Abstract
Pathological tau aggregation is a primary neuropathological feature of many neurodegenerative diseases. Intriguingly, despite the common presence of tau aggregates in these diseases the affected brain regions, clinical symptoms, and morphology, conformation, and isoform ratio present in tau aggregates varies widely. The tau-mediated disease mechanisms that drive neurodegenerative disease are still unknown. Tau interactome studies are critically important for understanding tauopathy. They reveal the interacting partners that define disease pathways, and the tau interactions present in neuropathological aggregates provide potential insight into the cellular environment and protein interactions present during pathological tau aggregation. Here we provide a combined analysis of 12 tau interactome studies of human brain tissue, human cell culture models and rodent models of disease. Together, these studies identified 2084 proteins that interact with tau in human tissue and 1152 proteins that interact with tau in rodent models of disease. Our combined analysis of the tau interactome revealed consistent enrichment of interactions between tau and proteins involved in RNA binding, ribosome, and proteasome function. Comparison of human and rodent tau interactome studies revealed substantial differences between the two species. We also performed a second analysis to identify the tau interacting proteins that are enriched in neurons containing granulovacuolar degeneration or neurofibrillary tangle pathology. These results revealed a timed dysregulation of tau interactions as pathology develops. RNA binding proteins, particularly HNRNPs, emerged as early disease-associated tau interactors and therefore may have an important role in driving tau pathology.
Similar content being viewed by others
Background
Tau is the central pathological protein in many types of neurodegenerative disease. Collectively, these diseases are referred to as “tauopathies” due to the key pathological role of tau. Tauopathies can be divided into primary tauopathies including corticobasal degeneration (CBD), Pick’s disease (PiD), progressive supranuclear palsy (PSP), argyrophilic grain disease (AGD), globular glial tauopathy (GGT), aging related tau astrogliopathy (ARTAG), primary age related tauopathy (PART), and chronic traumatic encephalopathy (CTE) or secondary tauopathies such as Alzheimer’s disease (AD). Despite the undisputed central role of tau in tauopathies, the mechanism(s) by which tau drives disease remains elusive. Additionally, it is unknown if tau drives disease via the same mechanism across all tauopathies. These critical knowledge gaps are partly due to the complexity of tau: there are 6 isoforms of tau in the human brain, all of which differently accumulate in tauopathies (e.g. aggregates of 3R tau isoforms accumulate in Pick’s disease, aggregates of 4R tau isoforms accumulate in PSP and CBD [37]. Furthermore, tau is the most post-translationally modified protein in neurodegenerative diseases. 118 post-translational modifications have been observed on tau in human AD brain tissue, 68 in CBD, 44 in GGT, 33 in PSP, 46 in PiD and 5 in AGD. This is very important from a disease mechanism perspective as many of these post-translational modifications differently influence the aggregation dynamics, toxicity, and protein–protein interactions of tau [46, 113]. In addition, the conformation of tau is different in each tauopathy, which further influences protein interactions and function [69]. Despite these challenges, recent studies have made significant progress in uncovering tau-mediated disease mechanisms, particularly by studying tau protein–protein interactions. Intriguingly, many of these studies have consistently observed pathological interactions between tau and RNA binding proteins in tauopathies. Here, we provide a comprehensive review of tau protein–protein interactions in tauopathies and highlight how the interaction between RNA binding proteins and tau may mediate disease in tauopathies.
The protein tau
The specific type of tau species is an important determinant of protein interactions and downstream pathological consequences. The MAPT gene (microtubule associated protein tau) encodes six protein isoforms commonly expressed in adult neurons [7, 104]. These vary structurally by the inclusion of 0, 1 or 2 N-terminal repeats (denoted as 0 N, 1 N and 2 N, exons 2 and 3) and the inclusion of three or four microtubule binding domains (denoted as 3R or 4R, exon 10) [7, 104]. In healthy neurons there is an equivalent amount of 3R and 4R tau. These isoforms have distinct but overlapping functions and protein–protein interactions [71]. Interestingly, central nervous system neurons extending into the periphery and peripheral neurons express a high molecular weight isoform of tau whose function remains unknown [34, 40]. This larger form of tau arises from the inclusion of a large exon (exon 4a) between the N-terminal repeats and proline rich region, and may limit tau aggregation propensity in the peripheral nervous system. Tau isoforms are regulated by a wide array of RNA-binding proteins and splicing factors (including HNRNPA1, HNRNPE2/E3, HNRNPG, HNRNPK, SRSF2, and PTB [20, 72, 111]). Tau is expressed robustly throughout the human brain with isoform specific expression at different development stages, different regions, and in select cell populations [17, 22, 73, 81, 104].
Understanding the different roles tau isoforms can play in a cell is critical to deciphering disease processes. For example, 4R isoforms show the greatest propensity to aggregate and lead to cognitive defects in animal models [95, 97], while, 0N3R tau isoforms lead to axonal transport defects and reduce the life-span of drosophila models [97]. Tau isoforms are differentially distributed within neurons, with 2 N tau isoforms showing enrichment in neuronal cell bodies whilst 0 N and 1 N tau isoforms are sorted to axons [13, 70] where they alter microtubule dynamics [90]. 1 N tau isoforms have also been found in the nucleus [70, 75] where tau has been reported to mediate ribosomal gene expression, which has been implicated in proteostasis disruption in tauopathy [31], and epigenetic and heterochromatin changes especially during stress events [49, 62, 76, 106]. Furthermore, tau isoforoms may interact differently with disease associated proteins such as APOE [71], which may lead to disease specific interactions where one tau isoform is altered.
Importantly tau also changes its localisation in neurons post injury or in disease states. Canoncially tau is believed to be enriched in axons of neurons but redistributes to the somato-dendritic compartment or dendritic spines when exposed to trauma, amyloid-beta, hyperphosphorylation by FYN kinase and other signals. It has also been shown that the mutant P301L tau mislocalises to dendritic spines in FTLD [53]. However, a recent study has cast doubt on whether tau mislocalises to the somato-dendritic compartments or is already abundant in this region prior to disease [91]. These disparities are possibly due to the difficulty in detecting tau isoforms or inappropriate localisation of tau in model systems. Regardless, it is important to consider the localisation of tau in the context of interactions as mislocalisation of tau will influence its interaction profile.
Tau isoforms are dysregulated in tauopathies
In physiological conditions, tau is a soluble and unstructured protein. However, in tauopathy it oligomerises and then forms aggregates with a range of conformations [38], resulting in distinct neuropathological lesions in each tauopathy [26]. Many tauopathies have a bias towards one tau isoform over the others [21, 38, 87, 103] and the isoform of tau appears to influence the rate of aggregation with 4R isoforms aggregating the most rapidly [121]. It is not yet understood why this is the case. For example, PiD displays increased aggregation of 3R tau [33] as compared AGD, CBD, GGT and PSP which all have a bias for developing 4R tau aggregates [26, 37, 103]. Neurofibrillary tangles in AD contain both 3R and 4R tau but the ratio of 3R:4R tau in tangles shifts based on the maturity and location of the aggregate [23].
Post-translational modifications of tau in tauopathies
Post-translational modifications (PTMs) of tau also significantly influence protein interactions, sub-cellular localisation, and aggregation propensity of tau [21, 66, 74]. Tau undergoes a vast range of PTMs including phosphorylation, ubiquitination, acetylation, methylation, glycosylation, glycation, oxidation and SUMOylation (for review see [5]). Identification of specific PTM sites involved in human disease historically occurred through antibody-based techniques; however, this approach is not a comprehensive method of identifying all PTMs. The use of mass spectrometry has allowed a more comprehensive analysis of PTMs of tau in post-mortem human tauopathy brain tissue, overcoming these limitations of disease models and antibody-based approaches. Recent mass spectrometry studies have identified numerous additional phosphorylation sites on tau, and further characterised tau’s less abundant PTMs including acetylation, methylation, and ubiquitination [58, 113]. Most studies analysing PTM sites present in human tauopathy brain tissue have focused on AD tissue, with a small number of studies examining CBD, PSP and Pick’s disease tissue [1, 10, 29, 30, 47, 50, 58, 78, 92, 115]. Limited studies have examined PTMs directly in post-mortem tissue in GGT and AGD [47, 58, 94], and no studies to date have analysed tau PTMs in PART. A summary of tau PTMs present in human brain tissue across a range of tauopathies is presented in Fig. 1 and Supplementary Table 1.
Summary of PTMs present on tau in AD and primary tauopathies. A PTMs observed in AD mapped to full length tau and B in primary tauopathies; AGD, CBD, GGT, PiD, PSP. Alternatively spliced regions are coded light orange = 1 N, dark orange = 2 N and light green = 3R, dark green 4R and purple for the proline rich region. Compiled from [1, 10, 29, 30, 36, 50, 58, 78, 92, 94, 113, 115]. Please note that while this compilation of tau PTMs was completed to the best of our knowledge, this is an evolving field and may not be comprehensive
Patterns of phosphorylation differ across tauopathies, with AD, CBD and PiD most frequently phosphorylated, followed by GGT and PSP (Fig. 1; Supplementary Table 1). Few phosphorylation sites have been identified in AGD (Fig. 1). The most frequent phosphorylation sites across these diseases cluster within the proline-rich region, with an additional common cluster in the C-terminal domain (Fig. 1). In contrast, acetylation predominantly occurs upon lysine residues within the microtubule binding regions. These same residues can also be involved in ubiquitination and methylation. Both AD and GGT have multiple lysine residues (K274, K280, K281) where acetylation and ubiquitination compete for common sites. Ubiquitination of tau is a feature of all tauopathies. Interestingly, ubiquitination in the C-terminal domain appears unique to AD (Fig. 1) and models show this PTM stabilizes binding between protofilaments within AD straight filaments [10]. Similarly, cryo-EM suggests K353 ubiquitination in CBD may stabilise packing between protofilaments, with competing acetylation at this site changing filament structure and interactions between filaments [10]. Further combined cryo-EM and mass spectrometry studies of different tauopathies could elucidate the role of specific PTMs in mediating tau fibrillization and aggregation in human disease.
Patterns of PTMs unique to certain tauopathies have previously been identified in the literature, such as the absence of phosphorylation at S262 in PiD, in contrast to its presence in AD, CBD, GGT and PSP [58]. This may relate to structural differences unique to tau filaments in PiD, a 3R tauopathy, with S262 falling within the core region in PiD instead of its location outside the core in AD and other tauopathies (Supplementary Table 1). In conjunction with a tight turn in PiD tau filaments at G261, it is hypothesised that the core location of S262 in PiD may reduce its accessibility to kinases [33]. However, the mechanisms behind other absent PTMs in PiD, such as T263 and S412 phosphorylation, and K281 acetylation, are yet to be explored, or their relevance in human disease clarified. Similarly, PSP’s PTM signature includes an absence of T50, S237 and S238 phosphorylation, contrasting AD, CBD, GGT and PiD (Supplementary Table 1). With a lack of specific biomarkers for sporadic forms of FTD [100], the identification of these nuances in PTMs between AD, CBD, GGT, PSP and PiD could support investigation of new biomarker assays that could diagnose non-AD tauopathies.
Temporal variations in PTMs also exist within tauopathies, with the phosphorylation pattern of tau sequentially changing during AD, with T231 increased significantly by Braak stage III/IV, but other residues (such as S199, S202/T205, S422) significantly increased only at Braak stage V/VI in human AD brains [89]. S396 phosphorylation was present in all tauopathies involving analysis of human post-mortem tissue (Supplementary Table 1). While there is a general perception in the field that phosphorylation of S396/S404 (identified using the PHF1 antibody) occurs later in the disease process, multiple recent studies show that phosphorylation of S396 is actually observed very early in disease in both PSP and AD, suggesting that it may be a feature of early stage disease [9, 78]. In vitro studies in HEK-293 cells suggest kinases involved in AD such as GSK3β require stepwise tau phosphorylation before S396 phosphorylation, with S404 and S400 requiring phosphorylation beforehand [68]. However, the impact of these different spatial and temporal patterns of PTMs of tau, and relevance to human disease remain under investigation. The identification of sequential patterns of PTMs in tauopathies holds the potential to help distinguish early from late-stage disease.
The unique conformations of tau aggregates in tauopathies
Tau undergoes multiple stages of aggregation during tauopathy progression from monomeric soluble tau to small oligomers to large aggregates. These changes complicate the understanding of how tau conveys toxicity as these different aggregate conformations likely have different interactomes. Oligomers are present earlier in disease progression, have higher propensity for seeding, and their small size allows greater mobility and potential to disrupt physiological tau interactions [42, 56, 64]. Oligomers are considered more toxic than large insoluble aggregates [11] as they generate more pathology than fibrils when seeded into mice [56]. Furthermore, Ghag et al. [42] convincingly demonstrated that tau oligomers are the toxic species in vitro, while tau fibrils are comparatively inert.
Recent cryo-EM studies have confirmed that the conformation of tau is different between tauopathies. For example, tau aggregates into straight or paired helical filaments in AD [3, 35]. In PiD, aggregates have a sharp folded beta-sheet that can align end-to-end to form a wide filament from two narrow filaments [33, 96]. Tau aggregates in CBD have a non-proteinaceous molecule in the centre of the folded structure [119]. Importantly, recombinant tau aggregates have a different conformation than aggregates extracted directly from human diseased brain tissue, highlighting a significant caveat associated with using recombinant tau in experimental studies [118]. Tau aggregates purified from AD, PiD, PSP and CBD post-mortem brain tissue replicate disease-specific pathology in both tissue culture and mouse models of disease, suggesting the disease-specific tau aggregates characterise each tauopathy [2, 19, 25, 27, 28, 51, 64, 88, 114]. Pathological aggregate ‘strains’ maintain their preference for cell types and regional specificity when injected into mouse brains [19, 51, 88], suggesting that the microenvironment and specific interactions of tau are important factors in driving pathology. Furthermore, whilst 3R or 4R aggregates can seed aggregates primarily composed of either isoform, they are much more efficient at seeding the aggregation of conformers of the same isoform [1, 51, 101, 117]. Together, the differences in regional vulnerability, preference for tau isoforms and distinct aggregate structures that are unique in each tauopathy suggest that different underlying mechanisms may drive disease in each tauopathy.
Tau interactome studies reveal consistent interaction with RNA binding proteins
Analysis of the tau interactome, particularly the analysis of the interactome of pathological tau strains, is an excellent way to define disease mechanisms that drive tauopathies. The protein interactions of tau are influenced by several factors such as cellular location, isoform, aggregation state, conformation, and the presence of PTMs. Therefore, analysis of strain- and disease-specific tau interactomes provides a unique opportunity to uncover disease mechanisms that drive tauopathies. While research in this area is still in its infancy, several studies have investigated the tau interactome in a variety of models and post-mortem human brain tissue. These studies have primarily used affinity purification-mass spectrometry or localised proteomics to examine tau interactions. Most of these studies have either analysed the total tau interactome or have limited their analysis to full length (2N4R) tau. Furthermore, tau interactome studies have been largely carried out in the context of AD (balanced 3R/4R tau or AD post-mortem human tissue) or examining the interactome of tau expressing mutations present in rare familial cases of FTLD (P301L or V337M) (summarised in Table 1).
Here, we have performed a comprehensive review followed by a combined analysis of tau interactome studies to identify and compare tau interactors in human post-mortem tissue, human cell culture models, and rodent models. Literature review identified twelve tau interactome studies that satisfied our search criteria and provided sufficient data for re-analysis (Table 1). To allow for direct comparison between studies we used a vote-counting approach and gene ID as the protein identifier. Tau interactors were filtered to those that appeared in at least 3/7 human studies and 2/5 rodent studies.
Four studies analysed tau interactions in AD post-mortem human brain tissue. Two assessed the interactome of total tau in AD brain homogenates vs cognitively normal age-matched controls [12, 54], one assessed the interactome of total tau in endoplasmic reticulum enriched fractions of AD vs cognitively normal age-matched controls [82], and one assessed the PHF-1 immunoreactive phosphorylated tau interactome in AD brain tissue [30]. 3 additional studies analysed the tau interactome in human cell models; Gunawardana et al. [48] analysed the tau interactome in SH-SY5Y neuroblastoma cells transfected with GFP tagged 2N4R tau (with or without P301L mutation), Wang et al. [110] analyzed the tau interactome in CRISPR engineered human neural progenitor ReNCell VM cells and Human Neuroblastoma IMR-32 cells expressing equal amounts of GFP tagged 3R and 4R tau, and Tracy et al. [105] analyzed the tau interactome in human induced pluripotent stem cells (IPSCs) expressing APEX-fused 2N4R tau (with and without either P301L or V337M mutations). Importantly, the interactomes of these tagged tau constructs appear broadly consistent with other studies.
Combined analysis of the 7 studies assessing tau interactions in human post-mortem tissue or human cell lines identified 2084 tau interacting proteins (Supplementary Table 2). Of these, 261 proteins were consistently found in > 3 studies. 72 of these proteins interacted with both phosphorylated tau and total tau, and 253 proteins also interacted with either P301L or V337M mutant tau (Supplementary Table 2). Protein–protein interaction network analysis of the 261 proteins that interacted with tau consistently across > 3 studies showed particularly significant enrichment of RNA binding proteins and ribosomal proteins (Fig. 2.A). The most consistent tau-interacting RNA binding proteins were heterogeneous nuclear ribonucleoproteins (HNRNPs), FUS, SFPQ and PTBP1 (Table 2, Supplementary Table 2). The consistent interaction between tau and RNA binding proteins across multiple studies is particularly interesting as RNA is a known component of tau aggregates in tauopathies [44, 45] and RNA (especially tRNAs) enhances the aggregation of tau [59, 120]. Furthermore, previous studies suggest that RNA can be required as an intermediate linker for select tau-protein interactions, particularly with ribosomal proteins [48]. However, it is important to note that this is not always the case; for example, several HNRNP-tau interactions are enhanced after RNA removal [48].
Tau interactome analyses reveal consistent tau-protein interactions across human brain and cell culture models. A PPI network of human tau interacting proteins consistent in > 3 of 7 studies. Network node colours are clustered by k-means set with 8 groups. String network is limited to physical interactions only. Clusters of interacting proteins are manually annotated (see Supplementary Table 2 for annotated proteins). B PPI network of rodent model tau interacting proteins. Network node colours are clustered by k-means set with 8 groups. String network is limited to physical interactions only. Clusters of interacting proteins are manually annotated (see Supplementary Table 3 for annotated proteins). C Zoomed in view of PPI network of RNA-binding proteins in human tissue D GO Molecular function enrichments of human and rodent models generated in R with the package clusterProfiler v4.2.2. Top 8 processes are listed. The background gene set for rodent and human studies included all proteins detected in IP studies (see Supplementary Table 4 for GO molecular function annotations). All PPI networks were generated with the STRING database (v11.5, https://string-db.org/)
Comparison of the P301L mutant and wild-type tau interactomes highlighted reduced interaction between tau and proteasome subunits and increased interaction between tau and 40/60S ribosomal subunits, translation initiation factors and HNRNPs [48]. This provides key evidence that different tau strains have different interactomes, which provides downstream avenues for future research investigating disease mechanisms. Two of the most consistent clusters of proteins in the human tau interactome are proteosome and ribosome proteins (Fig. 2.A, D). The proteosome is frequently dysregulated in tauopathy. Tau aggregates or tau overexpression lead to altered proteostasis and protease inhibition [18, 116] and small molecules activating the proteosome can reverse cognitive deficits associated with tau [86]. Almost all consistent tau interacting proteosome proteins also interact with phosphorylated tau, suggesting that these interactions may be enhanced at later stage pathology [60] and may reflect disease-associated tau inhibiting the proteosome in a manner similar to amyloid-beta and α-synuclein [80, 102]. The interaction between tau and the ribosome has been a growing area of interest in the study of tauopathy. Tau interacts with, and sequesters, ribosomal components – particularly regulatory components [32, 63], resulting in decreased protein synthesis in models of tauopathy [31]. Ash et al. [11] demonstrated that TIA1 can readily drive the aggregation of tau alongside other RNA binding proteins which required crowding agents to drive aggregate tau. Furthermore, tRNAs increase the rate of tau aggregation in vitro [120], and as such the ribosome may act as a seeding point for tau oligomerisation and aggregation. Interestingly ribosome proteins don’t appear robustly enriched in interactions with phosphorylated tau, suggesting tau may play a consistent role in ribosomal regulation that is altered with disease state and not required for late-stage pathology. Whether the interaction between tau and ribosomal proteins interrupts specific populations of translating ribosomes is yet to be fully explored.
Five studies have analysed the tau interactome in rodents. Liu et al. [70] assayed total tau, 3R tau, and 4R tau specific interactions in mice. Wang et al. [109] investigated full-length tau interactions of PS19 mice (expressing P301S mutant tau). Maziuk et al. [79] investigated total tau interactomes in wild type and rTg4510 mice (expressing P301L human mutant tau) and Sinsky et al. [99] explored the interactomes of 2N4R tau in wild type and transgenic humanised tau rats. Jiang et al. [57] assessed the interactions of inducible oligomerising tau constructs in mouse primary neurons. These rodent studies detected fewer total proteins than the human studies described above: with 244 of 1,152 (15.9%) detected proteins present in 2 or more rodent tau interactome studies (Supplementary Table 3). This inconsistency and lack of detection is likely a result of the wide variety of tau species studied, the low numbers of animals included in each study, and the fact that humanised mutant tau may not interact well with mouse RNA binding proteins and ribosomal proteins. Importantly, many of the consistent protein interactions with tau that were observed in human studies were not observed in rodent studies (Fig. 2.B). For example, while some studies did report interactions between tau and some HNRNP proteins (for example Hnrnpa2/b1 and a3 in rats [99] and Hnrnpa0, l, k, u, r and q in rTg4510 mice [79] and Hnrnpa0, a1 and a2b1 in inducible oligomeric tau expressing C7Bl6/J primary neurons), interactions between tau and RNA binding proteins was not a consistent feature of rodent studies. Additionally, interactions between tau and ribosomal proteins were not consistently observed in rodent studies (Fig. 2.B). Rodent studies showed consistent enrichment of tau interactions with heat shock proteins, and energy metabolism, synapse, and trafficking proteins (Fig. 2.B, D). Despite the inconsistencies noted between human and rodent studies of the tau interactome, some interactions were maintained between species. For example, interaction between tau and proteins involved in vesicle trafficking and the unfolded protein response were observed in both humans and rodents. This highlights some important caveats to consider when using rodent models of disease to study disease mechanisms present in human tauopathies.
Association of tau interacting proteins with pathology
One important unanswered question is whether protein interactions with tau influence the development or progression of tau pathology in tauopathies. Two recent studies have used localized proteomics to analyze the proteins present in neurons containing neurofibrillary tangles (NFTs) or granulovacuolar degeneration (GVD; pathology hypothesized to be the precursor to the development of NFTs) isolated from AD human brain tissue [30, 52]. Integration of these two studies with our combined analysis of the human tau interactome identified proteins that are both present in pathological aggregates and interact with tau, and are therefore more likely to have a mechanistic role in the development of tau pathology in AD.
165 of the 261 consistent tau interactors were present in NFTs [30], showing that many tau interacting proteins were associated with pathological aggregates in AD. 37 tau interacting proteins were significantly altered in GVD containing neurons and 64 tau interacting proteins were significantly altered in NFT containing neurons [52]. We then specifically analysed which phosphorylated tau interacting proteins were enriched in neurons containing GVD or NFTs. This analysis showed that neurons containing GVD were significantly enriched in phosphorylated tau interacting proteins (42 proteins; p = 1.64*10–5, Fisher exact test, Fig. 3.A, Supplementary Table 5), as were neurons containing NFTs (48 proteins; p = 1.63*10–4, Fisher exact test, Fig. 3.A, Supplementary Table 5). These phosphorylated tau interacting proteins that were enriched in GVDs or NFTs included many with functions associated with the ribosome, synapses, and vesicle trafficking (Fig. 3.B, Supplementary Table 6). Tau interacting proteins altered in neurons containing NFTs largely included proteins involved in protein degradation pathways (Fig. 3.B, C, D, Supplementary Table 6). Given the consistent interaction between tau and RNA binding proteins described above, we were particularly interested to analyse whether altered levels of RNA binding proteins were associated with tau pathology. The majority of RBPs showed a trend for lower levels in GVD and NFT containing neurons (Supplementary Table 2). Noteworthy exceptions included HNRNPK, HNRNPA1 and HNRNPA2/B1, which all showed a trend for increased expression in GVD containing neurons (Fig. 3.C). Interestingly, these three HNRNPs all interacted with phosphorylated tau and their trend for increased expression was only observed in neurons containing early-stage pathology; increased expression was no longer observed in neurons containing NFTs. The interaction between total tau and these three HNRNPs is highly consistent in human studies including both in cell models and post-mortem brain tissue (Table 2). This suggests that not all HNRNPs have the same actions in tauopathy and lends weight to the hypothesis that HNRNPK, HNRNPA1 and HNRNPA2/B1 may influence the development of early tau pathology. The differential enrichment of these HNRNPs in neurons with GVD pathology and NFTs suggests there may be multiple phases to the dysregulation of tau interactions in disease with early stress response proteins present in GVD.
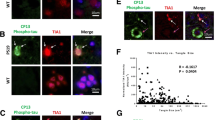
Analysis of proteins that interact with pTau and are altered in NFT or GVD containing neurons. A Venn diagram showing the overlap between pTau interacting proteins of Drummond et al. [30] and proteins with a trended change in expression in NFT or GVD containing neurons [52]. This trend was defined as a fold change in expression > 0.1 either up or down as p-values were not supplied with this dataset only FDR and most other proteomics studies fail to report FDR. B Enrichment plot summarising GO cellular compartment terms enriched for each dataset generated in R with clsuterProfiler v4.2.2. Nodes are divided into pie charts that show the proportion of genes shared for that category. Nodes are connected by shared genes to cluster terms into broader functions. Only the top 50 terms are shown. Clusters are annotated based on the majority of component pathways (Supplementary Table 6). C PPI network of proteins that are pTau interactors and altered in GVD and D NFT containing neurons. PPI networks were generated with the STRING database (v11.5, https://string-db.org/). String nodes are coloured by selected STRING annotation for those proteins (RNA binding and proteolysis). STRING network included physical and regulatory interactions
HNRNPs: general biology and dysregulation in tauopathy
Our comprehensive review of tau interactome studies suggest that the interaction between HNRNPs (and more broadly RNA binding proteins) and tau may have an important role in tauopathy disease mechanisms. HNRNPs have broad molecular functions including splicing regulation, mRNA transport and stability, and mediating mRNA stress responses via stress granule formation [15, 41]. Their activity and binding are targeted to sub-classes of mRNA (e.g. binding m6A mRNAs by HNRNPA2/B1 [4, 57]). Understanding the interaction of HNRNPs with tau in disease contexts is critical as these proteins have the immediate ability to impact multiple specific pathways within a cell if dysregulated. Mutations in HNRNPs are associated risk variants for neurodevelopmental and neurodegenerative disorders (including ALS/FTLD, multisystem proteinopathy, Bain-type and HNRNPR-related or HNRNPU-related disorders amongst others [43, 61]). Furthermore, HNRNPs readily aggregate in disease and can enhance the rate of conformational change for other disease associated proteins such as TDP43 [16, 24, 77, 93]. Importantly, reduction of RNA binding proteins TIA1 and HNRNPA2/B1 expression has been shown to reduce or prevent the formation of tau pathology [57, 107].
While there is consistent evidence of tau interacting with HNRNPs (and with RBPs more broadly), the consequences of these interactions in disease contexts are largely unexplored. One hypothesis is that tau binds to RBPs, the ribosome and RNAs as part of the stress granule response. TIA1 – an RNA binding protein – has been shown to regulate the toxicity and aggregation of tau. TIA1 exacerbates tau aggregation with RNA and drives the generation of toxic tau oligomers in vitro and in cell models and reduction of TIA1 is protective against tau toxicity [11, 107, 108]. In addition, tau accelerates the formation of TIA1 positive stress granules [107]. In vivo, TIA1 regulates the toxicity of tau oligomers. Reduction of TIA1 inhibits the spread and prevents toxicity of seeded tau aggregates, but interestingly, increased neurofibrillary tangle burden and neuroinflammation [8, 56, 65]. TIA1 and other RNA binding proteins co-aggregate with tau in small lesions in rTg4510 mice and AD post-mortem brain tissue but appear adjacent to (not in) larger aggregates [56, 79]. While this interaction is increased with pathology, it is important to note that tau also interacts with RBPs and the ribosome in cell models without significant pathology or stress [48, 105, 110], suggesting these interactions occur in physiological conditions but are increased and stabilised in pathological conditions which then leads to detrimental effects for the cell. However, none of the affinity purification-mass spectrometry studies above detected TIA1 as a tau interactor. The interaction between TIA1 and tau has only been observed in rTg4510 mice with moderate/severe phenotypes or mammalian cells overexpressing both TIA1 and 0N4R tau, suggesting that the interaction between TIA1 and tau may be either of low abundance or indirect.
Oligomeric tau has been shown to co-aggregate with Musashi 1 and 2 in inducible HEK-293 cells [85] and with tau aggregates in AD and FTD [84, 98]. These aggregates develop in the cytoplasm and nucleus, impede Musahsi 1/2 nuclear/cytoplasmic transport and destabilise the nuclear laminin leading to cellular damage. Interestingly, oligomeric tau induces astrocytic senescence and drives the release of HMGB1 which initiates further cellular senescence [39]. In our combined analysis of tau interactions, Musashi 2 and HMGB1 were found to interact with tau in one study—Tracy et al. [105]. These papers highlight the broad impact of tau on RNA binding proteins and potential to impact other cells in the brain if they can drive the release of HMGB1.
Only a small number of studies have focused on HNRNP interactions with tau. HNRNPA1 and HNRNPK co-localize with small tau inclusions but are not associated with large tau inclusions such as NFTs. A number of HNRNPs form their own separate insoluble aggregates as tau pathology progresses, such as HNRNPK in FTLD [14]. HNRNPK aggregates did not co-localize with large tau aggregates and were mutually exclusive of TDP43 pathology [14], but instead appeared in neighbouring neurons in vulnerable brain regions that did not contain tau inclusions. One possible explanation for this is that HNRNPs may interact with oligomeric tau species early in disease rather than fibrillar tau aggregates [79], a hypothesis explicitly supported by Jiang et al. [57]. HNRNPK modifies tau toxicity in drosophila models [6], suggesting that whilst HNRNPK aggregates do not appear to directly interact with tau in pathological inclusions, it is involved in mediating tau toxicity. Intracellular HNRNPK puncta were distinct from GS3BP2 stress granules, suggesting the aggregation of HNRNPK is separate from the TIA1-tau aggregation pathways, and TDP-43 positive inclusions [14]. Knock-down of HNRNPK in SH-SY5Y neuroblastoma cells resulted in similar splicing defects that are present in FTLD human brain tissue [14], suggesting that the accumulation of HNRNPs in tauopathy may result in loss of function. It is not yet known whether this loss of function is a result of stress granule formation or an earlier disruption of spliceosome function [54, 67, 83, 85].
HNRNPA2/B1 also mediates tau aggregation. Jiang et al. [57] showed that numerous HNRNPs interacted with tau oligomers, generated using an optogenetic approach in mouse primary neurons, including HNRNP A0, A1, A2B1, DL, F, H, K, L, M, R and UL1. Many ribosomal proteins also interacted with tau oligomers. Of these, HNRNPA2/B1 was the most significant tau oligomer interactor and interaction resulted in mislocalization of HNRNPA2/B1 from the nucleus to the cytoplasm in vitro, in mouse models of tauopathy and in human AD post-mortem brain tissue [57]. Importantly, reduction of HNRNPA2/B1 expression delayed tau oligomer formation and reduced toxicity. Curiously this interaction and tau oligomer toxicity was mediated by m6A RNA and knockdown of METTL3 (an m6A writer) delayed oligomer formation. This was correlated with increased prevalence of m6A labelled mRNA in AD with disease severity (fourfold in mouse models, 2.5 fold in AD brains) [57], suggesting that specific mRNA types are dysregulated with worsening severity in disease. Stable tau oligomer expression also elicits the aggregation of stress granule markers TIA1 and EIF3-η and PABP [57], but no enrichment for stress granule markers G3BP1, G3BP2, CAPRIN or USP10. This suggests there may be different subsets of stress granules that accumulate specific transcripts.This would cause tau:RNA binding protein interactions to sequester specific subsets of the mRNA pool and disproportionately impact the targeted pathways. Furthermore, HNRNPA2/B1 mediates the development of tau-containing stress granules and the interaction between tau oligomers and HNRNPA2/B1 alter protein synthesis [55]. Together, these results provide evidence for a concerted disruption of RNA metabolism by tau. Furthermore, the interactions between tau and RNA binding proteins may promote further dysregulation by over-activation of stress responses.
Conclusions
The impacts of tau dysregulation through oligomerisation and aggregation in disease are likely conveyed by protein–protein (or protein-RNA) interactions specific to the brain region, sub-cellular compartment and cell type affected. As tau accumulates it can sequester interacting proteins and these interacting proteins can potentially modify the development of pathology. As tau pathology accumulates over the lifetime of a patient it has the potential to act as a time-capsule, preserving information about the early micro-environment that generated these aggregates. Our combined analysis of the human tau interactome with proteins present in neurons containing GVD or NFTs provides new information about the disease mechanisms that may be involved in tauopathies. Whilst many of the studies done to date have been limited in scope (i.e., have targeted a single tau isoform or a single disease), together these studies consistently identify many novel and overlooked protein interactions that may mediate toxicity in tauopathies. Our analysis has highlighted the particular importance of the interaction between tau and RNA binding proteins (particularly HNRNPs) in tauopathies. The consistent detection of these interactions across multiple human studies suggests that they are bona fide tau interactors.
There are still many unanswered questions in this field that require further research. For example, the vast majority of tau interactome studies performed to date have only focused on AD or models expressing mutated tau present in rare cases of FTLD. Similar experiments in other tauopathies or models using different tau isoforms would help to elucidate tau isoform and mutant specific interactions that may reveal novel disease mechanisms. Future mechanistic studies are required to determine whether specific protein interactions with tau influence pathology development and if these interactions could be therapeutically targeted. Initial studies suggest that disease associated PTMs change the interactome of tau, however, larger systematic studies are required to examine the impact of individual PTMs on the tau interactome across the spectrum of tauopathies.
A more detailed exploration of tau-protein interactions has the potential to reveal underlying disease mechanisms of tauopathies and to identify novel targets for drug development. Much more study is needed to accommodate the complexity of tau that results from the interaction of multiple tau isoforms, multiple different superstructures of aggregates and dysregulation in unique cells and regions in different diseases. It is critical we develop models that can fully accommodate this complexity and begin to move beyond single isoform/single disease approaches. Studies to date which investigate the mechanisms driving tau aggregation through interacting partners highlight the importance of understanding these interactions. Our combined analysis of tau interactome studies presented here highlight the potential importance of the interaction between tau and RNA binding proteins, particularly HNRNPs, in driving tauopathy. As such, exploring the mechanistic role of HNRNPs in tauopathy is an exciting future avenue of research.
Availability of data and materials
All data generated or analysed during this study are included in this published article (and its supplementary information files).
Abbreviations
- AD:
-
Alzheimer’s disease
- AGD:
-
Argyrophilic grain disease
- ARTAG:
-
Aging related tau astrogliopathy
- CBD:
-
Corticobasal degeneration
- CTE:
-
Chronic traumatic encephalopathy
- FTLD:
-
Frontotemporal lobar degeneration
- GGT:
-
Globular glial tauopathy
- GVD:
-
Granulovacuolar degeneration
- HNRNPs:
-
Heterogeneous nuclear ribonucleoproteins
- IPSCs:
-
Induced pluripotent stem cells
- NFTs:
-
Neurofibrillary tangles
- PART:
-
Primary age related tauopathy
- PiD:
-
Pick’s disease
- PSP:
-
Progressive supranuclear palsy
- PTM:
-
Post-translational modification
- pTau:
-
Phosphorylated tau
- RBP:
-
RNA binding protein
References
Abreha MH, Dammer EB, Ping L, Zhang T, Duong DM, Gearing M, Lah JJ, Levey AI, Seyfried NT. Quantitative analysis of the brain ubiquitylome in Alzheimer’s Disease. Proteomics. 2018;18(20):e1800108–e1800108. https://doi.org/10.1002/pmic.201800108.
Ahmed Z, Cooper J, Murray TK, Garn K, McNaughton E, Clarke H, Parhizkar S, Ward MA, Cavallini A, Jackson S, Bose S, Clavaguera F, Tolnay M, Lavenir I, Goedert M, Hutton ML, O’Neill MJ. A novel in vivo model of tau propagation with rapid and progressive neurofibrillary tangle pathology: the pattern of spread is determined by connectivity, not proximity. Acta Neuropathol. 2014;127(5):667–83. https://doi.org/10.1007/s00401-014-1254-6.
Al-Hilaly YK, Pollack SJ, Vadukul DM, Citossi F, Rickard JE, Simpson M, Storey JMD, Harrington CR, Wischik CM, Serpell LC. Alzheimer’s Disease-like paired helical filament assembly from Truncated Tau protein is independent of disulfide crosslinking. J Mol Biol. 2017;429(23):3650–65. https://doi.org/10.1016/j.jmb.2017.09.007.
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–308. https://doi.org/10.1016/j.cell.2015.08.011.
Alquezar C, Arya S, Kao AW. Tau post-translational modifications: dynamic transformers of Tau function, degradation, and aggregation. Front Neurol. 2021;11(1826). https://doi.org/10.3389/fneur.2020.595532
Ambegaokar SS, Jackson GR. Functional genomic screen and network analysis reveal novel modifiers of tauopathy dissociated from tau phosphorylation. Hum Mol Genet. 2011;20(24):4947–77. https://doi.org/10.1093/hmg/ddr432.
Andreadis A. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim Biophys Acta. 2005;1739(2):91–103. https://doi.org/10.1016/j.bbadis.2004.08.010.
Apicco DJ, Ash PEA, Maziuk B, LeBlang C, Medalla M, Al Abdullatif A, Ferragud A, Botelho E, Ballance HI, Dhawan U, Boudeau S, Cruz AL, Kashy D, Wong A, Goldberg LR, Yazdani N, Zhang C, Ung CY, Tripodis Y, Kanaan NM, Ikezu T, Cottone P, Leszyk J, Li H, Luebke J, Bryant CD, Wolozin B. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat Neurosci. 2018;21(1):72–80. https://doi.org/10.1038/s41593-017-0022-z.
Aragão Gomes L, Uytterhoeven V, Lopez-Sanmartin D, Tomé SO, Tousseyn T, Vandenberghe R, Vandenbulcke M, von Arnim CAF, Verstreken P, Thal DR. Maturation of neuronal AD-tau pathology involves site-specific phosphorylation of cytoplasmic and synaptic tau preceding conformational change and fibril formation. Acta Neuropathol. 2021;141(2):173–92. https://doi.org/10.1007/s00401-020-02251-6.
Arakhamia T, Lee CE, Carlomagno Y, Duong DM, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, Seyfried NT, Petrucelli L, Fitzpatrick AWP. Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell. 2020;180(4):633-644.e612. https://doi.org/10.1016/j.cell.2020.01.027.
Ash PEA, Lei S, Shattuck J, Boudeau S, Carlomagno Y, Medalla M, Mashimo BL, Socorro G, Al-Mohanna LFA, Jiang L, Öztürk MM, Knobel M, Ivanov P, Petrucelli L, Wegmann S, Kanaan NM, Wolozin B. TIA1 potentiates tau phase separation and promotes generation of toxic oligomeric tau. Proc Natl Acad Sci. 2021;118(9):e2014188118. https://doi.org/10.1073/pnas.2014188118.
Ayyadevara S, Balasubramaniam M, Parcon PA, Barger SW, Griffin WST, Alla R, Tackett AJ, Mackintosh SG, Petricoin E, Zhou W, Shmookler Reis RJ. Proteins that mediate protein aggregation and cytotoxicity distinguish Alzheimer’s hippocampus from normal controls. Aging Cell. 2016;15(5):924–39. https://doi.org/10.1111/acel.12501.
Bachmann S, Bell M, Klimek J, Zempel H. Differential effects of the six human TAU isoforms: somatic retention of 2N-TAU and increased microtubule number induced by 4R-TAU. Front Neurosci. 2021;15(547). https://doi.org/10.3389/fnins.2021.643115
Bampton A, Gatt A, Humphrey J, Cappelli S, Bhattacharya D, Foti S, Brown A-L, Asi Y, Low YH, Foiani M, Raj T, Buratti E, Fratta P, Lashley T. HnRNP K mislocalisation is a novel protein pathology of frontotemporal lobar degeneration and ageing and leads to cryptic splicing. Acta Neuropathol. 2021;142(4):609–27. https://doi.org/10.1007/s00401-021-02340-0.
Bampton A, Gittings LM, Fratta P, Lashley T, Gatt A. The role of hnRNPs in frontotemporal dementia and amyotrophic lateral sclerosis. Acta Neuropathol. 2020;140(5):599–623. https://doi.org/10.1007/s00401-020-02203-0.
Batlle C, Yang P, Coughlin M, Messing J, Pesarrodona M, Szulc E, Salvatella X, Kim HJ, Taylor JP, Ventura S. hnRNPDL phase separation is regulated by alternative splicing and disease-causing mutations accelerate its aggregation. Cell Rep. 2020;30(4):1117-1128.e1115. https://doi.org/10.1016/j.celrep.2019.12.080.
Binder LI, Frankfurter A, Rebhun LI. The distribution of tau in the mammalian central nervous system. J Cell Biol. 1985;101(4):1371–8. https://doi.org/10.1083/jcb.101.4.1371.
Blaudin de Thé F-X, Lassus B, Schaler AW, Fowler SL, Goulbourne CN, Jeggo R, Mannoury la Cour C, Millan MJ, Duff KE. P62 accumulates through neuroanatomical circuits in response to tauopathy propagation. Acta Neuropathol Commun. 2021;9(1):177. https://doi.org/10.1186/s40478-021-01280-w.
Boluda S, Iba M, Zhang B, Raible KM, Lee VMY, Trojanowski JQ. Differential induction and spread of tau pathology in young PS19 tau transgenic mice following intracerebral injections of pathological tau from Alzheimer’s disease or corticobasal degeneration brains. Acta Neuropathol. 2015;129(2):221–37. https://doi.org/10.1007/s00401-014-1373-0.
Broderick J, Wang J, Andreadis A. Heterogeneous nuclear ribonucleoprotein E2 binds to tau exon 10 and moderately activates its splicing. Gene. 2004;331:107–14. https://doi.org/10.1016/j.gene.2004.02.005.
Buée L, Delacourte A. Comparative biochemistry of Tau in progressive supranuclear palsy, corticobasal degeneration, FTDP-17 and Pick’s Disease. Brain Pathol. 1999;9(4):681–93. https://doi.org/10.1111/j.1750-3639.1999.tb00550.x.
Bullmann T, Holzer M, Mori H, Arendt T. Pattern of tau isoforms expression during development in vivo. Int J Dev Neurosci. 2009;27(6):591–7. https://doi.org/10.1016/j.ijdevneu.2009.06.001.
Cherry JD, Esnault CD, Baucom ZH, Tripodis Y, Huber BR, Alvarez VE, Stein TD, Dickson DW, McKee AC. Tau isoforms are differentially expressed across the hippocampus in chronic traumatic encephalopathy and Alzheimer’s disease. Acta Neuropathol Commun. 2021;9(1):86. https://doi.org/10.1186/s40478-021-01189-4.
Chiang W-C, Lee M-H, Chen T-C, Huang J-R. Interactions between the intrinsically disordered regions of hnRNP-A2 and TDP-43 accelerate TDP-43’s conformational transition. Int J Mol Sci. 2020;21(16):5930. https://doi.org/10.3390/ijms21165930.
Chung D-EC, Carlomagno Y, Cook CN, Jansen-West K, Daughrity L, Lewis-Tuffin LJ, Castanedes-Casey M, DeTure M, Dickson DW, Petrucelli L. Tau exhibits unique seeding properties in globular glial tauopathy. Acta Neuropathol Commun. 2019;7(1):36. https://doi.org/10.1186/s40478-019-0691-9.
Chung D-EC, Roemer S, Petrucelli L, Dickson DW. Cellular and pathological heterogeneity of primary tauopathies. Mol Neurodegener. 2021;16(1):57. https://doi.org/10.1186/s13024-021-00476-x.
Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J, Probst A, Winkler DT, Reichwald J, Staufenbiel M, Ghetti B, Goedert M, Tolnay M. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci USA. 2013;110(23):9535–40. https://doi.org/10.1073/pnas.1301175110.
Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, Fraser G, Stalder AK, Beibel M, Staufenbiel M, Jucker M, Goedert M, Tolnay M. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11(7):909–13. https://doi.org/10.1038/ncb1901.
Dammer EB, Lee AK, Duong DM, Gearing M, Lah JJ, Levey AI, Seyfried NT. Quantitative phosphoproteomics of Alzheimer’s disease reveals cross-talk between kinases and small heat shock proteins. Proteomics. 2015;15(2–3):508–19. https://doi.org/10.1002/pmic.201400189.
Drummond E, Pires G, MacMurray C, Askenazi M, Nayak S, Bourdon M, Safar J, Ueberheide B, Wisniewski T. Phosphorylated tau interactome in the human Alzheimer’s disease brain. Brain. 2020. https://doi.org/10.1093/brain/awaa223.
Evans HT, Benetatos J, van Roijen M, Bodea L-G, Götz J. Decreased synthesis of ribosomal proteins in tauopathy revealed by non-canonical amino acid labelling. EMBO J. 2019;38(13):e101174–e101174. https://doi.org/10.15252/embj.2018101174.
Evans HT, Taylor D, Kneynsberg A, Bodea L-G, Götz J. Altered ribosomal function and protein synthesis caused by tau. Acta Neuropathol Commun. 2021;9(1):110. https://doi.org/10.1186/s40478-021-01208-4.
Falcon B, Zhang W, Murzin AG, Murshudov G, Garringer HJ, Vidal R, Crowther RA, Ghetti B, Scheres SHW, Goedert M. Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature. 2018;561(7721):137–40. https://doi.org/10.1038/s41586-018-0454-y.
Fischer I, Baas PW. Resurrecting the mysteries of big Tau. Trends Neurosci. 2020;43(7):493–504. https://doi.org/10.1016/j.tins.2020.04.007.
Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, Scheres SHW. Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature. 2017;547(7662):185–90. https://doi.org/10.1038/nature23002.
Forman MS, Zhukareva V, Bergeron C, Chin SSM, Grossman M, Clark C, Lee VMY, Trojanowski JQ. Signature Tau neuropathology in gray and white matter of corticobasal degeneration. Am J Pathol. 2002;160(6):2045–53. https://doi.org/10.1016/S0002-9440(10)61154-6.
Forrest SL, Kril JJ, Halliday GM. Cellular and regional vulnerability in frontotemporal tauopathies. Acta Neuropathol. 2019;138(5):705–27. https://doi.org/10.1007/s00401-019-02035-7.
Furukawa Y, Kaneko K, Nukina N. Tau protein assembles into isoform- and disulfide-dependent polymorphic fibrils with distinct structural properties. J Biol Chem. 2011;286(31):27236–46. https://doi.org/10.1074/jbc.M111.248963.
Gaikwad S, Puangmalai N, Bittar A, Montalbano M, Garcia S, McAllen S, Bhatt N, Sonawane M, Sengupta U, Kayed R. Tau oligomer induced HMGB1 release contributes to cellular senescence and neuropathology linked to Alzheimer’s disease and frontotemporal dementia. Cell Rep. 2021;36(3):109419. https://doi.org/10.1016/j.celrep.2021.109419.
Georgieff IS, Liem RK, Mellado W, Nunez J, Shelanski ML. High molecular weight tau: preferential localization in the peripheral nervous system. J Cell Sci. 1991;100(1):55–60. https://doi.org/10.1242/jcs.100.1.55.
Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135(8):851–67. https://doi.org/10.1007/s00439-016-1683-5.
Ghag G, Bhatt N, Cantu DV, Guerrero-Munoz MJ, Ellsworth A, Sengupta U, Kayed R. Soluble tau aggregates, not large fibrils, are the toxic species that display seeding and cross-seeding behavior. Protein Sci. 2018;27(11):1901–9. https://doi.org/10.1002/pro.3499.
Gillentine MA, Wang T, Hoekzema K, Rosenfeld J, Liu P, Guo H, Kim CN, De Vries BBA, Vissers LELM, Nordenskjold M, Kvarnung M, Lindstrand A, Nordgren A, Gecz J, Iascone M, Cereda A, Scatigno A, Maitz S, Zanni G, Bertini E, Zweier C, Schuhmann S, Wiesener A, Pepper M, Panjwani H, Torti E, Abid F, Anselm I, Srivastava S, Atwal P, Bacino CA, Bhat G, Cobian K, Bird LM, Friedman J, Wright MS, Callewaert B, Petit F, Mathieu S, Afenjar A, Christensen CK, White KM, Elpeleg O, Berger I, Espineli EJ, Fagerberg C, Brasch-Andersen C, Hansen LK, Feyma T, Hughes S, Thiffault I, Sullivan B, Yan S, Keller K, Keren B, Mignot C, Kooy F, Meuwissen M, Basinger A, Kukolich M, Philips M, Ortega L, Drummond-Borg M, Lauridsen M, Sorensen K, Lehman A, Lopez-Rangel E, Levy P, Lessel D, Lotze T, Madan-Khetarpal S, Sebastian J, Vento J, Vats D, Benman LM, McKee S, Mirzaa GM, Muss C, Pappas J, Peeters H, Romano C, Elia M, Galesi O, Simon MEH, van Gassen KLI, Simpson K, Stratton R, Syed S, Thevenon J, Palafoll IV, Vitobello A, Bournez M, Faivre L, Xia K, Acampado J, Ace AJ, Amatya A, Astrovskaya I, Bashar A, Brooks E, Butler ME, Cartner LA, Chin W, Chung WK, Daniels AM, Feliciano P, Fleisch C, Ganesan S, Jensen W, Lash AE, Marini R, Myers VJ, O’Connor E, Rigby C, Robertson BE, Shah N, Shah S, Singer E, Snyder LAG, Stephens AN, Tjernagel J, Vernoia BM, Volfovsky N, White LC, Hsieh A, Shen Y, Zhou X, Turner TN, Bahl E, Thomas TR, Brueggeman L, Koomar T, Michaelson JJ, O’Roak BJ, Barnard RA, Gibbs RA, Muzny D, Sabo A, Baalman Ahmed KL, Eichler EE, Siegel M, Abbeduto L, Amaral DG, Hilscher BA, Li D, Smith K, Thompson S, Albright C, Butter EM, Eldred S, Hanna N, Jones M, Coury DL, Scherr J, Pifher T, Roby E, Dennis B, Higgins L, Brown M, Alessandri M, Gutierrez A, Hale MN, Herbert LM, Schneider HL, David G, Annett RD, Sarver DE, Arriaga I, Camba A, Gulsrud AC, Haley M, McCracken JT, Sandhu S, Tafolla M, Yang WS, Carpenter LA, Bradley CC, Gwynette F, Manning P, Shaffer R, Thomas C, Bernier RA, Fox EA, Gerdts JA, Pepper M, Ho T, Cho D, Piven J, Lechniak H, Soorya LV, Gordon R, Wainer A, Yeh L, Ochoa-Lubinoff C, Russo N, Berry-Kravis E, Booker S, Erickson CA, Prock LM, Pawlowski KG, Matthews ET, Brewster SJ, Hojlo MA, Abada E, Lamarche E, Wang T, Murali SC, Harvey WT, Kaplan HE, Pierce KL, DeMarco L, Horner S, Pandey J, Plate S, Sahin M, Riley KD, Carmody E, Constantini J, Esler A, Fatemi A, Hutter H, Landa RJ, McKenzie AP, Neely J, Singh V, Van Metre B, Wodka EL, Fombonne EJ, Huang-Storms LY, Pacheco LD, Mastel SA, Coppola LA, Francis S, Jarrett A, Jacob S, Lillie N, Gunderson J, Istephanous D, Simon L, Wasserberg O, Rachubinski AL, Rosenberg CR, Kanne SM, Shocklee AD, Takahashi N, Bridwell SL, Klimczac RL, Mahurin MA, Cotrell HE, Grant CA, Hunter SG, Martin CL, Taylor CM, Walsh LK, Dent KA, Mason A, Sziklay A, Smith CJ, Earl RK, Nowakowski T, Bernier RA, Eichler EE, Study C, Consortium S. Rare deleterious mutations of HNRNP genes result in shared neurodevelopmental disorders. Genome Med. 2021;13(1):63. https://doi.org/10.1186/s13073-021-00870-6.
Ginsberg SD, Crino PB, Lee VMY, Eberwine JH, Trojanowski JQ. Sequestration of RNA in Alzheimer’s disease neurofibrillary tangles and senile plaques. Ann Neurol. 1997;41(2):200–9. https://doi.org/10.1002/ana.410410211.
Ginsberg SD, Galvin JE, Chiu TS, Lee VMY, Masliah E, Trojanowski JQ. RNA sequestration to pathological lesions of neurodegenerative diseases. Acta Neuropathol. 1998;96(5):487–94. https://doi.org/10.1007/s004010050923.
Goedert M, Spillantini MG. Ordered assembly of Tau protein and neurodegeneration. Adv Exp Med Biol. 2019;1184:3–21. https://doi.org/10.1007/978-981-32-9358-8_1.
Grinberg LT, Wang X, Wang C, Sohn PD, Theofilas P, Sidhu M, Arevalo JB, Heinsen H, Huang EJ, Rosen H, Miller BL, Gan L, Seeley WW. Argyrophilic grain disease differs from other tauopathies by lacking tau acetylation. Acta Neuropathol. 2013;125(4):581–93. https://doi.org/10.1007/s00401-013-1080-2.
Gunawardana CG, Mehrabian M, Wang X, Mueller I, Lubambo IB, Jonkman JEN, Wang H, Schmitt-Ulms G. The human Tau interactome: binding to the ribonucleoproteome, and impaired binding of the proline-to-leucine mutant at position 301 (P301L) to chaperones and the proteasome. Mol Cell Proteomics. 2015;14(11):3000–14. https://doi.org/10.1074/mcp.M115.050724.
Guo C, Jeong H-H, Hsieh Y-C, Klein H-U, Bennett DA, De Jager PL, Liu Z, Shulman JM. Tau activates transposable elements in Alzheimer’s Disease. Cell Rep. 2018;23(10):2874–80. https://doi.org/10.1016/j.celrep.2018.05.004.
Hanger DP, Byers HL, Wray S, Leung K-Y, Saxton MJ, Seereeram A, Reynolds CH, Ward MA, Anderton BH. Novel phosphorylation sites in Tau from Alzheimer brain support a role for Casein Kinase 1 in Disease pathogenesis*. J Biol Chem. 2007;282(32):23645–54. https://doi.org/10.1074/jbc.M703269200.
He Z, McBride JD, Xu H, Changolkar L, Kim S-J, Zhang B, Narasimhan S, Gibbons GS, Guo JL, Kozak M, Schellenberg GD, Trojanowski JQ, Lee VMY. Transmission of tauopathy strains is independent of their isoform composition. Nat Commun. 2020;11(1):7. https://doi.org/10.1038/s41467-019-13787-x.
Hondius DC, Koopmans F, Leistner C, Pita-Illobre D, Peferoen-Baert RM, Marbus F, Paliukhovich I, Li KW, Rozemuller AJM, Hoozemans JJM, Smit AB. The proteome of granulovacuolar degeneration and neurofibrillary tangles in Alzheimer’s disease. Acta Neuropathol. 2021;141(3):341–58. https://doi.org/10.1007/s00401-020-02261-4.
Hoover BR, Reed MN, Su J, Penrod RD, Kotilinek LA, Grant MK, Pitstick R, Carlson GA, Lanier LM, Yuan LL, Ashe KH, Liao D. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron. 2010;68(6):1067–81. https://doi.org/10.1016/j.neuron.2010.11.030.
Hsieh Y-C, Guo C, Yalamanchili HK, Abreha M, Al-Ouran R, Li Y, Dammer EB, Lah JJ, Levey AI, Bennett DA, De Jager PL, Seyfried NT, Liu Z, Shulman JM. Tau-mediated disruption of the spliceosome triggers cryptic RNA splicing and neurodegeneration in Alzheimer’s Disease. Cell Rep. 2019;29(2):301-316.e310. https://doi.org/10.1016/j.celrep.2019.08.104.
Jiang F, Tang X, Tang C, Hua Z, Ke M, Wang C, Zhao J, Gao S, Jurczyszyn A, Janz S, Beksac M, Zhan F, Gu C, Yang Y. HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA. J Hematol Oncol. 2021;14(1):54. https://doi.org/10.1186/s13045-021-01066-6.
Jiang L, Ash PEA, Maziuk BF, Ballance HI, Boudeau S, Abdullatif AA, Orlando M, Petrucelli L, Ikezu T, Wolozin B. TIA1 regulates the generation and response to toxic tau oligomers. Acta Neuropathol. 2019;137(2):259–77. https://doi.org/10.1007/s00401-018-1937-5.
Jiang L, Lin W, Zhang C, Ash PEA, Verma M, Kwan J, van Vliet E, Yang Z, Cruz AL, Boudeau S, Maziuk BF, Lei S, Song J, Alvarez VE, Hovde S, Abisambra JF, Kuo M-H, Kanaan N, Murray ME, Crary JF, Zhao J, Cheng J-X, Petrucelli L, Li H, Emili A, Wolozin B. Interaction of tau with HNRNPA2B1 and N6-methyladenosine RNA mediates the progression of tauopathy. Mol Cell. 2021;81(20):4209-4227.e4212. https://doi.org/10.1016/j.molcel.2021.07.038.
Kametani F, Yoshida M, Matsubara T, Murayama S, Saito Y, Kawakami I, Onaya M, Tanaka H, Kakita A, Robinson AC, Mann DMA, Hasegawa M. Comparison of common and disease-specific post-translational modifications of pathological Tau associated with a wide range of Tauopathies. Front Neurosci. 2020;14:581936. https://doi.org/10.3389/fnins.2020.581936.
Kampers T, Friedhoff P, Biernat J, Mandelkow EM, Mandelkow E. RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 1996;399(3):344–9. https://doi.org/10.1016/S0014-5793(96)01386-5.
Keck S, Nitsch R, Grune T, Ullrich O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer’s disease. J Neurochem. 2003;85(1):115–22. https://doi.org/10.1046/j.1471-4159.2003.01642.x.
Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, Kanagaraj AP, Carter R, Boylan KB, Wojtas AM, Rademakers R, Pinkus JL, Greenberg SA, Trojanowski JQ, Traynor BJ, Smith BN, Topp S, Gkazi A-S, Miller J, Shaw CE, Kottlors M, Kirschner J, Pestronk A, Li YR, Ford AF, Gitler AD, Benatar M, King OD, Kimonis VE, Ross ED, Weihl CC, Shorter J, Taylor JP. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–73. https://doi.org/10.1038/nature11922.
Klein H-U, McCabe C, Gjoneska E, Sullivan SE, Kaskow BJ, Tang A, Smith RV, Xu J, Pfenning AR, Bernstein BE, Meissner A, Schneider JA, Mostafavi S, Tsai L-H, Young-Pearse TL, Bennett DA, De Jager PL. Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat Neurosci. 2019;22(1):37–46. https://doi.org/10.1038/s41593-018-0291-1.
Koren SA, Hamm MJ, Meier SE, Weiss BE, Nation GK, Chishti EA, Arango JP, Chen J, Zhu H, Blalock EM, Abisambra JF. Tau drives translational selectivity by interacting with ribosomal proteins. Acta Neuropathol. 2019;137(4):571–83. https://doi.org/10.1007/s00401-019-01970-9.
Lasagna-Reeves CA, Castillo-Carranza DL, Sengupta U, Sarmiento J, Troncoso J, Jackson GR, Kayed R. Identification of oligomers at early stages of tau aggregation in Alzheimer’s disease. FASEB J. 2012;26(5):1946–59. https://doi.org/10.1096/fj.11-199851.
LeBlang CJ, Medalla M, Nicoletti NW, Hays EC, Zhao J, Shattuck J, Cruz AL, Wolozin B, Luebke JI. Reduction of the RNA binding protein TIA1 exacerbates neuroinflammation in tauopathy. Front Neurosci. 2020;14(285). https://doi.org/10.3389/fnins.2020.00285
Lefebvre T, Ferreira S, Dupont-Wallois L, Bussière T, Dupire M-J, Delacourte A, Michalski J-C, Caillet-Boudin M-L. Evidence of a balance between phosphorylation and O-GlcNAc glycosylation of Tau proteins—a role in nuclear localization. Biochim Biophys Acta. 2003;1619(2):167–76. https://doi.org/10.1016/S0304-4165(02)00477-4.
Lester E, Ooi FK, Bakkar N, Ayers J, Woerman AL, Wheeler J, Bowser R, Carlson GA, Prusiner SB, Parker R. Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron. 2021;109(10):1675-1691.e1679. https://doi.org/10.1016/j.neuron.2021.03.026.
Li T, Paudel HK. Glycogen synthase kinase 3β phosphorylates Alzheimer’s Disease-specific Ser396 of microtubule-associated protein Tau by a sequential mechanism. Biochemistry. 2006;45(10):3125–33. https://doi.org/10.1021/bi051634r.
Lippens G, Gigant B. Elucidating Tau function and dysfunction in the era of cryo-EM. J Biol Chem. 2019;294(24):9316–25. https://doi.org/10.1074/jbc.REV119.008031.
Liu C, Götz J. Profiling murine Tau with 0N, 1N and 2N isoform-specific antibodies in brain and peripheral organs reveals distinct subcellular localization, with the 1n isoform being enriched in the nucleus. PLoS ONE. 2014;8(12):e84849. https://doi.org/10.1371/journal.pone.0084849.
Liu C, Song X, Nisbet R, Götz J. Co-immunoprecipitation with Tau Isoform-specific Antibodies Reveals Distinct Protein Interactions and Highlights a Putative Role for 2N Tau in Disease. J Biol Chem. 2016;291(15):8173–88. https://doi.org/10.1074/jbc.M115.641902.
Liu Y, Szaro BG. hnRNP K post-transcriptionally co-regulates multiple cytoskeletal genes needed for axonogenesis. Development. 2011;138(14):3079–90. https://doi.org/10.1242/dev.066993.
LoPresti P, Szuchet S, Papasozomenos SC, Zinkowski RP, Binder LI. Functional implications for the microtubule-associated protein tau: localization in oligodendrocytes. Proc Natl Acad Sci USA. 1995;92(22):10369–73. https://doi.org/10.1073/pnas.92.22.10369.
Losev Y, Paul A, Frenkel-Pinter M, Abu-Hussein M, Khalaila I, Gazit E, Segal D. Novel model of secreted human tau protein reveals the impact of the abnormal N-glycosylation of tau on its aggregation propensity. Sci Rep. 2019;9(1):2254. https://doi.org/10.1038/s41598-019-39218-x.
Lu J, Li T, He R, Bartlett PF, Götz J. Visualizing the microtubule-associated protein tau in the nucleus. Sci China Life Sci. 2014;57(4):422–31. https://doi.org/10.1007/s11427-014-4635-0.
Maina MB, Bailey LJ, Wagih S, Biasetti L, Pollack SJ, Quinn JP, Thorpe JR, Doherty AJ, Serpell LC. The involvement of tau in nucleolar transcription and the stress response. Acta Neuropathol Commun. 2018;6(1):70–70. https://doi.org/10.1186/s40478-018-0565-6.
Martin EW, Thomasen FE, Milkovic NM, Cuneo MJ, Grace CR, Nourse A, Lindorff-Larsen K, Mittag T. Interplay of folded domains and the disordered low-complexity domain in mediating hnRNPA1 phase separation. Nucleic Acids Res. 2021;49(5):2931–45. https://doi.org/10.1093/nar/gkab063.
Martínez-Maldonado A, Ontiveros-Torres MÁ, Harrington CR, Montiel-Sosa JF, Prandiz RG-T, Bocanegra-López P, Sorsby-Vargas AM, Bravo-Muñoz M, Florán-Garduño B, Villanueva-Fierro I, Perry G, Garcés-Ramírez L, de la Cruz F, Martínez-Robles S, Pacheco-Herrero M, Luna-Muñoz J. Molecular processing of tau protein in progressive supranuclear palsy: neuronal and glial degeneration. J Alzheimers Dis. 2021;79(4):1517–31. https://doi.org/10.3233/JAD-201139.
Maziuk BF, Apicco DJ, Cruz AL, Jiang L, Ash PEA, da Rocha EL, Zhang C, Yu WH, Leszyk J, Abisambra JF, Li H, Wolozin B. RNA binding proteins co-localize with small tau inclusions in tauopathy. Acta Neuropathol Commun. 2018;6(1):71. https://doi.org/10.1186/s40478-018-0574-5.
McKinnon C, De Snoo ML, Gondard E, Neudorfer C, Chau H, Ngana SG, O’Hara DM, Brotchie JM, Koprich JB, Lozano AM, Kalia LV, Kalia SK. Early-onset impairment of the ubiquitin-proteasome system in dopaminergic neurons caused by α-synuclein. Acta Neuropathol Commun. 2020;8(1):17. https://doi.org/10.1186/s40478-020-0894-0.
McMillan P, Korvatska E, Poorkaj P, Evstafjeva Z, Robinson L, Greenup L, Leverenz J, Schellenberg GD, D’Souza I. Tau isoform regulation is region- and cell-specific in mouse brain. J Comp Neurol. 2008;511(6):788–803. https://doi.org/10.1002/cne.21867.
Meier S, Bell M, Lyons DN, Ingram A, Chen J, Gensel JC, Zhu H, Nelson PT, Abisambra JF. Identification of novel tau interactions with endoplasmic reticulum proteins in Alzheimer’s Disease brain. J Alzheimers Dis. 2015;48(3):687–702. https://doi.org/10.3233/JAD-150298.
Montalbano M, Jaworski E, Garcia S, Ellsworth A, McAllen S, Routh A, Kayed R. Tau modulates mRNA transcription, alternative polyadenylation profiles of hnRNPs, chromatin remodeling and spliceosome complexes. Front Mol Neurosci. 2021;14:742790. https://doi.org/10.3389/fnmol.2021.742790.
Montalbano M, McAllen S, Puangmalai N, Sengupta U, Bhatt N, Johnson OD, Kharas MG, Kayed R. RNA-binding proteins Musashi and tau soluble aggregates initiate nuclear dysfunction. Nat Commun. 2020;11(1):4305. https://doi.org/10.1038/s41467-020-18022-6.
Montalbano M, McAllen S, Sengupta U, Puangmalai N, Bhatt N, Ellsworth A, Kayed R. Tau oligomers mediate aggregation of RNA-binding proteins Musashi1 and Musashi2 inducing Lamin alteration. Aging Cell. 2019;18(6):e13035. https://doi.org/10.1111/acel.13035.
Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL, Duff KE. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med. 2016;22(1):46–53. https://doi.org/10.1038/nm.4011.
Nakano M, Riku Y, Nishioka K, Hasegawa M, Washimi Y, Arahata Y, Takeda A, Horibe K, Yamaoka A, Suzuki K, Tsujimoto M, Li Y, Yoshino H, Hattori N, Akagi A, Miyahara H, Iwasaki Y, Yoshida M. Unclassified four-repeat tauopathy associated with familial parkinsonism and progressive respiratory failure. Acta Neuropathol Commun. 2020;8(1):148. https://doi.org/10.1186/s40478-020-01025-1.
Narasimhan S, Guo JL, Changolkar L, Stieber A, McBride JD, Silva LV, He Z, Zhang B, Gathagan RJ, Trojanowski JQ, Lee VMY. Pathological Tau Strains from human brains recapitulate the diversity of Tauopathies in nontransgenic mouse brain. J Neurosci. 2017;37(47):11406–23. https://doi.org/10.1523/JNEUROSCI.1230-17.2017.
Neddens J, Temmel M, Flunkert S, Kerschbaumer B, Hoeller C, Loeffler T, Niederkofler V, Daum G, Attems J, Hutter-Paier B. Phosphorylation of different tau sites during progression of Alzheimer’s disease. Acta Neuropathol Commun. 2018;6(1):52. https://doi.org/10.1186/s40478-018-0557-6.
Panda D, Samuel JC, Massie M, Feinstein SC, Wilson L. Differential regulation of microtubule dynamics by three- and four-repeat tau: implications for the onset of neurodegenerative disease. Proc Natl Acad Sci. 2003;100(16):9548–53. https://doi.org/10.1073/pnas.1633508100.
Paterno G, Bell BM, Gorion K-MM, Prokop S, Giasson BI. Reassessment of neuronal Tau distribution in adult human brain and implications for Tau pathobiology. Acta Neuropathol Commun. 2022;10(1):94. https://doi.org/10.1186/s40478-022-01394-9.
Ping L, Kundinger SR, Duong DM, Yin L, Gearing M, Lah JJ, Levey AI, Seyfried NT. Global quantitative analysis of the human brain proteome and phosphoproteome in Alzheimer’s disease. Scientific Data. 2020;7(1):315. https://doi.org/10.1038/s41597-020-00650-8.
Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R. Intrinsically Disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep. 2018;22(6):1401–12. https://doi.org/10.1016/j.celrep.2018.01.036.
Samimi N, Sharma G, Kimura T, Matsubara T, Huo A, Chiba K, Saito Y, Murayama S, Akatsu H, Hashizume Y, Hasegawa M, Farjam M, Shahpasand K, Ando K, Hisanaga S-I. Distinct phosphorylation profiles of tau in brains of patients with different tauopathies. Neurobiol Aging. 2021;108:72–9. https://doi.org/10.1016/j.neurobiolaging.2021.08.011.
Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, Forster C, Yue M, Orne J, Janus C, Mariash A, Kuskowski M, Hyman B, Hutton M, Ashe KH. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309(5733):476–81. https://doi.org/10.1126/science.1113694.
Scheres SH, Zhang W, Falcon B, Goedert M. Cryo-EM structures of tau filaments. Curr Opin Struct Biol. 2020;64:17–25. https://doi.org/10.1016/j.sbi.2020.05.011.
Sealey MA, Vourkou E, Cowan CM, Bossing T, Quraishe S, Grammenoudi S, Skoulakis EMC, Mudher A. Distinct phenotypes of three-repeat and four-repeat human tau in a transgenic model of tauopathy. Neurobiol Dis. 2017;105:74–83. https://doi.org/10.1016/j.nbd.2017.05.003.
Sengupta U, Montalbano M, McAllen S, Minuesa G, Kharas M, Kayed R. Formation of toxic oligomeric assemblies of RNA-binding protein: musashi in Alzheimer’s disease. Acta Neuropathol Commun. 2018;6(1):113. https://doi.org/10.1186/s40478-018-0615-0.
Sinsky J, Majerova P, Kovac A, Kotlyar M, Jurisica I, Hanes J. Physiological Tau interactome in brain and its link to Tauopathies. J Proteome Res. 2020;19(6):2429–42. https://doi.org/10.1021/acs.jproteome.0c00137.
Swift IJ, Sogorb-Esteve A, Heller C, Synofzik M, Otto M, Graff C, Galimberti D, Todd E, Heslegrave AJ, van der Ende EL, Van Swieten JC, Zetterberg H, Rohrer JD. Fluid biomarkers in frontotemporal dementia: past, present and future. J Neurol Neurosurg Psychiatry. 2021;92(2):204. https://doi.org/10.1136/jnnp-2020-323520.
Tarutani A, Miyata H, Nonaka T, Hasegawa K, Yoshida M, Saito Y, Murayama S, Robinson AC, Mann DMA, Tomita T, Hasegawa M. Human tauopathy-derived tau strains determine the substrates recruited for templated amplification. Brain. 2021. https://doi.org/10.1093/brain/awab091.
Thibaudeau TA, Anderson RT, Smith DM. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat Commun. 2018;9(1):1097. https://doi.org/10.1038/s41467-018-03509-0.
Togo T, Sahara N, Yen S-H, Cookson N, Ishizawa T, Hutton M, de Silva R, Lees A, Dickson DW. Argyrophilic grain disease is a sporadic 4-repeat Tauopathy. J Neuropathol Exp Neurol. 2002;61(6):547–56. https://doi.org/10.1093/jnen/61.6.547.
Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C, Luk C, Gibbs JR, Dillman A, Hernandez DG, Arepalli S, Singleton AB, Cookson MR, Pittman AM, de Silva R, Weale ME, Hardy J, Ryten M. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum Mol Genet. 2012;21(18):4094–103. https://doi.org/10.1093/hmg/dds238.
Tracy TE, Madero-Pérez J, Swaney DL, Chang TS, Moritz M, Konrad C, Ward ME, Stevenson E, Hüttenhain R, Kauwe G, Mercedes M, Sweetland-Martin L, Chen X, Mok S-A, Wong MY, Telpoukhovskaia M, Min S-W, Wang C, Sohn PD, Martin J, Zhou Y, Luo W, Trojanowski JQ, Lee VMY, Gong S, Manfredi G, Coppola G, Krogan NJ, Geschwind DH, Gan L. Tau interactome maps synaptic and mitochondrial processes associated with neurodegeneration. Cell. 2022. https://doi.org/10.1016/j.cell.2021.12.041.
Ulrich G, Salvadè A, Boersema P, Calì T, Foglieni C, Sola M, Picotti P, Papin S, Paganetti P. Phosphorylation of nuclear Tau is modulated by distinct cellular pathways. Sci Rep. 2018;8(1):17702. https://doi.org/10.1038/s41598-018-36374-4.
Vanderweyde T, Apicco DJ, Youmans-Kidder K, Ash PEA, Cook C, Lummertz da Rocha E, Jansen-West K, Frame AA, Citro A, Leszyk JD, Ivanov P, Abisambra JF, Steffen M, Li H, Petrucelli L, Wolozin B. Interaction of tau with the RNA-binding protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep. 2016;15(7):1455–66. https://doi.org/10.1016/j.celrep.2016.04.045.
Vanderweyde T, Yu H, Varnum M, Liu-Yesucevitz L, Citro A, Ikezu T, Duff K, Wolozin B. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in Tauopathies. J Neurosci. 2012;32(24):8270. https://doi.org/10.1523/JNEUROSCI.1592-12.2012.
Wang P, Joberty G, Buist A, Vanoosthuyse A, Stancu I-C, Vasconcelos B, Pierrot N, Faelth-Savitski M, Kienlen-Campard P, Octave J-N, Bantscheff M, Drewes G, Moechars D, Dewachter I. Tau interactome mapping based identification of Otub1 as Tau deubiquitinase involved in accumulation of pathological Tau forms in vitro and in vivo. Acta Neuropathol. 2017;133(5):731–49. https://doi.org/10.1007/s00401-016-1663-9.
Wang X, Williams D, Müller I, Lemieux M, Dukart R, Maia IBL, Wang H, Woerman AL, Schmitt-Ulms G. Tau interactome analyses in CRISPR-Cas9 engineered neuronal cells reveal ATPase-dependent binding of wild-type but not P301L Tau to non-muscle myosins. Sci Rep. 2019;9(1):16238–16238. https://doi.org/10.1038/s41598-019-52543-5.
Wang Y, Wang J, Gao L, Stamm S, Andreadis A. An SRp75/hnRNPG complex interacting with hnRNPE2 regulates the 5′ splice site of tau exon 10, whose misregulation causes frontotemporal dementia. Gene. 2011;485(2):130–8. https://doi.org/10.1016/j.gene.2011.06.020.
Wang Q, Woltjer RL, Cimino PJ, Pan C, Montine KS, Zhang J, Montine TJ. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19:869–71.
Wesseling H, Mair W, Kumar M, Schlaffner CN, Tang S, Beerepoot P, Fatou B, Guise AJ, Cheng L, Takeda S, Muntel J, Rotunno MS, Dujardin S, Davies P, Kosik KS, Miller BL, Berretta S, Hedreen JC, Grinberg LT, Seeley WW, Hyman BT, Steen H, Steen JA. Tau PTM profiles identify patient heterogeneity and stages of Alzheimer’s Disease. Cell. 2020;183(6):1699-1713 e1613. https://doi.org/10.1016/j.cell.2020.10.029.
Woerman AL, Aoyagi A, Patel S, Kazmi SA, Lobach I, Grinberg LT, McKee AC, Seeley WW, Olson SH, Prusiner SB. Tau prions from Alzheimer’s disease and chronic traumatic encephalopathy patients propagate in cultured cells. Proc Natl Acad Sci USA. 2016;113(50):E8187–96. https://doi.org/10.1073/pnas.1616344113.
Wray S, Saxton M, Anderton BH, Hanger DP. Direct analysis of tau from PSP brain identifies new phosphorylation sites and a major fragment of N-terminally cleaved tau containing four microtubule-binding repeats. J Neurochem. 2008;105(6):2343–52. https://doi.org/10.1111/j.1471-4159.2008.05321.x.
Yu A, Fox SG, Cavallini A, Kerridge C, O’Neill MJ, Wolak J, Bose S, Morimoto RI. Tau protein aggregates inhibit the protein-folding and vesicular trafficking arms of the cellular proteostasis network. J Biol Chem. 2019;294(19):7917–30. https://doi.org/10.1074/jbc.RA119.007527.
Zareba-Paslawska J, Patra K, Kluzer L, Revesz T, Svenningsson P. Tau isoform-driven CBD pathology transmission in oligodendrocytes in humanized tau mice. Front Neurol. 2021;11(1825). https://doi.org/10.3389/fneur.2020.589471
Zhang W, Falcon B, Murzin AG, Fan J, Crowther RA, Goedert M, Scheres SHW. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. Elife. 2019;8:e43584. https://doi.org/10.7554/eLife.43584.
Zhang W, Tarutani A, Newell KL, Murzin AG, Matsubara T, Falcon B, Vidal R, Garringer HJ, Shi Y, Ikeuchi T, Murayama S, Ghetti B, Hasegawa M, Goedert M, Scheres SHW. Novel tau filament fold in corticobasal degeneration. Nature. 2020;580(7802):283–7. https://doi.org/10.1038/s41586-020-2043-0.
Zhang X, Lin Y, Eschmann NA, Zhou H, Rauch JN, Hernandez I, Guzman E, Kosik KS, Han S. RNA stores tau reversibly in complex coacervates. PLoS Biol. 2017;15(7):e2002183. https://doi.org/10.1371/journal.pbio.2002183.
Zhong Q, Congdon EE, Nagaraja HN, Kuret J. Tau isoform composition influences rate and extent of filament formation. J Biol Chem. 2012;287(24):20711–9. https://doi.org/10.1074/jbc.M112.364067.
Acknowledgements
Not applicable
Funding
This study was supported by funding from the Bluesand Foundation, Alzheimer’s Association (AARG-21–852072), NIH (AG060882) and funding to ForeFront, a collaborative research group dedicated to the study of frontotemporal dementia and motor neuron disease, from the National Health and Medical Research Council of Australia (NHMRC) program Grant (#1132524).
Author information
Authors and Affiliations
Contributions
TK, AH, and ED wrote the manuscript. TK performed the analysis and generated figures. All authors read and approved the final manuscript.
Authors’ information
Not applicable.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Table 1.
List of post-translational modifications detected on tau across AD, AGD, CBD, PiD and PSP. Supplementary Table 2. Summary of human tau interactome and localised pathology proteomics studies. Supplementary Table 3. Summary of rodent Tau interactome studies. Supplementary Table 4. Gene Ontology Molecular Function Gene Sets and Enrichment Statistics. Supplementary Table 5. Summary of pathology targeted localised proteomics studies. Supplementary Table 6. GO cellular component comparison gene sets used for figure 3B.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kavanagh, T., Halder, A. & Drummond, E. Tau interactome and RNA binding proteins in neurodegenerative diseases. Mol Neurodegeneration 17, 66 (2022). https://doi.org/10.1186/s13024-022-00572-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13024-022-00572-6