Abstract
Background
Langerhans cell histiocytosis (LCH) is a rare neoplastic disease that occurs in both children and adults, and BRAF V600E is detected in up to 64% of the patients. Several studies have discussed the associations between BRAF V600E mutation and clinicopathological manifestations, but no clear conclusions have been drawn regarding the clinical significance of the mutation in pediatric patients.
Results
We retrieved the clinical information for 148 pediatric LCH patients and investigated the BRAF V600E mutation using next-generation sequencing alone or with droplet digital PCR. The overall positive rate of BRAF V600E was 60/148 (41%). The type of sample (peripheral blood and formalin-fixed paraffin-embedded tissue) used for testing was significantly associated with the BRAF V600E mutation status (p-value = 0.000 and 0.000). The risk of recurrence declined in patients who received targeted therapy (p-value = 0.006; hazard ratio 0.164, 95%CI: 0.046 to 0.583). However, no correlation was found between the BRAF V600E status and gender, age, stage, specific organ affected, TP53 mutation status, masses close to the lesion or recurrence.
Conclusions
This is the largest pediatric LCH study conducted with a Chinese population to date. BRAF V600E in LCH may occur less in East Asian populations than in other ethnic groups, regardless of age. Biopsy tissue is a more sensitive sample for BRAF mutation screening because not all of circulating DNA is tumoral. Approaches with low limit of detection or high sensitivity are recommended for mutation screening to avoid type I and II errors.
Similar content being viewed by others
Background
Langerhans cell histiocytosis (LCH) is inflammatory neoplasia of the myeloid precursor cells. The disease is characterized by the clonal proliferation of CDa1 + /CD207 + dendritic cells, whose features are similar to those of epidermal Langerhans cells. LCH is the most common histiocytic disorder but is a rare neoplastic disease. LCH presents in patients of all ages but most prevalent in children, with 3.5 years old being the medium age of diagnosis [1]. Generally, the incidence rates are 4 ~ 8 children per million and 1 ~ 2 adults per million each year, similar to those of pediatric Hodgkin’s lymphoma [1,2,3,4]. The highest incidence rate is observed among infants less than one year of age [1]. LCH lesions can develop in almost all systems but have a particular affinity for the skeleton (80%), skin (33%) and pituitary gland (25%) in children [1, 5]. The clinical manifestations depend on the specific organs involved and the extent of involvement, varying remarkably from spontaneously regressing lesions in isolated organs to fatal diseases in multiple systems [3]. The diversity of the symptoms contributes to the high misdiagnosis rates (16%), further leading to delays in appropriate treatment [1, 6]. The classification of LCH is based on the number of lesions present and organs affected. Approximately two-thirds of children present with single-system involvement, which has an excellent prognosis and a 5-year survival rate of virtually 100%. However, multisystem LCH, involving two or more organs, frequently has an unpredictable course. LCH involving risk organs at diagnosis is considered a high-risk disease, especially with organ dysfunctions [1, 7].
BRAF is considered one of the most common and well-known mutated kinases in human cancer, with driver mutations in several cancers, including melanoma, colorectal cancer, thyroid cancer and non-small cell lung cancer [8,9,10,11]. Its V600E mutation accounts for more than 90% of BRAF-activating mutations [2]. Studies have shown that BRAF V600E mutations are present in up to 64% of LCH samples [12]. Further research demonstrated that the BRAF mutation in LCH lesions significantly elevates phospho-extracellular signal-regulated kinase (ERK) expression, suggesting the activation of the mitogen-activated protein kinase (MAPK) pathway, which further suppresses cell migration and augments cell survival [13, 14]. However, the clinical significance of BRAF V600E in LCH is still contradictory. The low incidence and misdiagnosis of LCH limits studies in a large number of LCH patients. This article presents a retrospective single-center study on a cohort of pediatric LCH patients aimed at determining the frequency of mutations and clarifying the associations between the clinical features and BRAF V600E mutation in LCH.
Results
Clinical information
A total of 148 patients diagnosed with LCH were included in this research (Fig. 1). All were pediatric and of Chinese origin. The pertinent characteristics of patients are summarized in Table 1. The cohort included 88 boys and 60 girls, with a gender ratio of 1.47. The median and mean onset ages were 2 and 3.3 years old (range 0–16 years old), respectively. The onset age for more than half of the patients (57%) was equal to or less than two years old. Patients with the highest incidence were those younger than 1-year-old (29%). Seventy patients (47%) had single-system involvement, specifically 44 patients with single-site lesions (SS-S) and 26 patients with multiple-site lesions (SS-M). Seventy-eight patients (53%) had multiple-system involvement, specifically 57 patients without risk-organ involvement (MS-RO−) and 21 patients with risk-organ involvement (MS-RO+). Bone (79%) was the most common location of LCH involvement, followed by the skin (41%), lungs (26%), lymph nodes (14%), liver (14%), spleen (8%), pituitary gland (7%), bone marrow (4%) and thymus (3%). The imageology for the involvement of various organs is presented in Fig. 2. In addition, 71 patients (48%) had masses thought to be associated with LCH, including eosinophilic granuloma (10/71), lymphadenopathy (10/71) or extra-lymphatic masses (bone, 56/71; neck, 2/71; gingiva, 1/71; soft tissue in the nasal cavity, 1/71; medulla oblongata, 1/71; thymus, 1/71). According to the latest follow-up data, 41 of 136 LCH patients (30%) had relapsed. After treatments using various protocols (chemotherapy, targeted therapy, surgery or no treatment), LCH improved in 127 of 136 patients (93%). Two LCH patients, who only had skin lesions and recovered without receiving any treatments, were also included. Nearly all of the patients in the cohort are still alive, with a 1-year overall survival rate of 100% and 5-year of 99%. Only one patient had LCH concurrent with acute lymphoid leukemia and eventually died of peripheral T-cell lymphoma, although her LCH symptoms improved.
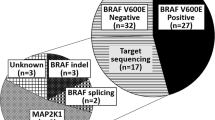
A flow chart of patient recruitment. A cohort of 166 LCH patients tested with next-generation sequencing (NGS) was recruited. Among them, the medical records of 148 LCH patients were available. Sixty of 148 patients showed BRAF V600E positivity in NGS. BRAF V600E mutation was identified in 60/148 (41%) pediatric LCH patients
Spectrum of clinical presentations in Langerhans Cell Histiocytosis (LCH). A–D Different skin lesions. E, F Cranial bone lesions on single and multiple sites. G LCH patient with cranial and mandibular bone lesions and swollen eyes. H, I Lung lesions. J Pituitary lesions (arrow). K Skin lesions at birth. L Nail lesions. M–O PET-CT images showed a single bone lesion involving the right tibia; multiple bone lesions; multiple-system lesions involving the bones, lung and lymph nodes (arrow)
Mutation analysis
The positive rates in next-generation sequencing (NGS) with LCH-panel were 28% (22/78) and 74% (62/84) in peripheral blood and formalin-fixed paraffin-embedded (FFPE) tissue samples, respectively. A group of genes was detected in our LCH cohort, including ARAF, BRAF, MAP2K1, KRAS, KIT, FGFR3, and ALK (unpublished data will be described in the next paper). BRAF V600E mutation was detected in 60 of 148 (41%) patients, based on the results of both LCH-panel NGS and droplet digital PCR (ddPCR) (Table 1). The NGS detection rate was 14% (11/78) in peripheral blood, with a V600E positive rate of 23% (18/78). In FFPE tissue samples, the NGS detection rate was 52% (44/84), with a V600E positive rate of 54% (45/84). Of the 14 patients in whom both types of samples were used for LCH-panel sequencing, the positive rates were 14% (2/14) for peripheral blood and 50% (7/14) for FFPE tissue, showing a low level of concordance between the two types of samples. Mutations in FFPE tissues were detected in bone and adjacent soft tissue (27/58), skin (15/22), lymph nodes (1/2), soft tissue on the neck (1/2), bone marrow (1/1) and soft tissue in the nasal cavity (1/1). No BRAF V600E mutation was detected in the gingiva (0/1) and thymus (0/1). NGS also revealed that 72 patients (49%) carried a TP53 mutation (c.98G > C, p.P33R).
Correlation of BRAF V600E mutation status and clinicopathologic characteristics
The type of sample sent for sequencing was significantly correlated to BRAF V600E (p-value = 0.000 (2.66E−04) in peripheral blood and p-value = 0.000 (4.15E−04) in FFPE tissue). V600E mutations were detected in peripheral blood at a lower percentage (odds ratio 0.274, 95% confidence interval (CI) 0.128–0.567) than in FFPE tissues (odds ratio 3.734, 95% CI 1.741–8.341). The sensitivity was 61% (11/18) and 98% (44/45) for peripheral blood and FFPE tissue, respectively. However, there was no statistically significant correlation between the BRAF V600E status and gender (p-value = 0.689 > 0.05), onset age (p-value = 0.575 > 0.05), stage (p-value = 0.154 > 0.05), specific organ affected (all p-values > 0.05), TP53 mutation status (p-value = 0.572 > 0.05), the presence of masses close to the lesion (p-value = 1.000 > 0.05) or recurrence (p-value = 0.407 > 0.05) (Fig. 3A) (Table 1). In our cohort, 52% (31/60) of patients harboring BRAF V600E were given targeted therapy, including but not limited to dabrafenib (30/31). The recurrence rate was 7% (2/29) in patients given targeted therapy and 33% (9/27) in patients that were not. Targeted therapy was statistically significantly associated with a decreased risk of recurrence in LCH patients (p-value = 0.006 < 0.05; hazard ratio 0.164, 95%CI: 0.046 to 0.583) (Fig. 3B).
Recurrence possibility for BRAF V600E status and targeted therapy. A The relationship between recurrence and the mutation statue of BRAF V600E (Log-rank (Mantel-Cox) test, p-value = 0.407 > 0.05, not statistically significant). B Patients who received targeted therapy had a lower risk of recurrence (Log-rank (Mantel-Cox) test, p-value = 0.006 < 0.05, statistically significant; hazard ratio 0.164, 95%CI: 0.046 to 0.583)
Discussion
BRAF V600E frequency in LCH
Since BRAF V600E mutation in tissues was detected in almost all studies evaluated, only the frequencies of detection in tissues were compared and discussed here. In all reported studies, the frequency ranged from 0 to 64%, with an overall frequency of 47% (Table 2). In the current study, BRAF V600E was detected in 54% of LCH tissues specimens, consistent with that in the references. For patients < 18 years old, regardless of whether ethnicity was taken into account, BRAF V600E mutation was more frequent in them than in adults. The frequency in our pediatric cohort (54%) was consistent with the overall pediatric frequency (53%). In addition, when we analyzed frequency according to geographical factors, patients from East Asia showed a lower frequency than other ethnic groups, regardless of age. We therefore hypothesize that the frequency of BRAF V600E mutation varies across ethnic groups. Previous studies have also shown that ethnic background appears to influence the risk of LCH development [15]. However, the reasons for these differences are uncertain. The recruitment criteria, sample size, techniques used or the definition of various factors may all contribute to these discrepancies. For example, only samples with a variant allele frequency (VAF) greater than 4% were considered mutant in Badalian-Very’s study [16], but only samples with a VAF less than 0.1% were considered wild-type in our study. The definitions of mutation are inconsistent between studies. If we apply 4% as the threshold for VAF to analyze NGS data from our tissues, 49% (41/84) of patients are classified as BRAF V600E-positive (52% when the threshold for VAF is 0.1%), which is much less frequent than in Badalian-Very’s study (57%). In further research, setting identical recruitment criteria, increasing the sample size, unifying a standard testing approach and clarifying the definition of important factors can be considered to figure out the discrepancies.
It has been reported that most (70%) LCH patients present with single-system involvement, ranging from 60 to 91.8% of patients, and several studies support this statement [1, 17,18,19,20,21,22]. However, in our cohort, only 48% of patients presented with single-system LCH. The uneven distribution of medical resources may explain the lower percentage in this study. Our hospital has advanced medical resources. Most of the patients travelled from less developed areas to seek medical advice in our hospital. Since most LCH patients with single-system involvement present mild symptoms and may not even require treatment, they are more likely to seek treatment in local hospitals. Thus, patients with severe disease have a higher probability of seeking medical treatment in our hospital, leading to a lower percentage of LCH patients with single-system involvement.
Analysis of associations
Since previous studies showed contradictory findings on the correlations between mutation status and clinicopathological features, we aimed to clarify this relationship. In our cohort, the type of sample used for sequencing was significantly associated with the BRAF V600E mutation status. The frequency of mutations detected in FFPE tissues (54%) was much higher than that in peripheral blood samples (23%). In particular, for 14 patients whose peripheral blood and FFPE samples were both available, the positive rate of NGS for FFPE (50%, 7/14) was higher than that for blood (14%, 2/14). Similarly, the frequencies of mutations detected in peripheral blood samples were lower than that detected in tissues in several studies [12, 23, 24]. The concordance indicates that tissue is a relatively more reliable sample for identifying mutations. However, peripheral blood is not a useless sample for LCH. Blood from different periods can be used to detect circulating BRAF V600E to monitor disease progression over time [12], especially when lesions are recovered. Since the DNA isolated from peripheral blood is not exclusively tumoral and only a small amount of DNA can escape from the lesion into the circulation, it is reasonable to observe a relatively low positive rate in the blood.
Our data showed that the BRAF V600E mutation status was not associated with the risk of recurrence. Intriguingly, previous studies reported that BRAF V600E mutation was correlated with an increased risk of recurrence [12, 13]. In our cohort, LCH patients without BRAF V600E mutation appeared more likely to experience recurrence. We are inclined to attribute this result to targeted therapy, as we found a statisticall significant association between targeted therapy and a decreased risk of recurrence in patients with LCH. The lower risk of recurrence in patients carrying the V600E mutation may be due to the fact that they received timely and effective treatment. Since this is a retrospective study and patients received targeted therapy without professional guidance and medical supervision, we lack robust evidence to draw definitive conclusions regarding the effectiveness of targeted therapy on LCH. However, this study dose shed some light on the effectiveness of dabrafenib in the treatment of LCH. Dabrafenib and vemurafenib are specific to BRAF V600E/K mutation and were approved by the US Food and Drug Administration (FDA) for use in melanoma [25,26,27]. The FDA also approved vemurafenib for Erdheim-Chester Disease, another rare type of histiocytic neoplasm involving BRAF V600 mutation [28]. In recent studies, LCH patients carrying V600E also reportedly improved after vemurafenib administration alone or in combination with dabrafenib [28,29,30]. Thus, vemurafenib and dabrafenib have great potentials as targeted therapies for the treatment of LCH. A well-designed randomized clinical trial of single-agent dabrafenib to determine its effectiveness in LCH is planned and will be conducted in our institution soon.
The finding of no correlation between the BRAF V600E and TP53 mutation status is inconsistent with the results of a previous study in which FFPE tissues were analyzed [31]. The lack of a correlation is probably due to the high number of patients without BRAF V600E in our cohort. Moreover, there were no statistically significant correlations between the mutation status and other parameters. It is possible that the lack of complete assessment of some lesions clearly described in the medical records may have affected the above statistical results. For example, masses considered to be associated with LCH were identified by clinical observation or radiography alone without a combined biopsy, so some masses close to bone lesions may have been classified as eosinophilic granuloma. Lymphadenectasis or the presence of liver lesions in some patients was diagnosed using abdominal ultrasound without biopsy. Hence, some symptoms may not be clearly classifiable or may not be caused by LCH.
Approaches for BRAF V600E screening
The method used to evaluate mutation status can affect the results [32]. As shown in Table 2, immunohistochemistry (IHC, 10/21), Sanger/direct sequencing (10/21), allele-specific PCR (9/21) and pyrosequencing (7/21) are popular approaches for BRAF V600E screening but only a few studies applied NGS (4/21) or ddPCR (3/21). In our cohort, the limit of detection (LOD, n% mutant allele in a background of wild-type alleles) was 0.12% in ddPCR and 0.1% in LCH-panel NGS. However, the LOD for BRAF mutation can vary considerably across methods: IHC (5%), Sanger sequencing (6.6%), pyrosequencing (5%), NGS (1–2%), allele-specific qPCR (0.5%) and ddPCR (0.0005%) [32,33,34,35]. When using a method with a high LOD, false-negative results are likely to occur. Thus, approaches with low LOD should be prioritized to reduce type II errors in the future.
Compared to other approaches, ddPCR is a relatively inexpensive and sensitive technique but it does not distinguish between BRAF V600E and V600D, leading to false-positive errors in the detection. Our study used both NGS and ddPCR to confirm the mutation status and avoid the misuse of V600E/K inhibitors for V600D patients. In fact, two patients showed positive for V600E in ddPCR, but they actually carried V600D, which was detected by NGS. Thus, to prevent false-positive errors, we recommend performing BRAF mutation screening for LCH patients using a method with high specificity.
Conclusions
This study detected the frequency of BRAF V600E mutation in FFPE tissue and peripheral blood samples from pediatric LCH patients of Chinese origin. The mutation is less frequent in children than in adults, and in East Asian populations than in other populations. We also found that tissue is a more reliable sample for genetic screening because only a small amount of DNA can escape from lesions into the circulation and not all of circulating DNA is tumoral. Blood sample is not favored for now, but it may have the potentials to contribute to the surveillance of disease progression in the future. The findings also suggest that dabrafenib is an effective drug for reducing the risk of relapse in LCH patients, though more rigorous studies are needed to verify its effectiveness. We recommend a low LOD or high sensitivity method for BRAF V600E mutation screening to avoid type I and II errors.
Methods
Patients and samples
A cohort of LCH patients admitted to the Department of Hematology, Children’s Hospital of Capital Institute of Pediatrics in China from 2018 to 2019 was assembled in this study. Genetic information for a total of 166 patients was retrieved. The clinical and laboratory data for 92 patients were retrieved from their medical records, and data for 56 patients were collected through follow-up phone calls. Eighteen patients were excluded due to a lack of reliable clinical information. Therefore, the study focused on the 148 patients whose clinical and genetic data were available. The LCH diagnosis for all patients included was established through routine immunohistochemical examinations with positive CD1a, S-100 protein and CD207/langerin results (Fig. 4), according to the diagnostic criteria from the Histiocyte Society [36]. Based on the organ/system involvements, LCH is classified as affecting single system at single/multiple site(s) (SS-S/SS-M) or involving multiple systems without/with risk organ (MS-RO−/MS-RO+), which was defined as the bone marrow, liver or spleen [2]. LCH with risk-organ involvement is considered as high-risk, while other presentations are low-risk. This study was approved by the research ethics committee of the Children’s Hospital of Capital Institute of Pediatrics (Identifier: SHERLLM2020005). The guardians of the pediatric patients also signed written informed consent forms for the investigation and publication of articles.
Next-generation sequencing
The NGS-LCH panel is commercially available by Running Gene Inc. (Beijing, China). A total of 29 genes involved in the RAF-ERK pathway and/or associated with LCH in previous studies were included in the panel (Additional file 1: Table S1 Gene list of LCH panel ).
Formalin-fixed paraffin-embedded (FFPE) tissue (n = 84) or/and peripheral blood (n = 78) samples from 148 LCH patients were collected, processed and sent for sequencing. Both FFPE tissue and peripheral blood samples were collected from 14 patients. Circulating DNA or DNA samples were isolated from the peripheral blood or FFPE tissues using the QIAamp Circulating Nucleic Acid Kit (#55114) or QIAamp FFPE DNA Tissue Kit (#56404, Qiagen, Hilden, Germany), respectively. The concentrations of extracted DNA were determined using NanoDrop One (#840-329700, Thermo Fisher Scientific, Waltham, MA) and the Qubit dsDNA HS Assay Kit (#Q32851, Invitrogen, Carlsbad, CA). Agarose gel electrophoresis or the DNA High Sensitivity Kit for Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA) was used for quality control. Qualified DNA samples were fragmented into 200 to 300 bp and then processed with the KAPA Hyper Prep Kit (Kapa Biosystems, Wilmington, MA) to build a DNA library. Customized probes were applied to perform a hybridization against the pooled libraries, and captured DNA fragments were enriched using PCR. The final products were sequenced on the Illumina HiSeq X10 platform (Illumina, San Diego, CA) with 150-bp paired-end reads. The raw average sequence depth was > 30,000 × for circulating DNA samples and > 500 × for tissue DNA samples.
Raw data were processed to fastq format by bcl2fastq v2.20 (Illumina, San Diego, CA). Software fastp v0.12.7 [37] was used for quality control and Burrows-Wheeler Alignment tool v0.7.16 [38] was used to map the pair-end reads to the human reference genome (GRCh37/hg19). The single nucleotide variants (SNVs) and insertions and deletions (INDELs) of targeted genes were called using GATK v3.7 [39] with MuTech2 [40]. Gene fusions were called using Genefuse version v0.5.0 [41]. Called variants were filtered in (reads > 5; depth > 500X) using snpEff v4.3 [42]. Filtered variants were annotated using an in-house annotation system based on public databases (1000 genomes project, ExAC, Exome Variant Server, dbSNP, COSMIC, My Cancer Genome, etc.). We identified several variants that were probably associated with LCH (unpublished data) but only focused on the BRAF V600E mutation here.
Droplet digital PCR
The results of NGS for 69 patients were validated using ddPCR. Briefly, 2 × ddPCR Supermix for Probes (#1863028, Bio-Rad, Hercules, CA), 20 × assay PrimePCR™ ddPCR™ Mutation Assay Kit BRAF WT for p.V600E and BRAF p.V600E (#1863100, Bio-Rad, Hercules, CA), ddH2O and DNA templates were mixed well in a 20 μL reaction system, following the manufacturer’s instructions. Each reaction mix was loaded into a Droplet Generation 8 cartridge. Droplet Generation oil was loaded into the bottom row of the cartridge. After a 2-min running on the QX200 Droplet Generator (Bio-Rad, Hercules, CA), the droplets in the top row of the cartridge were collected and transferred into a 96-well plate. PCR amplification was performed with the T100 Thermal Cycler (Bio-Rad, Hercules, CA) following the instructions for the ddPCR Supermix for Probes. Afterward, the droplets were read and analyzed using the QX200 Droplet Reader (Bio-Rad, Hercules, CA).
Statistical analysis
The frequency of BRAF mutations, along with the demographic information and assessed features for published cohorts of LCH patients bearing BRAF V600E mutations in tissue, were also summarized (Table 2). The frequency distributions of the clinicopathological data were collected and tabulated for patients with and without the BRAF V600E mutation (Table 1).
We hypothesized an association between each clinical characteristic and the mutation status (alpha cut-off of 0.05). The Chi-square test or Fisher’s exact test was used to determine the associations between clinical characteristics and the mutation status. Kaplan–Meier estimates and the Log-rank (Mantel-Cox) test were applied for recurrence and targeted therapy. P-values < 0.05 were considered statistically significant for all tests. Statistical analyses were performed with Microsoft Excel version 16.37 (Microsoft, Redmond, WA) and R (https://www.R-project.org/).
Availability of Data and Materials
The datasets analyzed during the current study are not publicly available because the data is used by another undergoing study but are available from the corresponding author on reasonable request.
Abbreviations
- ddPCR:
-
Droplet digital PCR
- ERK:
-
Extracellular signal-regulated kinase
- FDA:
-
Food and Drug Administration
- FFPE:
-
Formalin-fixed paraffin-embedded
- INDELs:
-
Insertions and deletions
- LCH:
-
Langerhans cell histiocytosis
- MAPK:
-
Mitogen-activated protein kinase
- NGS:
-
Next-generation sequencing
- SNVs:
-
Single nucleotide variants
- SS-S:
-
Single-system involvement at single site
- SS-M:
-
Single-system involvement at multiple sites
- MS-OR− :
-
Multiple-system without risk-organ involvement
- MS-OR+ :
-
Multiple-system with risk-organ involvement
References
Krooks J, Minkov M, Weatherall AG. Langerhans cell histiocytosis in children: history, classification, pathobiology, clinical manifestations, and prognosis. J Am Acad Dermatol. 2018;78(6):1035–44.
Thacker NH, Abla O. Pediatric Langerhans cell histiocytosis: state of the science and future directions. Clin Adv Hematol Oncol. 2019;17(2):122–31.
Allen CE, Merad M, McClain KL. Langerhans-cell histiocytosis. N Engl J Med. 2018;379(9):856–68.
Salotti JA, Nanduri V, Pearce MS, Parker L, Lynn R, Windebank KP. Incidence and clinical features of Langerhans cell histiocytosis in the UK and Ireland. Arch Dis Child. 2009;94(5):376–80.
Haupt R, Minkov M, Astigarraga I, Schafer E, Nanduri V, Jubran R, et al. Langerhans cell histiocytosis (LCH): guidelines for diagnosis, clinical work-up, and treatment for patients till the age of 18 years. Pediatr Blood Cancer. 2013;60(2):175–84.
Goyal G, Young JR, Koster MJ, Tobin WO, Vassallo R, Ryu JH, et al. The Mayo Clinic Histiocytosis Working Group Consensus Statement for the Diagnosis and Evaluation of Adult Patients With Histiocytic Neoplasms: Erdheim-Chester Disease, Langerhans Cell Histiocytosis, and Rosai-Dorfman Disease. Mayo Clin Proc. 2019;94(10):2054–71.
Gadner H, Minkov M, Grois N, Potschger U, Thiem E, Arico M, et al. Therapy prolongation improves outcome in multisystem Langerhans cell histiocytosis. Blood. 2013;121(25):5006–14.
Cicenas J, Tamosaitis L, Kvederaviciute K, Tarvydas R, Staniute G, Kalyan K, et al. KRAS, NRAS and BRAF mutations in colorectal cancer and melanoma. Med Oncol. 2017;34(2):26.
Elisei R, Ugolini C, Viola D, Lupi C, Biagini A, Giannini R, et al. BRAF(V600E) mutation and outcome of patients with papillary thyroid carcinoma: a 15-year median follow-up study. J Clin Endocrinol Metab. 2008;93(10):3943–9.
Rothschild SI. Targeted therapies in non-small cell lung cancer-beyond EGFR and ALK. Cancers. 2015;7(2):930–49.
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–54.
Berres ML, Lim KP, Peters T, Price J, Takizawa H, Salmon H, et al. BRAF-V600E expression in precursor versus differentiated dendritic cells defines clinically distinct LCH risk groups. J Exp Med. 2014;211(4):669–83.
Zeng K, Ohshima K, Liu Y, Zhang W, Wang L, Fan L, et al. BRAFV600E and MAP2K1 mutations in Langerhans cell histiocytosis occur predominantly in children. Hematol Oncol. 2016;35(4):845–51.
Hogstad B, Berres ML, Chakraborty R, Tang J, Bigenwald C, Serasinghe M, et al. RAF/MEK/extracellular signal-related kinase pathway suppresses dendritic cell migration and traps dendritic cells in Langerhans cell histiocytosis lesions. J Exp Med. 2018;215(1):319–36.
Ribeiro KB, Degar B, Antoneli CB, Rollins B, Rodriguez-Galindo C. Ethnicity, race, and socioeconomic status influence incidence of Langerhans cell histiocytosis. Pediatr Blood Cancer. 2015;62(6):982–7.
Badalian-Very G, Vergilio JA, Degar BA, MacConaill LE, Brandner B, Calicchio ML, et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116(11):1919–23.
Kim BE, Koh KN, Suh JK, Im HJ, Song JS, Lee JW, et al. Clinical features and treatment outcomes of Langerhans cell histiocytosis: a nationwide survey from Korea histiocytosis working party. J Pediatr Hematol Oncol. 2014;36(2):125–33.
Bubolz AM, Weissinger SE, Stenzinger A, Arndt A, Steinestel K, Bruderlein S, et al. Potential clinical implications of BRAF mutations in histiocytic proliferations. Oncotarget. 2014;5(12):4060–70.
Heritier S, Emile JF, Barkaoui MA, Thomas C, Fraitag S, Boudjemaa S, et al. BRAF mutation correlates with high-risk langerhans cell histiocytosis and increased resistance to first-line therapy. J Clin Oncol. 2016;34(25):3023–30.
Kambouchner M, Emile JF, Copin MC, Coulomb-Lhermine A, Sabourin JC, Della Valle V, et al. Childhood pulmonary Langerhans cell histiocytosis: a comprehensive clinical-histopathological and BRAF(V600E) mutation study from the French national cohort. Hum Pathol. 2019;89:51–61.
Tong C, Jia X, Jia Y, He Y. Langerhans cell histiocytosis in Chinese adults: absence of BRAF mutations and increased FOXP3(+) regulatory T cells. Int J Clin Exp Pathol. 2014;7(6):3166–73.
Zeng K, Wang Z, Ohshima K, Liu Y, Zhang W, Wang L, et al. BRAF V600E mutation correlates with suppressive tumor immune microenvironment and reduced disease-free survival in Langerhans cell histiocytosis. Oncoimmunology. 2016;5(7):e1185582.
Kobayashi M, Ando S, Kawamata T, Makiyama J, Yokoyama K, Imai Y, et al. Clinical features and outcomes of adult Langerhans cell histiocytosis: a single-center experience. Int J Hematol. 2020;112(2):185–92.
Satoh T, Smith A, Sarde A, Lu HC, Mian S, Trouillet C, et al. B-RAF mutant alleles associated with Langerhans cell histiocytosis, a granulomatous pediatric disease. PLoS ONE. 2012;7(4):e33891.
Maverakis E, Cornelius LA, Bowen GM, Phan T, Patel FB, Fitzmaurice S, et al. Metastatic melanoma—a review of current and future treatment options. Acta Derm Venereol. 2015;95(5):516–24.
Long GV, Hauschild A, Santinami M, Atkinson V, Mandala M, Chiarion-Sileni V, et al. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N Engl J Med. 2017;377(19):1813–23.
Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–65.
Diamond EL, Subbiah V, Lockhart AC, Blay JY, Puzanov I, Chau I, et al. Vemurafenib for BRAF V600-mutant erdheim-chester disease and langerhans cell histiocytosis: analysis of data from the histology-independent, phase 2 Open-label VE-BASKET Study. JAMA Oncol. 2018;4(3):384–8.
Charles J, Beani JC, Fiandrino G, Busser B. Major response to vemurafenib in patient with severe cutaneous Langerhans cell histiocytosis harboring BRAF V600E mutation. J Am Acad Dermatol. 2014;71(3):e97–9.
Stewart JR, Murzaku EC, Sode TT, Gordon KA. Cutaneous Langerhans cell histiocytosis with gastrointestinal involvement treated with dabrafenib. JAAD Case Rep. 2018;4(1):95–7.
McGinnis LM, Nybakken G, Ma L, Arber DA. Frequency of MAP2K1, TP53, and U2AF1 mutations in BRAF-mutated langerhans cell histiocytosis: further characterizing the genomic landscape of LCH. Am J Surg Pathol. 2018;42(7):885–90.
Ballester LY, Cantu MD, Lim KPH, Sarabia SF, Ferguson LS, Renee Webb C, et al. The use of BRAF V600E mutation-specific immunohistochemistry in pediatric Langerhans cell histiocytosis. Hematol Oncol. 2018;36(1):307–15.
Ihle MA, Fassunke J, Konig K, Grunewald I, Schlaak M, Kreuzberg N, et al. Comparison of high resolution melting analysis, pyrosequencing, next generation sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer. 2014;14:13.
Richter A, Grieu F, Carrello A, Amanuel B, Namdarian K, Rynska A, et al. A multisite blinded study for the detection of BRAF mutations in formalin-fixed, paraffin-embedded malignant melanoma. Sci Rep. 2013;3:1659.
Reid AL, Freeman JB, Millward M, Ziman M, Gray ES. Detection of BRAF-V600E and V600K in melanoma circulating tumour cells by droplet digital PCR. Clin Biochem. 2015;48(15):999–1002.
Chu T, D’Angio GJ, Favara BE, Ladisch S, Nesbit M, Pritchard J. Histiocytosis syndromes in children. Lancet. 1987;2(8549):41–2.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–9.
Chen S, Liu M, Huang T, Liao W, Xu M, Gu J. GeneFuse: detection and visualization of target gene fusions from DNA sequencing data. Int J Biol Sci. 2018;14(8):843–8.
Cingolani P, Platts A, le Wang L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92.
Liu X, Zhang Y, Zhou CX. High prevalence of BRAF V600E mutations in langerhans cell histiocytosis of head and neck in chinese patients. Int J Surg Pathol. 2019;27(8):836–43.
Sasaki Y, Guo Y, Arakawa F, Miyoshi H, Yoshida N, Koga Y, et al. Analysis of the BRAFV600E mutation in 19 cases of Langerhans cell histiocytosis in Japan. Hematol Oncol. 2017;35(3):329–34.
Hayase T, Saito S, Shioda Y, Imamura T, Watanabe K, Ohki K, et al. Analysis of the BRAF and MAP2K1 mutations in patients with Langerhans cell histiocytosis in Japan. Int J Hematol. 2020;112(4):560–7.
Go H, Jeon YK, Huh J, Choi SJ, Choi YD, Cha HJ, et al. Frequent detection of BRAF(V600E) mutations in histiocytic and dendritic cell neoplasms. Histopathology. 2014;65(2):261–72.
Alayed K, Medeiros LJ, Patel KP, Zuo Z, Li S, Verma S, et al. BRAF and MAP2K1 mutations in Langerhans cell histiocytosis: a study of 50 cases. Hum Pathol. 2016;52:61–7.
Brown NA, Furtado LV, Betz BL, Kiel MJ, Weigelin HC, Lim MS, et al. High prevalence of somatic MAP2K1 mutations in BRAF V600E-negative Langerhans cell histiocytosis. Blood. 2014;124(10):1655–8.
Pina-Oviedo S, Medeiros LJ, Li S, Khoury JD, Patel KP, Alayed K, et al. Langerhans cell histiocytosis associated with lymphoma: an incidental finding that is not associated with BRAF or MAP2K1 mutations. Mod Pathol. 2017;30(5):734–44.
Roden AC, Hu X, Kip S, Parrilla Castellar ER, Rumilla KM, Vrana JA, et al. BRAF V600E expression in Langerhans cell histiocytosis: clinical and immunohistochemical study on 25 pulmonary and 54 extrapulmonary cases. Am J Surg Pathol. 2014;38(4):548–51.
Haroche J, Charlotte F, Arnaud L, von Deimling A, Helias-Rodzewicz Z, Hervier B, et al. High prevalence of BRAF V600E mutations in Erdheim-Chester disease but not in other non-Langerhans cell histiocytoses. Blood. 2012;120(13):2700–3.
Sahm F, Capper D, Preusser M, Meyer J, Stenzinger A, Lasitschka F, et al. BRAFV600E mutant protein is expressed in cells of variable maturation in Langerhans cell histiocytosis. Blood. 2012;120(12):e28-34.
Mehes G, Irsai G, Bedekovics J, Beke L, Fazakas F, Rozsa T, et al. Activating BRAF V600E mutation in aggressive pediatric Langerhans cell histiocytosis: demonstration by allele-specific PCR/direct sequencing and immunohistochemistry. Am J Surg Pathol. 2014;38(12):1644–8.
Acknowledgements
We thank all patients and their families for participation and cooperation.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
SF cared and recruited patients, conceptualized the study, collected, evaluated and interpreted clinical data. LH designed the study, collected and analyzed clinical and genetic data, drafted the initial manuscript, reviewed and revised the manuscript. MY, DZ and JC cared and recruited patients, collected, evaluated and interpreted clinical data. YG, YS and HZ collected clinical data and contributed to genetic sequencing. ZC and XC conceptualized and supervised the study. RL cared and recruited patients, conceptualized and supervised the study, collected, evaluated and interpreted clinical data, and reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent for participation and publication
This study was approved by the research ethics committee of the Children’s Hospital of Capital Institute of Pediatrics (Identifier: SHERLLM2020005). The guardians of pediatric patients also signed written informed consent forms for the investigation and publication of articles.
Competing interests
Author LH, YG, YS, HZ and ZC are employed by Running Gene Inc. The other authors declare that the research was conducted in the absence of any other commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1
. Gene list of LCH panel.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Feng, S., Han, L., Yue, M. et al. Frequency detection of BRAF V600E mutation in a cohort of pediatric langerhans cell histiocytosis patients by next-generation sequencing. Orphanet J Rare Dis 16, 272 (2021). https://doi.org/10.1186/s13023-021-01912-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13023-021-01912-3