Abstract
Introduction and objective
Developmental dysplasia of the hip (DDH) is a musculoskeletal disorder. Genetic and epigenetic changes in C-X3-C motif chemokine receptor 1 (CX3CR1) may lead to disturbance in chondrocyte development and change the labrum dimensions, which indirectly result in hip joint instability. Considering the important role of this gene in cell migration, cell adhesion and bone and cartilage development, we aimed to evaluate the CX3CR1 gene methylation in DDH pathogenesis.
Methods
Our study comprised of forty-five DDH patients and forty-five healthy control subjects with healthy femoral neck cartilage. The healthy controls had total or hemiarthroplasty for the femoral neck fracture. Samples were collected from the femoral head (cartilage) of DDH patients and healthy controls. Genomic DNA was obtained from the samples, and DNA methylation of CX3CR1 gene was analyzed via metabisulfite method.
Results
Methylation analysis reveals no significant differences in promoter of CX3CR1 gene in cartilage samples from DDH patients and healthy control subjects (P = 0.33).
Conclusion
Methylation status of CX3CR1 gene showed no significant difference between the patient and control groups. Our results indicate that DNA methylation may not modulate this gene in this disease and other epigenetic mechanisms such as non-coding RNAs and histone modifications could be implicated.
Similar content being viewed by others
Introduction
Developmental dysplasia of the hip (DDH) is one of the musculoskeletal disorder that is common in infancy and shows clinical manifestations ranging from clicky or stable hips with ultrasound or radiological evidence of acetabular dysplasia to dislocated, subluxatable or dislocatable hips [1, 2]. The normal development of the hip is dependent on a normal development of both the femoral head and the acetabulum. The femoral head must be firm and stable in the hip socket to form spherically and concentrically. In a situation with loose interaction between the acetabulum and the femoral head or with deficiency of either component, the outcome would be lack of sphericity and incongruence for hip joint [3]. In DDH, hips could be dislocated, subluxated, malformed or unstable. Dislocation includes displacement of the femoral head from the acetabulum completely. Subluxation indicates dislocation between the femoral head and acetabulum with some contact. Malformation is any abnormality in the development of the acetabulum and/or femoral head and instability includes inability of the hip to resist an externally applied force without developing a dislocation or subluxation [1]. In cases with mild dysplasia, the symptoms may not have presented clinically at all or may present itself at adult life but in cases with severe dysplasia the symptoms are present clinically in early childhood or infancy [4]. The incidence of DDH is almost 1 in 1000 live births. Since there is a lack of uniformity for DDH definition, the true incidence is not known, but the incidence of entire spectrum of DDH is much higher [3, 5,6,7]. Actually, the DDH incidence is not constant and relays on many factors. These inconsistency comes from the variables such as DDH definition, differences in clinical methods and skills for detection, timing of evaluation and genetic differences [8, 9]. Furthermore, gender could play a part, since DDH is more common in girls than boys [8]. Generally, the risk factors for DDH include mechanical constriction of the fetus, abnormal position in the 3rd trimester, postnatal environment and genetic risk factors [10, 11]. The risk factors connected to intrauterine mechanical constraint are large birth weight breech presentation and oligohydramnios [8, 12, 13].
A risk factor that is recently reported for DDH development is epigenetics. Since the concordance rate in monozygotic twin for DDH development is approximately 40%, the remaining risk factors are environmental factors such as epigenetics. DDH could be considered as a multifactorial disease with genetic and non-genetic risk factors that are implicated in pathogenesis of DDH [14,15,16]. Epigenetic is a process which affects and regulates gene expression through different mechanisms such as non-coding RNAs, histone modifications and DNA methylation. DNA methylation is the most important epigenetic mechanisms which modulates gene expression through heterochromatin and euchromatin formation [17]. We previously reported that the growth/differentiation factor 5 (GDF5) gene is hypermethylated in DDH patients [18].
C-X3-C motif chemokine receptor 1 (CX3CR1) is a chemokine and transmembrane protein involved in the migration and adhesion of leukocytes. It is documented that this receptor has vital role in homeostasis of bone and cartilage. Genetic and epigenetic changes in CX3CR1 may lead to disturbance in development of chondrocyte and change the labrum dimensions which indirectly result in hip joint instability [19]. Furthermore, Hoshino and colleagues have revealed that CX3CR1-CX3CL1 interactions play critical roles differentiation of osteoclasts and osteoblasts [20]. Another study by Djouad and colleagues illustrated that human mesenchymal stem cells are expressing high level of CX3CR1 in comparison to chondrocytes [21]. These results suggest that the mesenchymal cells require chemokine receptors for receiving signals and migration and these receptors may implicate in the developing structure of the cartilage anlage of the acetabulum.
We aimed to evaluate the methylation status of the CX3CR1 gene in DDH pathogenesis. According to literature review, this is the first study that provides insight about methylation pattern of CX3CR1 gene in DDH pathogenesis.
Material and methods
DDH patients and healthy controls
DDH patients were consecutively recruited between 2017 and 2018 from the orthopedics clinic of Imam Khomeini Hospital of Tehran University of Medical Sciences. The healthy subjects are participated in the study at the same time. Diseases such as osteoarthritis, metabolic bone disease and DDH were excluded from the control group. Our study comprises of 90 individuals (45 healthy controls and 45 DDH patients), and the mean age ± SD of controls and patients was 42 ± 15.2 and 45 ± 12.6, respectively. The male-to-female ratio was equal in patient and control groups (5 males and 40 females). The control individuals had neither family history nor clinical evidence of arthritis or any inflammatory disorders. The control samples were taken from people who were under a hemi- or total arthroplasty operation because of femoral head fracture. These cartilage samples are actually obtained from individuals with healthy femoral head. The Human Research Ethics Committee of Tehran University of Medical Sciences approved this study (IR.TUMS.IKHC.REC.1397.179). Informed consent was obtained from all control and patient subjects.
Sample collection
Samples were collected from the femoral head cartilage of healthy control subjects and DDH patients. QIAamp DNA Mini Kit (Qiagen) was used to extract genomic DNA from the samples according to the manufacturer’s instruction. DNA from all the samples extracted and stored at − 20 °C. Purity and yield of DNA were determined using a NanoDrop spectrophotometer at 260 nm and 280 nm (NanoDrop 2000c Spectrophotometer, Thermo Fisher Scientific, Wilmington, DE, USA).
Bisulfite treatment
The extracted DNA was treated with bisulfite (EpiTect Plus DNA Bisulfite Kit). The principle of this method of treatment is that the methylated cytosines will remain unchanged, while the unmethylated cytosine will be converted to uracil. According to bisulfite treatment kit, the following notes were applied before starting.
30 ml of 96% ethanol was added to Buffer BW and stored at room temperature (15–25 °C). 27 ml of 96% ethanol is added to buffer BD and stored at 2–8 °C. 310 μl of RNase-free water is added to carrier RNA and stored at − 20 °C.
EpiTect Plus DNA Bisulfite Kit
800 μl of RNase-free water was added to each aliquot of Bisulfite Mix and then vortexed to completely dissolve the Bisulfite Mix (several minutes). 200 μl PCR tubes were used for bisulfite reactions based on the protocol. The tubes were closed and mixed for several minutes. The blue color of the DNA Protect Buffer is considered as a correct PH and sufficient mixture. Thermal cycler program was set up, the tubes were located in the thermal cycler, and the incubation time for each section was obtained from the protocol.
After PCR, the tubes were centrifuged and the reactions were transferred into microcentrifuge clean tubes (1.5 ml), and then, 310 μl of Buffer BL was added into each tube according to the protocol, mixed and centrifuged shortly. Afterward, 250 μl of 96% ethanol was added into each tube, mixed, vortexed and centrifuged.
Then, the reactions from tubes were transferred into spin columns which were located on collection tubes and centrifuged for 60 s. The flow-through was discarded from collection tubes, and spine tubes were placed on collection tubes again.
The Buffer BW (500 μl) was added into each spine column, and the tubes were centrifuged for 60 s. Again, the flow-through was discarded from collection tubes and spine tubes were placed into collection tubes.
The Buffer BD (500 μl) was added into each spin column and incubated at room temperature for 15 min and then centrifuged for 60 s. Afterward, the flow-through was discarded from collection tubes and spine tubes were placed into collection tubes.
The Buffer BW (500 μl) was added into each spin column and centrifuged for 60 s. Then, the flow-through was discarded from collection tubes and spine tubes were placed into collection tubes. (This step was repeated again.)
The 96% ethanol (250 μl) was added into each spin column and centrifuged for 60 s. Then, spin columns were placed into collection tubes (2 ml) and centrifuged for 60 s. In order to evaporate the liquid, the tubes were incubated on a heating block for 5 min at 60 °C.
The spin columns were placed into microcentrifuge clean tubes (1.5 ml). Then, the Buffer EB (15 μl) was added into each spin column, incubated at room temperature for 60 s. To elute the DNA, tubes were centrifuged. Finally, the purified DNA was stored for up to 24 h at 2–8 °C. In order to use samples for more works and further evaluation, DNA should be stored at − 20 °C.
PCR amplification
A 221 base-pair segment of DNA at 3p22 band was target for PCR amplification. The segment was part of the promoter region of CX3CR1 gene. The DNA sequence of CX3CR1 promoter was obtained from the UCSC (University of California, Santa Cruz) website (https://genome.ucsc.edu/). The bisulfite specific primers for PCR were designed using MethPrimer website (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi).
Each PCR tube contained DNA Protect Buffer (35 μl), bisulfite mix (85 μl), RNase-free water (variable) and bisulfite-treated DNA (maximum 20 μl). For samples with high concentration (ranged from 1 ng to 2 μg), the final volume of RNase-free water and DNA solution was 20 μl. The PCR conditions were as follows: denaturation (95 °C for 5 min), incubation (60 °C for 25 min), denaturation (95 °C for 5 min), incubation (60 °C for 85 min), denaturation (95 °C for 5 min) and incubation (60 °C for 175 min). For amplification validation, all DNA samples were gel electrophoresed after PCR amplification with 2% agarose gel. After confirmation with electrophoresis, the samples were sequenced (Macrogen, Seoul, Korea) and evaluated using Codon Code Aligner version 2 (Codon Code Corporation, Dedham, MA, USA) software.
Statistical analysis
Our data were analyzed with SPSS version 22.0, and graph was generated using GraphPad Prism version 5 for windows (GraphPad Software, La Jolla, CA USA, www. graphpad.com). All data are shown as mean ± standard deviation (S.D.) and also analyzed for normal distribution using the Kolmogorov–Smirnov test. Mann–Whitney test and independent sample t test were applied. Value less than 0.05 was considered statistically significant.
Results
Methylation status of CX3CR1
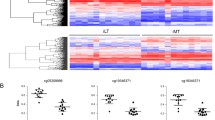
Methylation level of CX3CR1 gene in healthy control subjects and DDH patients is illustrated in Table 1 and Fig. 1. Methylation analysis reveals no significant differences in promoter of CX3CR1 gene between healthy control subjects and DDH patients (P = 0.33, Fig. 1, Table 1).
Discussion
DDH is a multifactorial disease that genetic factors and non-genetic factors such as epigenetics are implicated in its etiopathogenesis [3]. Monozygotic and dizygotic twin studies demonstrated that genetic factors are involved in the disease, but also indicated that there are others susceptible factors such as epigenetic and environmental factors [22, 23]. Actually, DDH is a complex disorder and genetic and epigenetic factors are contributed to its pathogenesis. Recently, researchers are interested toward the interaction and collaboration between environmental and genetic factors which finally modify the epigenome [24, 25]. In order to determine the DDH etiology, epigenetic mechanisms such as DNA methylation have a unique importance.
According to our knowledge, we could not find any study evaluating the methylation status of CX3CR1 gene in DDH pathogenesis, while there are some studies showing that the CX3CR1 could be considered as a genetic susceptible risk factor for DDH [26,27,28,29]. On the other hand, there are studies demonstrating that the CX3CR1 gene could be regulated through DNA methylation in human glioma tumors [30], myeloid cells [31] and CD8+ T cells [32].
These studies propose that the CX3CR1 gene could be a susceptible risk factor for DDH and also could be regulated through DNA methylation. Nonetheless, there is no report of evaluating methylation status of CX3CR1 in DDH. Our study showed no significant difference in methylation level of CX3CR1 gene between DDH patients and healthy controls. Our results indicate that this gene may not be regulated through DNA methylation mechanism in this disease and could be regulated via other epigenetic mechanisms such as microRNAs and histone modifications.
There are several issues which need to be addressed. In case of DNA methylation, it is not completely clear that DNA methylation abnormalities, which are reported through different studies [25, 33,34,35,36,37], are actually consequence of the disease or could be considered as a disease causes. Since the DNA methylation could be affected by disease stage and chronic inflammatory situation of disease, it seems that these abnormalities are a disease consequence rather than the disease cause. In order to understand the role of DNA methylation in disease pathogenesis, we have to compare the methylation status of a gene or genome at the early and late stage of diseases.
In summary, epigenetic modifications such as non-coding microRNAs, histone modifications, and DNA methylation could be considered as a diagnostic and prognostic marker in DDH. These alterations could be a promising therapeutic approach in near future for DDH patients. Fortunately, our understanding about epigenetic changes in DDH has been improved and gives us a chance to prevent or control the disease. Although methylation may not participate in modulating CX3CR1 gene in DDH pathogenesis, role of other epigenetic mechanisms should be explored.
Availability of data and materials
Data are available upon request.
References
Aronsson DD, Goldberg MJ, Kling TF, Roy DR. Developmental dysplasia of the hip. Pediatrics. 1994;94(2):201–8.
Cole W. Screening for congenital dislocation of the hip in Australia. J Paediatr Child Health. 1991;27(3):143–6.
Shaw BA, Segal LS. Evaluation and referral for developmental dysplasia of the hip in infants. Pediatrics. 2016;138(6):e20163107.
David T, Poynor M, Simm S, Parris MR, Hawnaur J, Rigg E, Mccrae F. Reasons for late detection of hip dislocation in childhood. The Lancet. 1983;322(8342):147–9.
Tredwell SJ. Neonatal screening for hip joint instability. Its clinical and economic relevance. Clin Orthop Relat Res 1992(281):63–68.
Gross R, Wisnefske M, Hitch M. The Otto Aufranc Award Paper. Infant hip screening. The Hip 1982:50–67.
Barlow T. Early diagnosis and treatment of congenital dislocation of the hip. J Bone Jt Surg Br. 1962;44(2):292–301.
Chan A, McCaul KA, Cundy PJ, Haan EA, Byron-Scott R. Perinatal risk factors for developmental dysplasia of the hip. Arch Dis Child Fetal Neonatal Ed. 1997;76(2):F94–100.
Bialik V, Bialik GM, Blazer S, Sujov P, Wiener F, Berant M. Developmental dysplasia of the hip: a new approach to incidence. Pediatrics. 1999;103(1):93–9.
Barr L, Rehm A. Should all twins and multiple births undergo ultrasound examination for developmental dysplasia of the hip? A retrospective study of 990 multiple births. Bone Jt J. 2013;95(1):132–4.
Mulpuri K, Song KM, Goldberg MJ, Sevarino K. Detection and nonoperative management of pediatric developmental dysplasia of the hip in infants up to six months of age. JAAOS J Am Acad Orthop Surg. 2015;23(3):202–5.
Wald N, Terzian E, Vickers P, Weatherall JC. Congenital talipes and hip malformation in relation to amniocentesis: a case-control study. The Lancet. 1983;322(8344):246–9.
Lapunzina P, Camelo JSL, Rittler M, Castilla EE. Risks of congenital anomalies in large for gestational age infants. J Pediatr. 2002;140(2):200–4.
Ghosh S, Fryer AA, Hoban PR, Wynn-Jones C, Maffulli N: Fibrillin 1 gene with R2726W mutation is absent in patients with primary protrusio acetabuli and developmental dysplasia of the hip. Med Sci Monit Int Med J Exp Clin Res 2009; 15(5):CR199–202.
Kapoor B, Dunlop C, Wynn-Jones C, Fryer AA, Strange RC, Maffulli N. Vitamin D and oestrogen receptor polymorphisms in developmental dysplasia of the hip and primary protrusio acetabuli–a preliminary study. J Negat Results Biomed. 2007;6(1):1–5.
Aicale R, Tarantino D, Maccauro G, Peretti G, Maffulli N: Genetics in orthopaedic practice. 2019.
Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484.
Baghdadi T, Nejadhosseinian M, Shirkoohi R, Mostafavi RT, Tamehri SS, Saffari M, Mortazavi SJ: DNA hypermethylation of GDF5 in developmental dysplasia of the hip (DDH). Mol Genet Genom Med 2019:e887–e887.
Feldman GJ, Parvizi J, Sawan H, Erickson JA, Peters CL. Linkage mapping and whole exome sequencing identify a shared variant in CX3CR1 in a large multi-generation family. J Arthroplasty. 2014;29(9):238–41.
Hoshino A, Ueha S, Hanada S, Imai T, Ito M, Yamamoto K, Matsushima K, Yamaguchi A, Iimura T. Roles of chemokine receptor CX3CR1 in maintaining murine bone homeostasis through the regulation of both osteoblasts and osteoclasts. J Cell Sci. 2013;126(4):1032–45.
Djouad F, Delorme B, Maurice M, Bony C, Apparailly F, Louis-Plence P, Canovas F, Charbord P, Noël D, Jorgensen C. Microenvironmental changes during differentiation of mesenchymal stem cells towards chondrocytes. Arthritis Res Ther. 2007;9(2):R33.
Stevenson DA, Mineau G, Kerber RA, Viskochil DH, Schaefer C, Roach JW. Familial predisposition to developmental dysplasia of the hip. J Pediatr Orthop. 2009;29(5):463–6.
Weinstein SL. Natural history of congenital hip dislocation (CDH) and hip dysplasia. Clin Orthop Relat Res. 1987;225:62–76.
Quaden DH, De Winter LM, Somers V. Detection of novel diagnostic antibodies in ankylosing spondylitis: an overview. Autoimmun Rev. 2016;15(8):820–32.
Mahmoudi M, Aslani S, Nicknam MH, Karami J, Jamshidi AR. New insights toward the pathogenesis of ankylosing spondylitis; genetic variations and epigenetic modifications. Mod Rheumatol. 2017;27(2):198–209.
Feldman G, Offemaria A, Sawan H, Parvizi J, Freeman TA. A murine model for developmental dysplasia of the hip: ablation of CX3CR1 affects acetabular morphology and gait. J Transl Med. 2017;15(1):233.
Basit S, Alharby E, Albalawi AM, Khoshhal KI: Whole genome SNP genotyping in a family segregating developmental dysplasia of the hip detected runs of homozygosity on chromosomes 15q13.3 and 19p13.2. Congn Anomal 2018;58(2):56–61.
Feldman GJ, Parvizi J, Sawan H, Erickson JA, Peters CL. Linkage mapping and whole exome sequencing identify a shared variant in CX3CR1 in a large multi-generation family. J Arthroplasty. 2014;29(9 Suppl):238–41.
Feldman GJ, Parvizi J, Levenstien M, Scott K, Erickson JA, Fortina P, Devoto M, Peters CL. Developmental dysplasia of the hip: linkage mapping and whole exome sequencing identify a shared variant in CX3CR1 in all affected members of a large multigeneration family. J Bone Miner Res. 2013;28(12):2540–9.
Locatelli M, Boiocchi L, Ferrero S, Martinelli Boneschi F, Zavanone M, Pesce S, Allavena P, Maria Gaini S, Bello L, Mantovani A. Human glioma tumors express high levels of the chemokine receptor CX3CR1. Eur Cytokine Netw. 2010;21(1):27–33.
Vento-Tormo R, Alvarez-Errico D, Rodriguez-Ubreva J, Ballestar E. Gains of DNA methylation in myeloid terminal differentiation are dispensable for gene silencing but influence the differentiated phenotype. FEBS J. 2015;282(9):1815–25.
Shin MS, You S, Kang Y, Lee N, Yoo SA, Park K, Kang KS, Kim SH, Mohanty S, Shaw AC et al: DNA methylation regulates the differential expression of CX3CR1 on human IL-7Ralphalow and IL-7Ralphahigh effector memory CD8+ T cells with distinct migratory capacities to the fractalkine. J Immunol (Baltimore, Md : 1950) 2015;195(6):2861–2869.
Aslani S, Mahmoudi M, Karami J, Jamshidi AR, Malekshahi Z, Nicknam MH. Epigenetic alterations underlying autoimmune diseases. Autoimmunity. 2016;49(2):69–83.
Aslani S, Mahmoudi M, Garshasbi M, Jamshidi AR, Karami J, Nicknam MH. Evaluation of DNMT1 gene expression profile and methylation of its promoter region in patients with ankylosing spondylitis. Clin Rheumatol. 2016;35(11):2723–31.
Karami J, Mahmoudi M, Amirzargar A, Gharshasbi M, Jamshidi A, Aslani S, Nicknam M. Promoter hypermethylation of BCL11B gene correlates with downregulation of gene transcription in ankylosing spondylitis patients. Genes Immun. 2017;18(3):170.
Foma AM, Aslani S, Karami J, Jamshidi A, Mahmoudi M. Epigenetic involvement in etiopathogenesis and implications in treatment of systemic lupus erythematous. Inflamm Res. 2017;66(12):1057–73.
Aslani S, Jafari N, Javan MR, Karami J, Ahmadi M, Jafarnejad M. Epigenetic modifications and therapy in multiple sclerosis. NeuroMol Med. 2017;19(1):11–23.
Acknowledgements
We are extremely grateful to those who participated in our study.
Funding
The work was supported by a grant from Deputy of Research, Tehran University of Medical Sciences. The funder of the study had no role in study design, data collection, data analysis, data interpretation or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Author information
Authors and Affiliations
Contributions
MN, HH and RS contributed to acquisition of data, interpretation of data, drafting the article and final approval of the article. JK and SMJM were involved in the conception and design of the study, analysis and interpretation of data, revising the article critically for important intellectual content and final approval of the article. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was performed based on the Declaration of Helsinki guidelines and was approved by the ethics committee at the Tehran University of Medical Sciences (Approval ID: IR.TUMS.IKHC.REC.1397.179).
Consent for publication
The written informed consent was signed by all patients before enrolling in the study.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Nejadhosseinian, M., Haerian, H., Shirkoohi, R. et al. Evaluation of CX3CR1 gene DNA methylation in developmental dysplasia of the hip (DDH). J Orthop Surg Res 17, 436 (2022). https://doi.org/10.1186/s13018-022-03324-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-022-03324-w