of human neural tube defects (NTDs) and reported that methylation of the microRNA hsa-let-7 g directly effects its expression (176). Furthermore, the methylation levels for hsa-let-7 g were significantly correlated with folate concentration. Thus, the correlation between aberrant methylation of hsa-let-7 g and folate metabolism could indicate that, by improving early-pregnancy nutrition, NTDs could be avoided through a mechanism which involves adequate dietary supply of methyl-donors for miRNA modification within fetal cells [176]. In addition, growing evidence indicates that small RNAs in sperm can mediate the intergenerational transmission of paternally phenotypes [177, 178]. For example, Chen and coworkers found that small RNA modifications in sperm were involved in encoding paternity information [150]. There, the authors found that depletion of mouse tRNA methyltransferase DNMT2 prevented the high-fat diet (HFD)-induced elevation of RNA modifications (m5C, m2G) in the 30-40nt RNA fraction of sperm, and this blocked the epigenetic transmission of phenotypic traits for HFD-induced metabolic disorder that would have otherwise been detected in the offspring. Indeed, Dnmt2-mediated m5C modifications is crucial to endow small RNAs within sperm with specialized secondary structures and biological properties, and this is a powerful example of how small RNA modifications are critical to the epigenetic inheritance of paternal traits [150]. Thus, in these examples of NTD and sperm function, small RNA modifications influence the developmental homeostasis and organismal survival of mammalian offspring.
Prediction and surveillance
In 2006, Seidel and colleagues applied a chromatography method to discover that the modified nucleosides in urine samples have high sensitivity and specificity as potential biomarkers to identify a variety of cancers in patients [179]. Recently, with the rapid development of detection techniques, the great potential of small RNA modification as a biomarker of disease is being realized. For example, Su and workers used an improved RNA sequencing detection technique of Thermostable Group II Intron Reverse Transcriptase (TGIRT) and discovered the presence of m1A, m1G and N2, N2- dimethylguanosine (m2 2G) modifications on tsRNAs and rsRNAs in bladder cancer cells [180]. Yan and colleagues applied a highly efficient liquid chromatography-tandem mass spectrometry method and discovered that small RNA modifications in the liver cells of diabetic mice were significantly altered compared to control treatment, highlighting the correlation between small RNA modifications and diabetes [181]. Zhang and coworkers found that 2′-O-methylcytidine (Cm), m7G, 2′-O-methylguanosine (Gm), and m2 2G modifications in 15–25 nt RNA from cells within the cerebral cortex of Alzheimer's disease patients were significantly increased, compared with normotypic control samples [182]. Konno and colleagues reported that methylation of some miRNAs was elevated in tumor samples compared to normal tissues. Of note in one particular study, the methylation levels for miR-17-5p in serum were sensitive enough to distinguish patients with pancreatic cancer from healthy individuals [69]. Furthermore, small RNA modifications have been suggested to be reliable as a potential biomarker for male. There, studying RNA samples from patients with asthenozoospermia and teratozoospermia relative to controls, Guo and colleagues used a high-throughput sequencing platform to detect RNA modifications and identified 13 RNA modification signatures on total sperm RNA, as well as 16 RNA modification signatures on sperm RNA fragments of varying sizes. Particularly, the modifications m1G, m5C, m2G and m1A were found to be significantly correlated with clinically-graded sperm motility measures [183].
In addition to their potential value as diagnostic markers, the presence and relative abundance of small RNA modifications may also serve as prognostic biomarkers. For example, in patients diagnosed with glioma, low miRNA-181a-5p expression and cytosine methylation levels were associated with poor survival prognosis (reported as median survival rates of 12.4 months and 8.5 months, respectively), while glioma patients with high levels of unmethylated miRNA-181a-5p were found to have a better survival prognosis (median 16.5 months) [184]. Guzzi and colleagues reported that the dysregulation of the terminal oligoguanine (TOG) at 5′-terminal end of tRFs, which is regulated by pseurouridine driven by PUS7 activity, is linked to leukemia transformation and reduced survival rates in patients and increases the risk of progressing from myelodysplastic syndrome (MDS) to acute myeloid leukemia [185].
Taken together, these findings suggest that small RNA modifications may be informative as markers that reflect the pathogenesis and progression of human disease.
Therapy and other applications
Given the examples of direct effects for small RNA modifications on human diseases such as bladder cancer and armed with the knowledge that evolutionarily conserved mechanisms drive small RNA modifications within cells, researchers are now exploiting these discoveries to design novel RNA-based treatments for human disorders [186]. For example, it has been found that Nm modified siRNAs are significantly more stable in serum so that it persists longer as an effective treatment to inhibit Enterovirus Type 71 (EV71) replication [187]. In another example that demonstrates their stability when delivered into animals, siRNAs modified by thiophosphate, Nm and other modifications, stimulant-related analytes were delivered intravenously into rats and, 24 h later, these modified RNA species could still be detected in rat blood and urine samples by liquid chromatography-high-resolution/high-precision mass spectrometry [188]. Nm modifications in plant miRNAs can extend their half-lives, and so this must be taken into consideration regarding the use of modified RNAs, their potential to be ingested by humans and the impact of RNA treatments in plant horticulture that has consequences on human physiology in those that adopt predominantly plant-based diets [189, 190]. In a related example, the tRNA methyltransferase known as TrmH which is required for G18 tRNA Nm, is not present in most bacteria, however, specific Nm modification to guanosine in bacterial tRNA position 18 is required to inhibit Toll-like receptor 7-mediated immune activation during a human host–pathogen response episode [191, 192]. Thus, Nm modification may be a feature of active selection in symbiotic and pathogenic species, such as in regulating the recognition of autologous and non-autologous-derived RNAs [15].
Recently, as indicated above, Su and colleagues reported that m1A modifications are highly enriched in 22-nucleotide long 3′ tRNA fragments and is dependent on its methylase TRMT6/61A [193]. In bladder cancer cells, high TRMT6/61A expression is observed, m1A modification levels of tRFs is increased and these molecular findings correlated with abnormal regulation of tRF target genes, such as those critical to the unfolded protein response [193]. Thus, small RNAs can regulate gene expression through base modifications, and this highlights their potential as a therapeutic avenue for the design of treatments to conditions such as bladder cancer (summarized in Fig. 5).
Applications of small RNA modifications. Current applications of small RNA modifications are described in the context of disease prevention (such as in neural tube defects (NTDs), lipid metabolic disorders), cancer treatment (such as bladder cancer), disease prediction and surveillance (such as for cancer, metabolic diseases, neurodegenerative disease and male infertility). This figure was created with BioRender.com
Detection techniques
Detection techniques of small RNAs
As researchers discover the critical roles for small RNAs in physiological and pathological processes, an increasing number of sequencing techniques to detect small RNAs with high sensitivity and specificity have been reported. However, the complex landscape of small RNA modification presents as a challenge for high-throughput analysis of small RNAs because such modifications interfere with the preparation of RNA-seq libraries and can limit their detection. Table 3 lists the current approaches to improving small RNA sequencing by overcoming specific RNA modifications.
Detection techniques of small RNA modifications
Several RNA-seq methods involve sequencing of the cDNA intermediate of RNA, and the conversion of RNA to cDNA can lead to loss of detection of small RNA modification. Table 4 lists the currently reported detection methods for small RNA modification sequencing. Recent studies have described two innovative methods for detecting m6A modifications on mRNA, including m6A-SAC-seq and eTAM-seq. The m6A-SAC-seq method uses the Dim1/KsgA family of dimethyltransferases, which transfer the methyl group from S-adenosyl-L-methionine (SAM) to adenosines, resulting in the formation of m6A, followed by N6,N6-dimethyladenosine (m62A) in consecutive methylation reactions [202]. eTAM-seq relies on global A deamination, which enables the detection of m6A as persistent A [203]. Nonetheless, neither of these two methods has been applied to detect small RNA modifications yet. With further modifications, both of these methods would be utilized in detecting small RNA modifications in the future. In summary, these technologies provide basic scientific tools and methods for comprehensive analysis of small RNA modifications and biological studies.
Conclusion and perspective
As a recently discovered class of regulatory RNAs, small RNAS (also known as sncRNAs) can negatively or positively regulate the expression of tumor markers through different molecular pathways and intracellular signaling mechanisms. These functions for small RNAs depend on their sequence, their three-dimensional structure and their extent of RNA modification. As highlighted in this review, emerging evidence suggests that RNA modifications are essential not only in their capacity to influence the biogenesis and function of small RNAs, but their potential value as biomarkers for human diseased states is also noted [15]. Novel detection technologies including high-throughput LC–MS/MS and sequencing-based approaches have accelerated our ability to identify, quantify and define the roles of small RNA modifications in homeostasis and diseases [137, 181, 195, 220, 221]. However, in contrast to the pace of the discovery of diverse types of small RNAs, which has been relatively rapid [14], research into the significance and biological impact of small RNA modification in health and disease remains in its infancy. One of the most important tasks in the future is to address these technical challenges that enable researchers in the field of sncRNA biology to capture and study all known modified sncRNA sequences with high sensitivity and specificity. It is anticipated that these advances will lead to new insights into the physiological roles of modified small RNAs and accelerate sncRNA drug discovery as viable treatments for human health conditions.
Availability of data and materials
Not applicable.
References
Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, et al. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe. 2019;26(2):217.
Das AS, Alfonzo JD, Accornero F. The importance of RNA modifications: From cells to muscle physiology. Wiley Interdisciplinary Reviews-RNA. 2022;13(4).
Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, et al. Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol Cell. 2018;71(6):986.
Lavi U, Fernandez-Munoz R, Darnell JE Jr. Content of N-6 methyl adenylic acid in heterogeneous nuclear and messenger RNA of HeLa cells. Nucleic Acids Res. 1977;4(1):63–9.
Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022;50(D1):D231–5.
Nombela P, Miguel-Lopez B, Blanco S. The role of m(6)A, m(5)C and Psi RNA modifications in cancer: novel therapeutic opportunities. Mol Cancer. 2021;20(1).
Shi HL, Wei JB, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–50.
Satterwhite ER, Mansfield KD. RNA methyltransferase METTL16: targets and function. Wiley Interdisciplinary Reviews-RNA. 2022;13(2).
Qu JW, Yan HM, Hou YF, Cao W, Liu Y, Zhang EF, et al. RNA demethylase ALKBH5 in cancer: from mechanisms to therapeutic potential. J Hematol Oncol. 2022;15(1).
Grosshans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451(7177):414–6.
Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43(6):880–91.
Babski J, Maier LK, Heyer R, Jaschinski K, Prasse D, Jager D, et al. Small regulatory RNAs in Archaea. RNA Biol. 2014;11(5):484–93.
Xiong Q, Zhang Y, Li J, Zhu Q. Small non-coding RNAs in human cancer. Genes (Basel). 2022;13(11).
Shi J, Zhou T, Chen Q. Exploring the expanding universe of small RNAs. Nat Cell Biol. 2022;24(4):415–23.
Zhang X, Cozen AE, Liu Y, Chen Q, Lowe TM. Small RNA modifications: integral to function and disease. Trends Mol Med. 2016;22(12):1025–34.
Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79.
Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14.
Czech B, Munafo M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, et al. piRNA-guided genome defense: from biogenesis to silencing. Annu Rev Genet. 2018;52:131–57.
Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20(2):89–108.
Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–55.
Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet. 2015;16(2):98–112.
Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24(17):1832–60.
Wilkinson ME, Charenton C, Nagai K. RNA splicing by the spliceosome. Annu Rev Biochem. 2020;89:359–88.
Scott MS, Ono M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie. 2011;93(11):1987–92.
Barciszewska MZ, Szymanski M, Erdmann VA, Barciszewski J. 5S ribosomal RNA. Biomacromol. 2000;1(3):297–302.
Hahne JC, Lampis A, Valeri N. Vault RNAs: hidden gems in RNA and protein regulation. Cell Mol Life Sci. 2021;78(4):1487–99.
Horos R, Buscher M, Kleinendorst R, Alleaume AM, Tarafder AK, Schwarzl T, et al. The small non-coding vault RNA1–1 acts as a riboregulator of autophagy. Cell. 2019;176(5):1054–6712.
Guglas K, Kolodziejczak I, Kolenda T, Kopczynska M, Teresiak A, Sobocinska J, et al. YRNAs and YRNA-derived fragments as new players in cancer research and their potential role in diagnostics. Int J Mol Sci. 2020;21(16).
Kowalski MP, Krude T. Functional roles of non-coding Y RNAs. Int J Biochem Cell Biol. 2015;66:20–9.
Chen Q, Zhang X, Shi J, Yan M, Zhou T. Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem Sci. 2021;46(10):790–804.
Xu D, Qiao D, Lei Y, Zhang C, Bu Y, Zhang Y. Transfer RNA-derived small RNAs (tsRNAs): versatile regulators in cancer. Cancer Lett. 2022;546: 215842.
Guglas K, Kołodziejczak I, Kolenda T, Kopczyńska M, Teresiak A, Sobocińska J, et al. YRNAs and YRNA-derived fragments as new players in cancer research and their potential role in diagnostics. Int J Mol Sci. 2020;21(16).
Nicolas FE, Hall AE, Csorba T, Turnbull C, Dalmay T. Biogenesis of Y RNA-derived small RNAs is independent of the microRNA pathway. FEBS Lett. 2012;586(8):1226–30.
Xia L, Guo H, Wu X, Xu Y, Zhao P, Yan B, et al. Human circulating small non-coding RNA signature as a non-invasive biomarker in clinical diagnosis of acute myeloid leukaemia. Theranostics. 2023;13(4):1289–301.
Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11(10):1268–71.
Meng C, Wei Z, Zhang Y, Yan L, He H, Zhang L, et al. Regulation of cytochrome P450 3A4 by small vault RNAb derived from the non-coding vault RNA1 of multidrug resistance-linked vault particle. Mol Med Rep. 2016;14(1):387–93.
Chu C, Yu L, Wu B, Ma L, Gou LT, He M, et al. A sequence of 28S rRNA-derived small RNAs is enriched in mature sperm and various somatic tissues and possibly associates with inflammation. J Mol Cell Biol. 2017;9(3):256–9.
Falaleeva M, Stamm S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. BioEssays. 2013;35(1):46–54.
Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15(7):1233–40.
Desrosiers RC, Friderici KH, Rottman FM. Characterization of Novikoff hepatoma mRNA methylation and heterogeneity in the methylated 5’ terminus. Biochemistry. 1975;14(20):4367–74.
Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71(10):3971–5.
Chenkiang S, Nevins JR, Darnell JE. N-6-methyl-adenosine in adenovirus type-2 nuclear-RNA is conserved in the formation of messenger-RNA. J Mol Biol. 1979;135(3):733–52.
Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci. 2013;38(4):204–9.
Yi YC, Chen XY, Zhang J, Zhu JS. Novel insights into the interplay between m(6)A modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1).
Schumann U, Shafik A, Preiss T. METTL3 gains R/W access to the epitranscriptome. Mol Cell. 2016;62(3):323–4.
Liu JZ, Yue YN, Han DL, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N-6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–5.
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–89.
Pendleton KE, Chen BB, Liu KQ, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824.
Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol. 2017;33:319–42.
Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32(5–6):415–29.
Jia GF, Fu Y, Zhao X, Dai Q, Zheng GQ, Yang Y, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–7.
Zheng GQ, Dahl JA, Niu YM, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29.
Li F, Zhao D, Wu J, Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m(6)A mononucleotide reveals an aromatic cage for m(6)A recognition. Cell Res. 2014;24(12):1490–2.
Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10(11):927–9.
Alarcon CR, Goodarzi H, Lee H, Liu XH, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–308.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–95.
Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519(7544):482–5.
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18.
Zhu H, Sun B, Zhu L, Zou G, Shen Q. N6-methyladenosine induced miR-34a-5p promotes TNF-alpha-induced nucleus pulposus cell senescence by targeting SIRT1. Front Cell Dev Biol. 2021;9: 642437.
Chen ZY, Chen X, Lei TY, Gu Y, Gu JY, Huang JL, et al. Integrative analysis of NSCLC identifies LINC01234 as an oncogenic lncRNA that interacts with HNRNPA2B1 and regulates miR-106b biogenesis. Mol Ther. 2020;28(6):1479–93.
Sun WQ, Li Y, Ma DY, Liu Y, Xu Q, Cheng DM, et al. ALKBH5 promotes lung fibroblast activation and silica-induced pulmonary fibrosis through miR-320a-3p and FOXM1. Cell Mol Biol Lett. 2022;27(1).
Wang HS, Deng QQ, Lv ZY, Ling YY, Hou X, Chen ZJ, et al. N6-methyladenosine induced miR-143–3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18(1).
Berulava T, Rahmann S, Rademacher K, Klein-Hitpass L, Horsthemke B. N6-adenosine methylation in MiRNAs. Plos One. 2015;10(2).
Yuan S, Tang H, Xing JY, Fan XQ, Cai XY, Li Q, et al. Methylation by NSun2 represses the levels and function of MicroRNA 125b. Mol Cell Biol. 2014;34(19):3630–41.
Yang L, Ma YM, Han WX, Li WW, Cui L, Zhao XH, et al. Proteinase-activated receptor 2 promotes cancer cell migration through RNA methylation-mediated repression of miR-125b. J Biol Chem. 2015;290(44):26627–37.
Klinge CM, Piell KM, Tooley CS, Rouchka EC. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci Rep. 2019;9.
Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mRNA degradation by m(6)A modification. Trends Genet. 2020;36(3):177–88.
Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction between N(6)-methyladenosine (m(6)A) modification and noncoding RNAs in cancer. Mol Cancer. 2020;19(1):94.
Konno M, Koseki J, Asai A, Yamagata A, Shimamura T, Motooka D, et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat Commun. 2019;10.
Zhao CP, Ling XL, Xia YX, Yan BX, Guan QL. LncRNA UCA1 promotes SOX12 expression in breast cancer by regulating m(6)A modification of miR-375 by METTL14 through DNA methylation. Cancer Gene Ther. 2022;29(7):1043–55.
Xu Y, Ye S, Zhang N, Zheng S, Liu H, Zhou K, et al. The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun (Lond). 2020;40(10):484–500.
Pan X, Hong X, Li S, Meng P, Xiao F. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp Mol Med. 2021;53(1):91–102.
Yue Y, Deng P, Xiao H, Tan MD, Wang H, Tian L, et al. N6-methyladenosine-mediated downregulation of miR-374c-5p promotes cadmium-induced cell proliferation and metastasis by targeting GRM3 in breast cancer cells. Ecotoxicol Environ Saf. 2022;229.
Peng W, Li J, Chen R, Gu Q, Yang P, Qian W, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38(1):393.
Li K, Gao S, Ma L, Sun Y, Peng ZY, Wu J, et al. Stimulation of let-7 maturation by metformin improved the response to tyrosine kinase inhibitor therapy in an m6a dependent manner. Front Oncol. 2022;11.
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18(1):181.
Rong L, Xu Y, Zhang K, Jin L, Liu X. HNRNPA2B1 inhibited SFRP2 and activated Wnt-beta/catenin via m6A-mediated miR-106b-5p processing to aggravate stemness in lung adenocarcinoma. Pathol Res Pract. 2022;233: 153794.
Li SS, Lu XX, Zheng DY, Chen WZ, Li YZ, Li F. Methyltransferase-like 3 facilitates lung cancer progression by accelerating m6A methylation-mediated primary miR-663 processing and impeding SOCS6 expression. J Cancer Res Clin Oncol. 2022;148(12):3485–99.
Ling Q, Wu SY, Liao XZ, Liu CY, Chen Y. Anesthetic propofol enhances cisplatin-sensitivity of non-small cell lung cancer cells through N6-methyladenosine-dependently regulating the miR-486–5p/RAP1-NF-kappa B axis. BMC Cancer. 2022;22(1).
Zhou GW, Yan KQ, Liu JK, Gao LJ, Jiang XZ, Fan YD. FTO promotes tumour proliferation in bladder cancer via the FTO/miR-576/CDK6 axis in an m6A-dependent manner. Cell Death Discovery. 2021;7(1).
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1):110.
Yan RC, Dai WW, Wu RX, Huang HB, Shu MF. Therapeutic targeting m6A-guided miR-146a-5p signaling contributes to the melittin-induced selective suppression of bladder cancer. Cancer Lett. 2022;534.
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6)-methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–43.
Liu J, Jiang K. METTL3-mediated maturation of miR-589-5p promotes the malignant development of liver cancer. J Cell Mol Med. 2022;26(9):2505–19.
Chen P, Li S, Zhang K, Zhao R, Cui J, Zhou W, et al. N(6)-methyladenosine demethylase ALKBH5 suppresses malignancy of esophageal cancer by regulating microRNA biogenesis and RAI1 expression. Oncogene. 2021;40(37):5600–12.
Liu Z, Wu K, Gu S, Wang W, Xie S, Lu T, et al. A methyltransferase-like 14/miR-99a-5p/tribble 2 positive feedback circuit promotes cancer stem cell persistence and radioresistance via histone deacetylase 2-mediated epigenetic modulation in esophageal squamous cell carcinoma. Clin Transl Med. 2021;11(9): e545.
Lin RR, Zhan M, Yang LH, Wang H, Shen H, Huang S, et al. Deoxycholic acid modulates the progression of gallbladder cancer through N-6-methyladenosine-dependent microRNA maturation. Oncogene. 2020;39(26):4983–5000.
Gong YQ, Jiang QS, Liu LJ, Liao QY, Yu J, Xiang Z, et al. METTL3-mediated m6A modification promotes processing and maturation of pri-miRNA-19a to facilitate nasopharyngeal carcinoma cell proliferation and invasion. Physiol Genomics. 2022;54(9):337–49.
Bi X, Lv X, Liu D, Guo H, Yao G, Wang L, et al. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021;28(3–4):335–49.
Bi XH, Lv X, Liu DJ, Guo HT, Yao G, Wang LJ, et al. METTL3 promotes the initiation and metastasis of ovarian cancer by inhibiting CCNG2 expression via promoting the maturation of pri-microRNA-1246. Cell Death Discovery. 2021;7(1).
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10(1):1858.
Hou Y, Zhang Q, Pang W, Hou L, Liang Y, Han X, et al. YTHDC1-mediated augmentation of miR-30d in repressing pancreatic tumorigenesis via attenuation of RUNX1-induced transcriptional activation of Warburg effect. Cell Death Differ. 2021;28(11):3105–24.
Lin S, Zhu Y, Ji C, Yu W, Zhang C, Tan L, et al. METTL3-induced miR-222-3p upregulation inhibits STK4 and promotes the malignant behaviors of thyroid carcinoma cells. J Clin Endocrinol Metab. 2022;107(2):474–90.
Wang P, Wang Z, Zhang M, Wu Q, Shi F, Yuan S. KIAA1429 and ALKBH5 oppositely influence aortic dissection progression via regulating the maturation of Pri-miR-143-3p in an m6A-dependent manner. Front Cell Dev Biol. 2021;9: 668377.
Zhang BY, Han L, Tang YF, Zhang GX, Fan XL, Zhang JJ, et al. METTL14 regulates M6A methylation-modified primary miR-19a to promote cardiovascular endothelial cell proliferation and invasion. Eur Rev Med Pharmacol Sci. 2020;24(12):7015–23.
Zha X, Xi XT, Fan XY, Ma MJ, Zhang YP, Yang YN. Overexpression of METTL3 attenuates high-glucose induced RPE cell pyroptosis by regulating miR-25-3p/PTEN/Akt signaling cascade through DGCR8. Aging-Us. 2020;12(9):8137–50.
Xia H, Wu Y, Zhao J, Li W, Lu L, Ma H, et al. The aberrant cross-talk of epithelium-macrophages via METTL3-regulated extracellular vesicle miR-93 in smoking-induced emphysema. Cell Biol Toxicol. 2022;38(1):167–83.
Li X, Xiong WQ, Long XF, Dai X, Peng Y, Xu Y, et al. Inhibition of METTL3/m(6)A/miR126 promotes the migration and invasion of endometrial stromal cells in endometriosis. Biol Reprod. 2021;105(5):1221–33.
Zhang CJ, Wang Y, Peng YN, Xu HJ, Zhou XL. METTL3 regulates inflammatory pain by modulating m(6)A-dependent pri-miR-365-3p processing. FASEB J. 2020;34(1):122–32.
Zhu H, Sun B, Zhu L, Zou GY, Shen Q. N6-methyladenosine induced miR-34a-5p promotes TNF-alpha-induced nucleus pulposus cell senescence by targeting SIRT1. Front Cell Dev Biol. 2021;9.
Zhang R, Qu YY, Ji ZJ, Hao CS, Su YM, Yao YY, et al. METTL3 mediates Ang-II-induced cardiac hypertrophy through accelerating pri-miR-221/222 maturation in an m6A-dependent manner. Cell Mol Biol Lett. 2022;27(1).
Gong R, Wang X, Li H, Liu S, Jiang Z, Zhao Y, et al. Loss of m(6)A methyltransferase METTL3 promotes heart regeneration and repair after myocardial injury. Pharmacol Res. 2021;174: 105845.
Zhao K, Yang CX, Zhang J, Sun W, Zhou B, Kong XQ, et al. METTL3 improves cardiomyocyte proliferation upon myocardial infarction via upregulating miR-17–3p in a DGCR8-dependent manner. Cell Death Discovery. 2021;7(1).
Zhang L, Zhao X, Wang J, Jin YW, Gong MX, Ye YY, et al. METTL3 suppresses neuropathic pain via modulating N6-methyladenosine-dependent primary miR-150 processing. Cell Death Discovery. 2022;8(1).
Liu EP, Lv L, Zhan YH, Ma Y, Feng JJ, He YL, et al. METTL3/N6-methyladenosine/ miR-21-5p promotes obstructive renal fibrosis by regulating inflammation through SPRY1/ERK/NF-kappa B pathway activation. J Cell Mol Med. 2021;25(16):7660–74.
Si W, Li Y, Ye S, Li Z, Liu Y, Kuang W, et al. Methyltransferase 3 mediated miRNA m6A methylation promotes stress granule formation in the early stage of acute ischemic stroke. Front Mol Neurosci. 2020;13:103.
Dimitrova DG, Teysset L, Carre C. RNA 2'-O-methylation (Nm) modification in human diseases. Genes (Basel). 2019;10(2).
Han X, Wang M, Zhao YL, Yang Y, Yang YG. RNA methylations in human cancers. Semin Cancer Biol. 2021;75:97–115.
Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, et al. Nm-seq maps 2’-O-methylation sites in human mRNA with base precision. Nat Methods. 2017;14(7):695–8.
Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3’-end uridylation activity in Arabidopsis. Curr Biol. 2005;15(16):1501–7.
Yu B, Yang ZY, Li JJ, Minakhina S, Yang MC, Padgett RW, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932–5.
Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17(14):1265–72.
Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2 ’-O-methylation of PIWI-interacting RNAs at their 3 ’ ends. Genes Dev. 2007;21(13):1603–8.
Chan CM, Zhou C, Brunzelle JS, Huang RH. Structural and biochemical insights into 2’-O-methylation at the 3’-terminal nucleotide of RNA by Hen1. Proc Natl Acad Sci USA. 2009;106(42):17699–704.
Ren GD, Xie M, Zhang SX, Vinovskis C, Chen XM, Yu B. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5’ RNA fragments generated by AGO1 cleavage. Proc Natl Acad Sci USA. 2014;111(17):6365–70.
Ren GD, Chen XM, Yu B. Uridylation of miRNAs by HEN1 SUPPRESSOR1 in Arabidopsis. Curr Biol. 2012;22(8):695–700.
Zhao YY, Yu Y, Zhai JX, Ramachandran V, Dinh TT, Meyers BC, et al. The Arabidopsis Nucleotidyl Transferase HESO1 Uridylates Unmethylated Small RNAs to Trigger Their Degradation. Curr Biol. 2012;22(8):689–94.
Ji L, Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22(4):624–36.
Yildirim I, Kierzek E, Kierzek R, Schatz GC. Interplay of LNA and 2’-O-methyl RNA in the structure and thermodynamics of RNA hybrid systems: a molecular dynamics study using the revised AMBER force field and comparison with experimental results. J Phys Chem B. 2014;118(49):14177–87.
Kumar S, Mapa K, Maiti S. Understanding the effect of locked nucleic acid and 2’-O-methyl modification on the hybridization thermodynamics of a miRNA-mRNA pair in the presence and absence of AfPiwi protein. Biochemistry. 2014;53(10):1607–15.
Yang ZY, Ebright YW, Yu B, Chen XM. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2 ’ OH of the 3 ’ terminal nucleotide. Nucleic Acids Res. 2006;34(2):667–75.
Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–4.
Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129(1):69–82.
Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJT, Roovers EF, et al. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J. 2010;29(21):3688–700.
Kirino Y, Mourelatos Z. 2’-O-methyl modification in mouse piRNAs and its methylase. Nucleic Acids Symp Ser (Oxf). 2007;51:417–8.
Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13(9):1397–401.
Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2 ’-O-methylated at their 3 ’ termini. Nat Struct Mol Biol. 2007;14(4):347–8.
Tu B, Liu L, Xu C, Zhai J, Li S, Lopez MA, et al. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 2015;11(4): e1005119.
Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2’-O-methylation of Piwi- interacting RNAs at their 3’ ends. Genes Dev. 2007;21(13):1603–8.
Abe M, Naqvi A, Hendriks GJ, Feltzin V, Zhu YQ, Grigoriev A, et al. Impact of age-associated increase in 2 ’-O-methylation of miRNAs on aging and neurodegeneration in Drosophila. Genes Dev. 2014;28(1):44–57.
Lim SL, Qu ZP, Kortschak RD, Lawrence DM, Geoghegan J, Hempfling AL, et al. HENMT1 and piRNA stability are required for adult male germ cell transposon repression and to define the spermatogenic program in the mouse. PLoS Genet. 2015;11(10): e1005620.
Liang H, Jiao Z, Rong W, Qu S, Liao Z, Sun X, et al. 3’-Terminal 2’-O-methylation of lung cancer miR-21-5p enhances its stability and association with Argonaute 2. Nucleic Acids Res. 2020;48(13):7027–40.
Angelova MT, Dimitrova DG, Da Silva B, Marchand V, Jacquier C, Achour C, et al. tRNA 2’-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila. Nucleic Acids Res. 2020;48(4):2050–72.
He Q, Yang L, Gao K, Ding P, Chen Q, Xiong J, et al. FTSJ1 regulates tRNA 2’-O-methyladenosine modification and suppresses the malignancy of NSCLC via inhibiting DRAM1 expression. Cell Death Dis. 2020;11(5):348.
Yi Y, Li Y, Meng Q, Li Q, Li F, Lu B, et al. A PRC2-independent function for EZH2 in regulating rRNA 2’-O methylation and IRES-dependent translation. Nat Cell Biol. 2021;23(4):341–54.
Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32(8):2453–63.
Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38(5):1415–30.
Kedersha NL, Rome LH. Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J Cell Biol. 1986;103(3):699–709.
Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4(2):255–61.
Sajini AA, Choudhury NR, Wagner RE, Bornelov S, Selmi T, Spanos C, et al. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat Commun. 2019;10(1):2550.
Carissimi C, Laudadio I, Lorefice E, Azzalin G, De Paolis V, Fulci V. Bisulphite miRNA-seq reveals widespread CpG and non-CpG 5-(hydroxy)methyl-Cytosine in human microRNAs. RNA Biol. 2021;18(12):2226–35.
Cheray M, Etcheverry A, Jacques C, Pacaud R, Bougras-Cartron G, Aubry M, et al. Cytosine methylation of mature microRNAs inhibits their functions and is associated with poor prognosis in glioblastoma multiforme. Mol Cancer. 2020;19(1).
Rosace D, Lopez J, Blanco S. Emerging roles of novel small non-coding regulatory RNAs in immunity and cancer. RNA Biol. 2020;17(8):1196–213.
Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33(18):2020–39.
Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24(15):1590–5.
Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015;34(18):2350–62.
Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19(9):900–5.
Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, Andersson-Rolf A, Selmi T, Blanco S, et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Rep. 2017;8(1):112–24.
Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534(7607):335–40.
Zhang YF, Zhang XD, Shi JC, Tuorto F, Li X, Liu YS, et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol. 2018;20(5):535.
Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49(5):341–51.
Borchardt EK, Martinez NM, Gilbert WV. Regulation and function of RNA pseudouridylation in human cells. Ann Rev Genetics. 2020;54:309–36.
Ge JH, Yu YT. RNA pseudouridylation: new insights into an old modification. Trends Biochem Sci. 2013;38(4):210–8.
Hammal T, Ferre-D’Amare AR. Pseudouridine synthases. Chem Biol. 2006;13(11):1125–35.
Han L, Kon Y, Phizicky EM. Functional importance of Psi(38) and Psi(39) in distinct tRNAs, amplified for tRNA(Gln(UUG)) by unexpected temperature sensitivity of the s(2)U modification in yeast. RNA. 2015;21(2):188–201.
Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44(4):660–6.
Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 2002;514(1):17–25.
Wu GW, Yu AT, Kantartzis A, Yu YT. Functions and mechanisms of spliceosomal small nuclear RNA pseudouridylation. Wiley Interdisciplinary Reviews-RNA. 2011;2(4):571–81.
Yu AT, Ge JH, Yu YT. Pseudouridines in spliceosomal snRNAs. Protein Cell. 2011;2(9):712–25.
Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA Biol. 2010;7(2):192–204.
Li XY, Zhu P, Ma SQ, Song JH, Bai JY, Sun FF, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol. 2015;11(8):592-U93.
Nir R, Hoernes TP, Muramatsu H, Faserl K, Kariko K, Erlacher MD, et al. A systematic dissection of determinants and consequences of snoRNA-guided pseudouridylation of human mRNA. Nucleic Acids Res. 2022;50(9):4900–16.
Guzzi N, Ciesla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173(5):1204.
Song JH, Zhuang Y, Zhu CX, Meng HW, Lu B, Xie BT, et al. Differential roles of human PUS10 in miRNA processing and tRNA pseudouridylation. Nat Chem Biol. 2020;16(2):160.
Kurimoto R, Chiba T, Ito Y, Matsushima T, Yano Y, Miyata K, et al. The tRNA pseudouridine synthase TruB1 regulates the maturation of let-7 miRNA. Embo J. 2020;39(20).
Furuichi Y, LaFiandra A, Shatkin AJ. 5’-Terminal structure and mRNA stability. Nature. 1977;266(5599):235–9.
Shimotohno K, Kodama Y, Hashimoto J, Miura KI. Importance of 5’-terminal blocking structure to stabilize mRNA in eukaryotic protein synthesis. Proc Natl Acad Sci U S A. 1977;74(7):2734–8.
Pandolfini L, Barbieri I, Bannister AJ, Hendrick A, Andrews B, Webster N, et al. METTL1 promotes let-7 microRNA processing via m7G methylation. Mol Cell. 2019;74(6):1278-90 e9.
Guy MP, Phizicky EM. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014;11(12):1608–18.
Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, Bohnsack MT. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14(9):1138–52.
Lin SB, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell. 2018;71(2):244.
Dunn DB. The occurrence of 1-methyladenine in ribonucleic acid. Biochim Biophys Acta. 1961;46:198–200.
Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N-1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530(7591):441.
Li XY, Xiong XS, Wang K, Wang LX, Shu XT, Ma SQ, et al. Transcriptome-wide mapping reveals reversible and dynamic N-1-methyladenosine methylome. Nat Chem Biol. 2016;12(5):311.
Su ZL, Monshaugen I, Wilson B, Wang FB, Klungland A, Ougland R, et al. TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat Commun. 2022;13(1).
Wang L, Shangguan S, Xin Y, Chang S, Wang Z, Lu X, et al. Folate deficiency disturbs hsa-let-7 g level through methylation regulation in neural tube defects. J Cell Mol Med. 2017;21(12):3244–53.
Rodgers AB, Morgan CP, Leu NA, Bale TL. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci U S A. 2015;112(44):13699–704.
Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400.
Seidel A, Brunner S, Seidel P, Fritz GI, Herbarth O. Modified nucleosides: an accurate tumour marker for clinical diagnosis of cancer, early detection and therapy control. Br J Cancer. 2006;94(11):1726–33.
Su Z, Monshaugen I, Klungland A, Ougland R, Dutta A. Characterization of novel small non-coding RNAs and their modifications in bladder cancer using an updated small RNA-seq workflow. Front Mol Biosci. 2022;9: 887686.
Yan M, Wang Y, Hu Y, Feng Y, Dai C, Wu J, et al. A high-throughput quantitative approach reveals more small RNA modifications in mouse liver and their correlation with diabetes. Anal Chem. 2013;85(24):12173–81.
Zhang X, Trebak F, Souza LAC, Shi J, Zhou T, Kehoe PG, et al. Small RNA modifications in Alzheimer’s disease. Neurobiol Dis. 2020;145: 105058.
Guo H, Shen X, Hu H, Zhou P, He T, Xia L, et al. Alteration of RNA modification signature in human sperm correlates with sperm motility. Mol Hum Reprod. 2022;28(9).
Cheray M, Etcheverry A, Jacques C, Pacaud R, Bougras-Cartron G, Aubry M, et al. Cytosine methylation of mature microRNAs inhibits their functions and is associated with poor prognosis in glioblastoma multiforme. Mol Cancer. 2020;19(1):36.
Guzzi N, Muthukumar S, Ciesla M, Todisco G, Ngoc PCT, Madej M, et al. Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic syndrome. Nat Cell Biol. 2022;24(3):299–306.
Bouchie A. Companies in footrace to deliver RNAi. Nat Biotechnol. 2012;30(12):1154–7.
Deng JX, Nie XJ, Lei YF, Ma CF, Xu DL, Li BA, et al. The highly conserved 5 ' untranslated region as an effective target towards the inhibition of Enterovirus 71 replication by unmodified and appropriate 2 '-modified siRNAs. J Biomed Sci. 2012;19.
Thomas A, Walpurgis K, Delahaut P, Kohler M, Schanzer W, Thevis M. Detection of small interfering RNA (siRNA) by mass spectrometry procedures in doping controls. Drug Test Anal. 2013;5(11–12):853–60.
Yang J, Farmer LM, Agyekum AA, Hirschi KD. Detection of dietary plant-based small RNAs in animals. Cell Res. 2015;25(4):517–20.
Hirschi KD, Pruss GJ, Vance V. Dietary delivery: a new avenue for microRNA therapeutics? Trends Biotechnol. 2015;33(8):431–2.
Jockel S, Nees G, Sommer R, Zhao Y, Cherkasov D, Hori H, et al. The 2’-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J Exp Med. 2012;209(2):235–41.
Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, et al. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J Exp Med. 2012;209(2):225–33.
Su Z, Monshaugen I, Wilson B, Wang F, Klungland A, Ougland R, et al. TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat Commun. 2022;13(1):2165.
Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods. 2015;12(9):879–84.
Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, et al. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods. 2015;12(9):835–7.
Watkins CP, Zhang W, Wylder AC, Katanski CD, Pan T. A multiplex platform for small RNA sequencing elucidates multifaceted tRNA stress response and translational regulation. Nat Commun. 2022;13(1):2491.
Wang H, Huang R, Li L, Zhu J, Li Z, Peng C, et al. CPA-seq reveals small ncRNAs with methylated nucleosides and diverse termini. Cell Discov. 2021;7(1):25.
Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y, et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol. 2021;23(4):424–36.
Hu JF, Yim D, Ma D, Huber SM, Davis N, Bacusmo JM, et al. Quantitative mapping of the cellular small RNA landscape with AQRNA-seq. Nat Biotechnol. 2021;39(8):978–88.
Honda S, Morichika K, Kirino Y. Selective amplification and sequencing of cyclic phosphate-containing RNAs by the cP-RNA-seq method. Nat Protoc. 2016;11(3):476–89.
Kugelberg U, Natt D, Skog S, Kutter C, Ost A. 5 XP sRNA-seq: efficient identification of transcripts with and without 5 phosphorylation reveals evolutionary conserved small RNA. RNA Biol. 2021;18(11):1588–99.
Hu L, Liu S, Peng Y, Ge R, Su R, Senevirathne C, et al. m(6)A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat Biotechnol. 2022;40(8):1210–9.
Xiao YL, Liu S, Ge R, Wu Y, He C, Chen M, et al. Transcriptome-wide profiling and quantification of N(6)-methyladenosine by enzyme-assisted adenosine deamination. Nat Biotechnol. 2023.
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767–72.
Clark WC, Evans ME, Dominissini D, Zheng G, Pan T. tRNA base methylation identification and quantification via high-throughput sequencing. RNA. 2016;22(11):1771–84.
Marchand V, Blanloeil-Oillo F, Helm M, Motorin Y. Illumina-based RiboMethSeq approach for mapping of 2’-O-Me residues in RNA. Nucleic Acids Res. 2016;44(16): e135.
Krogh N, Birkedal U, Nielsen H. RiboMeth-seq: profiling of 2’-O-me in RNA. Methods Mol Biol. 2017;1562:189–209.
Incarnato D, Anselmi F, Morandi E, Neri F, Maldotti M, Rapelli S, et al. High-throughput single-base resolution mapping of RNA 2΄-O-methylated residues. Nucleic Acids Res. 2017;45(3):1433–41.
Aschenbrenner J, Werner S, Marchand V, Adam M, Motorin Y, Helm M, et al. Engineering of a DNA polymerase for direct m(6) A sequencing. Angew Chem Int Ed Engl. 2018;57(2):417–21.
Schaefer M. RNA 5-methylcytosine analysis by bisulfite sequencing. Methods Enzymol. 2015;560:297–329.
Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37(2): e12.
Legrand C, Tuorto F, Hartmann M, Liebers R, Jacob D, Helm M, et al. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017;27(9):1589–96.
Khoddami V, Yerra A, Mosbruger TL, Fleming AM, Burrows CJ, Cairns BR. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc Natl Acad Sci U S A. 2019;116(14):6784–9.
Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–62.
Enroth C, Poulsen LD, Iversen S, Kirpekar F, Albrechtsen A, Vinther J. Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res. 2019;47(20): e126.
Marchand V, Ayadi L, Ernst FGM, Hertler J, Bourguignon-Igel V, Galvanin A, et al. AlkAniline-Seq: profiling of m(7) G and m(3) C RNA modifications at single nucleotide resolution. Angew Chem Int Ed Engl. 2018;57(51):16785–90.
Cui J, Liu Q, Sendinc E, Shi Y, Gregory RI. Nucleotide resolution profiling of m3C RNA modification by HAC-seq. Nucleic Acids Res. 2021;49(5): e27.
Marchand V, Pichot F, Neybecker P, Ayadi L, Bourguignon-Igel V, Wacheul L, et al. HydraPsiSeq: a method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res. 2020;48(19): e110.
Zhang N, Shi S, Jia TZ, Ziegler A, Yoo B, Yuan X, et al. A general LC-MS-based RNA sequencing method for direct analysis of multiple-base modifications in RNA mixtures. Nucleic Acids Res. 2019;47(20): e125.
Cai WM, Chionh YH, Hia F, Gu C, Kellner S, McBee ME, et al. A platform for discovery and quantification of modified ribonucleosides in RNA: application to stress-induced reprogramming of tRNA modifications. RNA Modification. 2015;560:29–71.
Su D, Chan CTY, Gu C, Lim KS, Chionh YH, McBee ME, et al. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat Protoc. 2014;9(4):828–41.
Acknowledgements
Manuscript editor Julian Heng (Remotely Consulting, Australia) provided professional English-language editing of this article (Manuscript Certificate No. Remotely Consulting_Certificate of Service_3Ci0Ng9U).
Funding
This work was supported by the National Natural Science Foundation of China (82203447), the Postdoctoral Research Project, West China Hospital, Sichuan University (18HXBH068), and Natural Science Foundation of Sichuan, China (2022NSFSC1424).
Author information
Authors and Affiliations
Contributions
Q.X. and Y.Z. contributed to conceptualization; writing-original draft preparation; and writing-review and editing. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
None declared.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xiong, Q., Zhang, Y. Small RNA modifications: regulatory molecules and potential applications. J Hematol Oncol 16, 64 (2023). https://doi.org/10.1186/s13045-023-01466-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13045-023-01466-w