Abstract
Objective
Delayed fracture healing increases the suffering of patients. An in-depth investigation of the pathogenesis of delayed fracture healing may offer new direction for the prevention and treatment.
Methods
The study included 63 normal healing tibial fractures and 58 delayed healing tibial fractures patients. Long non-coding RNA (lncRNA)TRPM2-AS, microRNA-545-3p (miR-545-3p), bone morphogenetic protein 2 (Bmp2) mRNA and osteogenic differentiation markers, including runt-related transcription factor 2 (Runx2), osteocalcin (Ocn), and alkaline phosphatase (Alp) mRNA expression were determined by Real-time quantitative reverse transcription-polymerase chain reaction in serum and MC3T3-E1 cells. The prediction potential of TRPM2-AS in delayed healing fracture patients was verified by receiver operating characteristic curves. The binding relationship of TRPM2-AS/miR-545-3p/Bmp2 was evaluated by dual luciferase reporter gene assay. Cell proliferation and apoptosis were detected by CCK-8 and flow cytometry.
Results
TRPM2-AS was remarkably down-regulated in patients with delayed fracture healing and could better predict the fracture healing status. TRPM2-AS downregulation inhibited osteogenic markers mRNA expression, restrained proliferation, and promoted apoptosis of MC3T3-E1 cells (p < 0.05). In delayed fracture healing, miR-545-3p was dramatically up-regulated and was negatively regulated by TRPM2-AS. Reducing miR-545-3p eliminate the negative effect of TRPM2-AS down-regulation on osteoblast proliferation and differentiation (p < 0.05). miR-545-3p targets Bmp2, which plays a positive role in osteoblast differentiation (p < 0.05).
Conclusion
This study found that TRPM2-AS has the potential to be a diagnostic marker for delayed fracture healing and revealed that the TRPM2-AS/miR-545-3p/Bmp2 axis affects fracture healing by regulating osteoblast.
Similar content being viewed by others
Introduction
Fractures are the most common traumatic disease in orthopedic clinic. In recent years, the increasing number of fractures has become a global public health problem. In most cases, bone structure and function is fully restored after healing. However, abnormal healing occurs in about 10% of fracture patients [1, 2]. As society ages, the physiological function of patients declines, increasing the likelihood of delayed healing and making prevention more difficult. Studies have shown that 20% of elderly patients with fractures require long-term rehabilitation and care, causing significant physical and psychological suffering for patients and placing a heavy burden on the healthcare system and society [3]. Despite this, the current success rate for treating impaired fracture healing remains low. Therefore, exploring effective mechanisms to promote delayed fracture healing is crucial to reduce the incidence of poor fracture healing.
Osteoblasts and osteoclasts regulate the balance of new bone formation and bone reconstruction in fracture healing, which are critical cells affecting this process [4, 5]. Osteoclasts primarily mediate bone resorption, breaking down necrotic or aged bone tissue into raw materials for new bone [6]. Osteoblasts mediate bone formation, synthesizing these raw materials into new bone cells [7]. Osteoblast dysfunction is often considered the direct cause of delayed fracture healing, and bone formation largely depends on the number and activity of osteoblasts. In bone tissue engineering, MC3T3-E1 cells are extensively used to study the differentiation, proliferation, and molecular mechanisms of osteoblasts. They have a well-established research system as a model of osteogenesis [8]. Therefore, exploring the mechanisms affecting osteoblast activity and differentiation using MC3T3-E1 cells may provide new directions and strategies for treating and preventing poor fracture healing.
Long non-coding RNA (lncRNA) can participate in various physiological and pathological processes such as cell proliferation, differentiation and apoptosis [9]. It was previously indicated that lncRNAs regulate the differentiation and function of osteoblasts. A report confirmed that LncRNA TUG1 can promote the differentiation and proliferation of osteoblasts by sponging miR-22-5p, thus promoting fracture healing [10]. LncRNA AC132217.4 has been found to affect the conduction of IGF-AKT signaling pathway, which plays an important regulatory role in the healing and regeneration process after bone injury [11]. This suggests that lncRNAs may have essential regulatory effects in bone formation, resorption, remodeling and repair by participating in the regulated proliferation and differentiation of osteoblasts.
In a study on osteogenic differentiation-related competitive endogenous RNA networks [12], the aberrantly expressed LncRNA TRPM2-AS drew the attention of the present study. Studies have discovered that the LncRNA TRPM2-AS is extensively participated in tumor development and plays an essential regulatory part in the malignant behavioral activities of cancer cells [13]. For example, LncRNA TRPM2-AS is key in chemoresistance of prostate cancer by targeting miR-497-5p [14]. In breast cancer, LncRNA TRPM2-AS has also been found to act as a competitive endogenous source of miR-140-3p, which regulates cancer cell activity [15]. Notably, a study on LncRNA TRPM2-AS’s involvement in the regulation of osteosarcoma cell proliferation and cell apoptosis [16] suggested that LncRNA TRPM2-AS might also regulate osteoblasts. At present, the function of LncRNA TRPM2-AS in fracture healing is not well understood. Therefore, this study believes that it is innovative to explore the role and mechanism of LncRNA TRPM2-AS in fracture healing.
This study first analyzed the expression of LncRNA TRPM2-AS in serum of patients with delayed fracture healing. Then, cell experiments were conducted to examine the role and mechanism of LncRNA TRPM2-AS expression in osteoblast proliferation and apoptosis. The aim is to provide a new therapeutic target for promoting clinical delayed fracture healing.
Materials and methods
Participants of this research
In this study, 200 patients with tibial fractures who underwent immobilization treatment in the First People’s Hospital of Jingzhou from 12 April 2018 to 24 November 2020 were selected as study subjects. Exclusion criteria: (1) history of previous fracture or orthopedic surgery; (2) significant organ dysfunction, malignant tumor; (3) infectious lesions, acute gastrointestinal inflammation, immune and hematological diseases; (4) bone metabolic diseases. After review at 4 months postoperatively, 121 patients were screened for inclusion in this study and grouped according to the following diagnostic criteria [17] for fracture healing and the specific fracture healing status of the patients. Among them, 63 patients were in the normal healing group: those who formed bone scabs within 4 months according to the process of fracture healing. 58 patients were in the delayed healing group: those who showed signs of healing for more than 4 months had bone atrophy at the broken end of the fracture, or had visible fracture lines on the X-ray line with no or minimal bone scabs. The present work was approved by the Ethics Committee of the First People’s Hospital of Jingzhou and was conducted under its supervision (registration number 201799).
Cell culture
The mouse pre-osteoblast cell line MC3T3-E1 used in the study was purchased from BeNa Culture collection (Beijing, China). Before the experiment, MC3T3-E1 cells were thawed and then cultured in DMEM (2,248,316; Biological Industries, China) medium containing fetal bovine serum (FBS, 10%, C04001-500; Biological Industries, China), penicillin and streptomycin (1%, SV30010; HyClone Co., Logan, UT, USA,) at 37 °C and 5% CO2. The medium was changed every 3 days. Cell differentiation was induced when the cell growth density reached 80%.
Cell transfection
The following plasmids were constructed before transfection:
-
(1)
si-TRPM2-AS plasmid: silencing TRPM2-AS.
-
(2)
si-NC plasmid: negative control for TRPM2-AS.
-
(3)
oe-TRPM2-AS plasmid: overexpression of TRPM2-AS.
-
(4)
miR-mimic plasmid: up-regulation of miR-545-3p.
-
(5)
miR-inhibitor plasmid: down-regulation of miR-545-3p.
-
(6)
miR-NC plasmid: negative control for miR-545-3p.mp2.
-
(7)
Bmp2-NC plasmid: negative control for Bmp2mp2.
-
(8)
si-Bmp2 plasmid: down-regulation of Bmp2.
Select logarithmically grown MC3T3-E1 cells for transfection and co-transfect the cells with the required plasmids and transfection reagents according to the instruction manual of Lipofectamine2000 reagent (11,668,027; Invitrogen, Carlsbad, USA). One hour before transfection, change the DMEM culture medium to serum-free medium. Six hours after transfection, replace the medium with fresh DMEM medium containing 10% FBS. Continue to culture the cells for 48 h and then collect them. Meanwhile, use normally cultured osteoblasts without any transfection as the control group.
CCK8 assay for cell viability
The MC3T3-E1 cells from the transfected and control groups in the logarithmic growth phase were selected. The old fluid in the culture flask was aspirated and washed with PBS. After washing, trypsin was added, and digestion was terminated by adding an equal amount of DMEM culture medium containing 10% FBS once the cells had rounded. The cells were centrifuged at room temperature at 1000 rpm for 5 min, and the supernatant was discarded, and the cells were resuspended by adding the above DMEM culture medium. The counted cells were inoculated into 96-well plates. Cell supernatants were discarded and 10 μL of CCK-8 solution (CCK-8 kit, C0038; Beyotime, Beijing, China) was added to each well at the indicated time points at 0, 24, 48, and 72 h of cell incubation, respectively. After 2 h of incubation, absorbance (OD450) was monitored at 450 nm using Enzyme-linked immunoassay.
Flow cytometry analysis of cell apoptosis
The MC3T3-E1 cells were collected from each group. First, 0.25% trypsin was added to digest the cells, then DMEM medium containing 10% FBS was added to prepare a cell suspension. The cells were washed twice with PBS. The cell density was adjusted to 2.5 × 106 /mL, 400 μL of cell suspension was taken. They were then added 10 μL of Annexin-FITC and 5 μL of propidium iodide (PI) (Annexin V-EGFP/PI Kit, CA1020-50 T; Solebo Technology Co., Ltd., Beijing, China) and incubated for 10 min away from light. Finally, the apoptosis rate was detected by flow cytometry.
Osteoblast differentiation
The MC3T3-E1 cells were inoculated in 6-well plates and induced for 15 d using osteogenic induction medium prepared with L-glycerophosphate (5 mM), dexamethasone (100 nM) and ascorbic acid (50 g/L) (Osteogenic induction medium, HUXUC-90021; Cyagen Biosciences, Guangzhou, China). The MC3T3-E1 cells on day 0, day 7 and day 15 of osteogenic differentiation induction were collected.
Sample collection
In the early morning of the fourth postoperative week, 5 mL of peripheral venous blood was drawn from the patient under the condition that the patient fulfils a 12-h fast. The venous blood was kept in an anticoagulation tube and centrifuged at 3000 rpm for 15 min to take the supernatant night (serum). Ensure that the relevant tests are completed within 48 h of serum collection and the remaining serum is stored at − 80 °C.
Real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
The expression of TRPM2-AS, miR-545-3p, bone morphogenetic protein 2 (Bmp2) mRNA was detected in the serum of the subjects and in MC3T3-E1 cells of each group and the mRNA expression of bone differentiation-related proteins was detected in MC3T3-E1 cells of each group, including runt-related transcription factor 2 (Runx2), osteocalcin (Ocn), alkaline phosphatase (Alp) mRNA. Total RNA was extracted from the cells using Trizol reagent (15,596,026; Thermo Fisher Scientific, Carlsbad, USA), and the concentration of RNA was determined by ND-2000 nanodrop (Thermo Fisher Scientific, Waltham, USA). Complementary deoxyribonucleic acid (cDNA) was synthesized by reverse transcription of the screened RNA using SuperScriptTM II kit (12,594,100, Thermo Fisher Scientific, Waltham, USA). Then the cDNA was used as the template for amplification. GAPDH was used as an internal reference for LncRNA and mRNA, and U6 was used as a miRNA internal reference, respectively. The relative expression levels mp2were calculated by 2−ΔΔCt method (The primers are shown in Table S1).
Luciferase report assay
-
1
First, the binding sites between TRPM2-AS and miR-545-3p were predicted using the lncRNASNP database (http: //gong_lab.hzau.edu.cn /lncRNASNP3#!/). Then, the TRPM2-AS wild-type plasmid (WT-TRPM2-AS) and mutant plasmid (MT-TRPM2-AS) were constructed by cloning the sequences with and without the binding site into the pmirGLO vector, respectively. The transfection reagent Lipofectamine2000 was used to co-transfect these plasmids with miR-NC or miR-545-3p mimic and inhibitor into MC3T3-E1 cells. A blank control group was also set up under the same conditions. After 48 h, TRPM2-AS luciferase activity was measured.
-
2
The binding site between miR-545-3p and Bmp2 was predicted using the TargetScan (http://www.targetscan.org/vert_72/) database. A wild-type (WT-Bmp2) plasmid containing the mutation site and a mutant plasmid without the mutation site (MT-Bmp2) were constructed. miR-NC or miR-545-3p mimic and inhibitor were then co-transfected with the two plasmids into MC3T3-E1 cells using Lipofectamine2000 reagent.
Statistical analysis
Continuous variables in the table and figure is represented by mean ± standard deviation (mean ± SD) and were analyzed with SPSS 22.0 and Graphpad prism 6. 0. Two-group and multi-group comparisons were obtained using Student’s t-test and one-way ANOVA followed by Tukey post-hoc tests, respectively. The diagnostic validity of TRPM2-AS in patients with delayed fracture healing was determined using the receiver operating characteristic curve (ROC). A p-value of less than 0.05 was considered statistically significant.
Results
Expression and prediction of lncRNA TRPM2-AS in delayed fracture healing
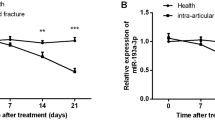
Firstly, the baseline data of patients included in normal fracture healing and delayed fracture healing were analyzed and compared, and it was found that the baseline difference between the two groups was not significant (p > 0.05, Table S2), which ensured the scientific validity of the experiment. The RT-qPCR was carried out on the serum of 63 patients with normal fracture healing and 58 patients with delayed healing who were included in the study. The results, demonstrated in Fig. 1A, indicated that TRPM2-AS expression was dramatically lower in the delayed group when compared with that in the normal group. Moreover, the constructed ROC showed that the area under the curve (AUC) of TRPM2-AS to distinguish patients with delayed fracture healing was 0.899 (95%CI 0.842–0.957), the specificity was 89.66%, and the sensitivity was 80.95% (Fig. 1B).
LncRNA TRPM2-AS expression in the serum of 63 patients with normal fracture healing and 58 patients with delayed fracture healing. A ROC demonstrates the diagnosis of LncRNA TRPM2-AS in patients with delayed fracture healing B (n = 3 independent experiments, and the data are presented as the mean ± SD)
Targeting relationship between lncRNA TRPM2-AS and miR-545-3p
The results were obtained by RT-qPCR assay of serum from patients included in the two groups, which indicated that miR-545-3p expression in patients with delayed fracture healing was markedly higher than that in patients with normal healing (Fig. 2A).miR-545-3p was significantly negatively correlated with TRPM2-AS (r = − 0.648, p < 0.0001, Fig. 2B) in patients with delayed fracture healing. According to the binding sites between TRPM2-AS and miR-545-3p, the gene fragments of WT-TRPM2-AS and MT-TRPM2-AS were designed (Table S3). Detection of TRPM2-AS luciferase activity in 4 groups of MC3T3-E1 cells showed that miR-545-3p negatively regulated WT-TRPM2-AS luciferase activity (Fig. 2C). In addition, knockdown of miR-545-3p did not significantly affect TRPM2-AS expression (Fig. 2D), while knockdown of TRPM2-AS significantly promoted miR-545-3p expression (Fig. 2E).
miR-545-3p expression in the serum of 63 patients with normal fracture healing and 58 patients with delayed fracture healing. A. Correlation between miR-545-3p and LncRNA TRPM2-AS in 58 patients with delayed fracture healing B Dual luciferase gene reporter assay for targeting relationship between miR-545-3p and LncRNA TRPM2-AS in MC3T3-E1 cells C Effect of knockdown of miR-545-3p on LncRNA TRPM2-AS in MC3T3-E1 cells D Effect of knockdown of LncRNA TRPM2-AS on miR-545-3p in MC3T3-E1 cells E (n = 3 independent experiments, and the data are presented as the mean ± SD)
Effects of lncRNA TRPM2-AS/miR-545-3p on osteoblasts
The MC3T3-E1 cells on day 0, day 7 and day 15 of osteogenic differentiation induction were collected,, respectively.. The differentiation results of induced MC3T3-E1 showed that the levels of osteogenic markers Alp, Runx2 and Ocn mRNA gradually increased with the extension of differentiation time (Fig. 3A), and the expression of TRPM2-AS increased progressively with time of cellular differentiation (Fig. 3B), whereas the expression of miR-545-3p decreased progressively with time (Fig. 3C). Samples of MC3T3-E1 cells on the 15th day of osteogenic differentiation induction were collected to explore the effect of TRPM2-AS on mRNAs of osteogenic differentiation-associated proteins, with three replicates in each group. It was revealed that knockdown of TRPM2-AS significantly reduced Alp, Runx2, and Ocn mRNA levels, which was alleviated by transfection with miR-545-3p inhibitor (Fig. 3D). In addition, CCK8 and apoptosis experiments on MC3T3-E1 cells subjected to different transfection treatments revealed that knockdown of TRPM2-AS accelerated inhibited MC3T3-E1 cells proliferation (Fig. 3E) and MC3T3-E1 cells apoptosis (Fig. 4A, flow cytometry is shown in Fig. 4B), while transfection with miR-545-3p inhibitor improved both phenomena.
Changes of osteoblast differentiation-related protein markers of MC3T3-E1 cells at different induction times A. Expression changes of LncRNA TRPM2-AS B and miR-545-3p in MC3T3-E1 cells at different induction times C. Effects of LncRNA TRPM2-AS knockdown on osteoblast differentiation protein markers D and proliferation E of MC3T3-E1 cells. (n = 3 independent experiments, and the data are presented as the mean ± SD)
Regulation of BMP2 on osteoblasts
Serum detection of the two groups showed that the expression of Bmp2 mRNA in patients with delayed fracture healing was significantly lower than that in patients with normal healing (Fig. 5A) In serum samples from patients with delayed fracture healing, Bmp2 mRNA expression was found to be dramatically negatively associated with miR-545-3p (r = − 0.612, p < 0.0001, Fig. 5B). In addition, The RT-qPCR assay of MC3T3-E1 cells on day 0, day 7 and day 15 undergoing osteogenic differentiation induction revealed that Bmp2 mRNA expression gradually increased with the increase of osteoblast differentiation time (Fig. 5C). To explore the targeting relationship between miR-545-3p and Bmp2, dual luciferase gene reporting detection was performed, and the specific sequence was shown in Table S4. The examination of Bmp2 luciferase activity in four groups of MC3T3-E1 cells indicated that the luciferase activity of Bmp2 decreased with the upregulation of miR-545-3p (Fig. 5D). In addition, it was found that the expression of Bmp2 mRNA was significantly decreased after knocking down the expression of TRPM2-AS, while knocking down miR-545-3p significantly upregulated Bmp2 mRNA expression (Fig. 5E) in MC3T3-E1 cells. Further exploration revealed that knockdown of Bmp2 mRNA expression reversed the promoting effect of miR-545-3p downregulation on Alp, Runx2, and Ocn mRNA levels in MC3T3-E1 cells undergoing osteogenic differentiation induction day 15 (Fig. 6A). Knockdown of Bmp2 mRNA expression also reversed the effects of miR-545-3p downregulation on MC3T3-E1 cells proliferation (Fig. 6B) and apoptosis (Fig. 7A, flow cytometry is shown in Fig. 7B).
Bmp2 mRNA expression in the serum of 63 patients with normal fracture healing and 58 patients with delayed fracture healing. A. Correlation between miR-545-3p and Bmp2 in 58 patients with delayed fracture healing B Changes of Bmp2 mRNA expression in MC3T3-E1 cells at different induction times C Dual luciferase gene reports detect the targeting relationship between miR-545-3p and Bmp2 in MC3T3-E1 cells D Effect of LncRNA TRPM2-AS and miR-545-3p knockdown on Bmp2 mRNA expression in MC3T3-E1 cells E (n = 3 independent experiments, and the data are presented as the mean ± SD)
Discussion
The process of fracture healing involves the interaction of multiple cells, cytokines, and genes, but due to the complexity of the etiology and the variety of fracture types, more fracture patients have some degree of impaired healing, which leads to delayed fracture healing and affects the physical and mental health of the patients [18]. Therefore, finding serum markers that can predict delayed fracture healing can guide timely clinical interventions to improve patients’ prognosis. In recent years, the application of LncRNA in predicting the prognosis of fracture surgery has been widely reported [19, 20]. For example, LncRNA HAGLR is significantly downregulated in rats with delayed fracture healing and has the potential to be a marker for predicting this condition [21]. Wang et al. found that lncRNA RORAD is involved in fracture healing regulation and could become a marker for pre-surgery diagnosis to predict bone healing in patients [22]. In this study, LncRNA TRPM2-AS was found to be significantly down-regulated in patients with delayed fracture healing, and the ROC results suggest that LncRNA TRPM2-AS has a high predictive value for patients with delayed healing. The above evidence suggests that LncRNA TRPM2-AS has the potential to be a potential marker for prognostic assessment after fracture surgery.
Relevant studies have shown that osteoblasts are the main bone-forming cells, indispensable for bone remodeling and healing. Enhancing osteoblast activity appropriately promotes bone healing [11, 23]. It was found that lncRNAs play a crucial regulatory role in the formation of osteoblasts. To explore the effects of LncRNA TRPM2-AS on osteoblasts, relevant experiments were conducted in this study. The results revealed that the downregulation of LncRNA TRPM2-AS resulted in a significant decrease in the proliferative capacity of osteoblasts and a significant increase in the rate of apoptosis. In addition, the expression of the osteogenic differentiation markers Alp, Runx2, and Ocn was significantly inhibited by the downregulation of LncRNA TRPM2-AS. Guo et al. found that LncRNA SNHG1 up-regulation can inhibit the proliferation and differentiation of osteoblasts and promote cell apoptosis [24]. LncRNA KCNQ1OT1 was found to activate the Wnt/β-catenin pathway, enhancing osteoblast proliferation and differentiation, thereby promoting fracture healing [25]. The above evidence suggests that LncRNA TRPM2-AS is involved in regulating the life activities of osteoblasts.
Studies have confirmed that lncRNAs can negatively regulate miRNAs as competitive endogenous RNA, participating in various cellular activities. Through a complex regulatory network, miRNAs play important regulatory roles in multiple aspects of bone-related diseases, including stem tendon injury [26, 27], osteoarthritis [28], human rheumatoid arthropathy [29, 30], osteoporosis [31], and tumor bone metastasis [32]. For example, LncRNA HOXA11-AS inhibits osteoblast proliferation and induce apoptosis by targeting miR-124-3p, thereby promoting fracture healing [33]. LncRNA HOTAIR is involved in the regulation of fracture healing in osteoporosis rats by negatively regulating miR-17-5p [34]. To explore the mechanism of LncRNA TRPM2-AS in osteoblasts, the downstream miRNAs of LncRNA TRPM2-AS were excavated in this study. Among them, miR-545-3p has attracted our attention. Previous studies have found that miR-545-3p can hinder osteogenic differentiation. lncRNA SERPINB9P1 can up-regulate SIRT6 expression through sponging miR-545-3p, thus promoting osteogenic differentiation of bone marrow mesenchymal stem cells [35]. lncRNA TUG1 also affects the expression of CNR2 by negatively regulating miR-545-3p and promotes the proliferation and differentiation of osteoblasts [36]. This study also found that miR-545-3p, as the downstream miRNA of LncRNA TRPM2-AS, is negatively regulated by LncRNA TRPM2-AS, and can reverse the negative effects of LncRNA TRPM2-AS knockdown on osteoblasts. This suggests that miR-545-3p plays an important role in LncRNA TRPM2-AS regulation of osteoblast proliferation and apoptosis.
This study found a targeting relationship between miR-545-3p and Bmp2, and negatively regulated the expression of Bmp2 mRNA. It was found that BMP2 knockdown inhibited the differentiation and growth of osteoblasts. BMP2, as the most important transcription factor for osteogenic differentiation, is a key signaling pathway in bone formation [37, 38]. It has been reported that BMP2 can play a role by activating Smad proteins and other signal transduction pathways to stimulate the expression of many target genes, including RUNX2 [39]. Chen. et al. found that miR-214-3p binds to the 3’UTR region of BMP1 to reduce its mRNA level and inhibit the maturation and differentiation of bone marrow mesenchymal stem cells [40]. The above evidence suggests that miR-545-3p plays an inhibitory role in delayed fracture healing by targeting Bmp2.
The novelty of this study lies in the fact that the role and mechanism of TRPM2-AS in fracture healing are proposed for the first time, which provides more theoretical basis for the pathogenesis of delayed fracture healing and the selection of therapeutic targets. Of course, there are some limitations in this study, such as due to time and funding, no animal experiments were performed to verify the modulatory effect of TRPM2-AS targeting miR-545-3p/Bmp2 on fracture healing. In the future, it will be further verified by expanding the sample range and sample size and combining with in vivo experiments. In addition, the present study only preliminarily investigated the effect of TRPM2-AS on osteoblast activity, however, it did not investigate its effect on osteoclast activity. Osteoclasts are also critical cells for fracture healing, regulating the balance of new bone formation and bone reconstruction. In follow-up studies, the effect of TRPM2-AS/miR-545-3p on osteoclast activity will be further investigated.
Conclusion
In conclusion, the results of the present study suggest that LncRNA TRPM2-AS has the potential to be a predictive marker of delayed fracture healing. It may contribute to delayed healing by mediating the miR-545-3p/Bmp2 axis, which enhances osteoblast differentiation and proliferation while suppressing osteoblast apoptosis.
Data availability
Corresponding authors may provide data and materials.
Availability of data and materials
Corresponding authors may provide data and materials.
References
Grigoryan M, Lynch JA, Fierlinger AL, et al. Quantitative and qualitative assessment of closed fracture healing using computed tomography and conventional radiography. Acad Radiol. 2003;10(11):1267–73.
Zhou W, Lin Z, Xiong Y, et al. Dual-targeted nanoplatform regulating the bone immune microenvironment enhances fracture healing. ACS Appl Mater Interfaces. 2021;13(48):56944–60.
Zhang SZ, Lu ZF, Xu YJ, et al. STEEL participates in fracture healing through upregulating angiogenesis-related genes by recruiting PARP 1. Eur Rev Med Pharmacol Sci. 2018;22(12):3669–75.
Han Y, Liu C, Lei M, et al. LncRNA TUG1 was upregulated in osteoporosis and regulates the proliferation and apoptosis of osteoclasts. J Orthop Surg Res. 2019;14(1):416.
Mi B, Chen L, Xiong Y, et al. Osteoblast/osteoclast and immune cocktail therapy of an exosome/drug delivery multifunctional hydrogel accelerates fracture repair. ACS Nano. 2022;16(1):771–82.
Li KL, Lu JG, Chen XH, et al. The role of the allantoin in promoting fracture healing in osteoclast-deficient zebrafish. Yi Chuan. 2023;45(4):341–53.
Zhang Z, Hu P, Wang Z, et al. BDNF promoted osteoblast migration and fracture healing by up-regulating integrin β1 via TrkB-mediated ERK1/2 and AKT signalling. J Cell Mol Med. 2020;24(18):10792–802.
Gibon E, Batke B, Jawad MU, et al. MC3T3-E1 osteoprogenitor cells systemically migrate to a bone defect and enhance bone healing. Tissue Eng Part A. 2012;18(9–10):968–73.
Bourgery M, Ekholm E, Fagerlund K, et al. Multiple targets identified with genome wide profiling of small RNA and mRNA expression are linked to fracture healing in mice. Bone Rep. 2021;15: 101115.
Li W, Li L, Cui R, et al. Bone marrow mesenchymal stem cells derived exosomal Lnc TUG1 promotes bone fracture recovery via miR-22-5p/Anxa8 axis. Hum Cell. 2023;36(3):1041–53.
Zhang C, Wu S, Chen E, et al. ALX1-transcribed LncRNA AC132217.4 promotes osteogenesis and bone healing via IGF-AKT signaling in mesenchymal stem cells. Cell Mol Life Sci. 2022;79(6):328.
Liu J, Yao Y, Huang J, et al. Comprehensive analysis of lncRNA-miRNA-mRNA networks during osteogenic differentiation of bone marrow mesenchymal stem cells. BMC Genomics. 2022;23(1):425.
Li F, Chen X. Contribution and underlying mechanisms of lncRNA TRPM2-AS in the development and progression of human cancers. Pathol Res Pract. 2023;251: 154887.
Shi T, Li R, Duan P, et al. TRPM2-AS promotes paclitaxel resistance in prostate cancer by regulating FOXK1 via sponging miR-497-5p. Drug Dev Res. 2022;83(4):967–78.
Sun T, Song Y, Yu H, et al. Identification of lncRNA TRPM2-AS/miR-140-3p/PYCR1 axis’s proliferates and anti-apoptotic effect on breast cancer using co-expression network analysis. Cancer Biol Ther. 2019;20(6):760–73.
Cai Y, Yang Y, Zhang X, et al. TRPM2-AS promotes the malignancy of osteosarcoma cells by targeting miR-15b-5p/PPM1D axis. Cell Cycle. 2022;21(8):835–50.
Park YK, Lee DY, Hur JW, et al. Delayed hinge fracture after plate-augmented, cervical open-door laminoplasty and its clinical significance. Spine J. 2014;14(7):1205–13.
Garg J, Ghoshal G, Sharma G, et al. Self emulsifying delivery system of Cissus Quadrangularis: evidence of enhanced efficacy and promising pharmacokinetic profile in the management of osteoporosis. AAPS PharmSciTech. 2024;25(5):107.
Liu L, Yuan Y. Downregulation of miR-221-3p by LncRNA TUG1 promoting the healing of closed tibial fractures in mice. Biomed Res Int. 2022;2022:1624446.
Li C, Qian YH. Inflammation-dependent activation of NCOA2 associates with p300 and c-MYC/Max heterodimer to transactivate RUNX2-AS1 and mediate RUNX2 downstream bone differentiation genes in the pathology of septic nonunion. Cytokine. 2022;158: 155992.
Pan LX, Ding W. LncRNA HAGLR accelerates femoral neck fracture healing through negatively regulating miRNA-19a-3p. Eur Rev Med Pharmacol Sci. 2020;24(8):4080–7.
Chen S, Ma H, Li M, et al. Long noncoding RNA NORAD promotes fracture healing through interacting with osteoblast differentiation via targeting miR-26a. Biomed Res Int. 2023;2023:9950037.
Wang J, Liu D, Guo B, et al. Role of biphasic calcium phosphate ceramic-mediated secretion of signaling molecules by macrophages in migration and osteoblastic differentiation of MSCs. Acta Biomater. 2017;51:447–60.
Guo X, Zhang J, Han X, et al. LncRNA SNHG1 delayed fracture healing via modulating miR-181a-5p/PTEN Axis. J Invest Surg. 2022;35(6):1304–12.
Gu H, Li Z, Lv XF, et al. LncRNA KCNQ1OT1 delayed fracture healing through the Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2019;23(11):4575–83.
Giordano L, Porta GD, Peretti GM, et al. Therapeutic potential of microRNA in tendon injuries. Br Med Bull. 2020;133(1):79–94.
Gargano G, Oliviero A, Oliva F, et al. Small interfering RNAs in tendon homeostasis. Br Med Bull. 2021;138(1):58–67.
Oliviero A, Della Porta G, Peretti GM, et al. MicroRNA in osteoarthritis: physiopathology, diagnosis and therapeutic challenge. Br Med Bull. 2019;130(1):137–47.
Gargano G, Oliva F, Oliviero A, et al. Small interfering RNAs in the management of human rheumatoid arthritis. Br Med Bull. 2022;142(1):34–43.
Gargano G, Asparago G, Spiezia F, et al. Small interfering RNAs in the management of human osteoporosis. Br Med Bull. 2023;148(1):58–69.
Lee KS, Lee J, Kim HK, et al. Extracellular vesicles from adipose tissue-derived stem cells alleviate osteoporosis through osteoprotegerin and miR-21-5p. J Extracell Vesicles. 2021;10(12): e12152.
Wang J, Du X, Wang X, et al. Tumor-derived miR-378a-3p-containing extracellular vesicles promote osteolysis by activating the Dyrk1a/Nfatc1/Angptl2 axis for bone metastasis. Cancer Lett. 2022;526:76–90.
Wang XN, Zhang LH, Cui XD, et al. lncRNA HOXA11-AS is involved in fracture healing through regulating mir-124-3p. Eur Rev Med Pharmacol Sci. 2017;21(21):4771–6.
Su Y, Meng X, Wang W, et al. LncRNA HOTAIR regulates fracture healing in osteoporotic rats through inhibition on MiR-17-5p. Minerva Med. 2021;112(4):525–7.
Wu M, Dai M, Liu X, et al. lncRNA SERPINB9P1 regulates SIRT6 mediated osteogenic differentiation of BMSCs via miR-545-3p. Calcif Tissue Int. 2023;112(1):92–102.
Hao R, Wang B, Wang H, et al. lncRNA TUG1 promotes proliferation and differentiation of osteoblasts by regulating the miR-545-3p/CNR2 axis. Braz J Med Biol Res. 2020;53(11): e9798.
Prall WC, Haasters F, Heggebö J, et al. Mesenchymal stem cells from osteoporotic patients feature impaired signal transduction but sustained osteoinduction in response to BMP-2 stimulation. Biochem Biophys Res Commun. 2013;440(4):617–22.
Lienau J, Schmidt-Bleek K, Peters A, et al. Insight into the molecular pathophysiology of delayed bone healing in a sheep model. Tissue Eng Part A. 2010;16(1):191–9.
Sheu TJ, Zhou W, Fan J, et al. Decreased BMP2 signal in GIT1 knockout mice slows bone healing. Mol Cell Biochem. 2014;397(1–2):67–74.
Chen J, Yang Y. LncRNA HAGLR absorbing miR-214-3p promotes BMP2 expression and improves tibial fractures. Am J Transl Res. 2021;13(10):11065–80.
Acknowledgements
Not applicable.
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
T. Z and J.L. M designed this study. R.J. K, L.N. H, and D.J. J conducted the experiment and analyzed the data. R.J. K, L.N. H, T. Z, J.L. M and D.J. J wrote the manuscript. All authors revised the manuscript. All authors reviewed and approved for publication.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by The Ethics Committee of the First People’s Hospital of Jingzhou and followed the principles outlined in the Declaration of Helsinki. In addition, informed consent has been obtained from the participants involved.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kang, R., Huang, L., Zeng, T. et al. Long non-coding TRPM2-AS regulates fracture healing by targeting miR-545-3p/Bmp2. J Orthop Surg Res 19, 466 (2024). https://doi.org/10.1186/s13018-024-04969-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-024-04969-5