Abstract
Background
Also known as clubfoot, idiopathic congenital talipes equinovarus (ICTEV) is the most common pediatric deformity and occurs in 1 in every 1000 live births. Even though it has been widely researched, the etiology of ICTEV remains poorly understood and is often described as being based on a multifactorial genesis. Genetic and environmental factors seem to have a major role in the development of this disease. Thus, the aim of this review is to analyze the available literature to document the current evidence on ICTEV etiology.
Methods
The literature on ICTEV etiology was systematically reviewed using the following inclusion criteria: studies of any level of evidence, reporting clinical or preclinical results, published in the last 20 years (1998–2018), and dealing with the etiology of ICTEV.
Results
A total of 48 articles were included. ICTEV etiology is still controversial. Several hypotheses have been researched, but none of them are decisive. Emerging evidence suggests a role of several pathways and gene families associated with limb development (HOX family; PITX1-TBX4), the apoptotic pathway (caspases), and muscle contractile protein (troponin and tropomyosin), but a major candidate gene has still not been identified. Strong recent evidence emerging from twin studies confirmed major roles of genetics and the environment in the disease pathogenesis.
Conclusions
The available literature on the etiology of ICTEV presents major limitations in terms of great heterogeneity and a lack of high-profile studies. Although many studies focus on the genetic background of the disease, there is lack of consensus on one or multiple targets. Genetics and smoking seem to be strongly associated with ICTEV etiology, but more studies are needed to understand the complex and multifactorial genesis of this common congenital lower-limb disease.
Similar content being viewed by others
Background
Congenital talipes equinovarus (CTEV) is a foot deformity characterized by hindfoot varus, forefoot (metatarsus) adductus, an augmented midfoot arch (cavus), and equinus. This pediatric malformation can be classified according to its clinical presentation. It can be secondary or syndromic when its presentation is associated with another congenital disease (20% of cases). However, it may also occur as an isolated birth defect with no other malformations (80% of cases), which introduces the concept of idiopathic CTEV (ICTEV). The etiology of CTEV is largely unknown. Secondary CTEV is usually a manifestation of distal arthrogryposis (DA), congenital myotonic dystrophy, myelomeningocele, or other congenital diseases. While the clinical presentation may be similar to the idiopathic form, secondary CTEV seems to derive from neuromuscular [1] and fetal abnormalities [2] involved in its etiopathogenesis, thus making ICTEV and syndromic CTEV rather different in clinical presentation, treatment, and proposed etiopathogenetic mechanism [3, 4].
ICTEV is one of the most common pediatric deformities. The epidemiological studies published over the last 55 years suggest a birth prevalence in the range of 0.5 to 2.0 cases/1000 live births, which results in an estimated 7–43 cases of clubfoot/year/million population, depending mainly on the birth rate [5]. The higher prevalence seems to be associated with social-demographic, genetic, and environmental risk factors, which explain its prevalence among low- and middle-income countries [5] and closed societies like the Maori population [6]. It affects males more than females [7] with a male-to-female ratio of 2:1, which is similar across different ethnic groups [8,9,10,11]. Kruse et al. proposed a reason for the gender difference in 2008 [12], but the phenotypic variability in affected individuals is still unknown.
Several treatments have been proposed throughout the centuries, but today, the gold-standard treatment is the Ponseti method [13, 14]. In syndromic cases, current evidence supports the Ponseti method or other more invasive surgical procedures [15]. The aim of this review is to analyze the available literature to provide an update on the evidence related to ICTEV etiology.
Materials and methods
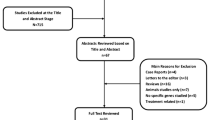
We conducted this systematic review according to the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [16]. Two medical electronic databases (PubMed and Science Direct) were searched by a single author (CE) on March 20, 2018. The research string used was “(clubfoot OR congenital talipes equinovarus OR clubfeet) AND (pathology OR embryology OR etiology OR etiopathogenesis OR genetics OR pathophysiology).” A total of n = 1590 articles were found. After excluding duplicates, n = 974 articles were selected.
The initial titles and abstracts were screened using the following inclusion criteria: studies of any level of evidence reporting clinical or preclinical results published in the last 20 years (1998–2018) and dealing with the etiology of ICTEV. Exclusion criteria were articles written in other languages or studies with a focus on secondary/syndromic CTEV, such as distal arthrogryposis, myelomeningocele, and Moebius syndrome. We also excluded all the remaining duplicates, articles dealing with other topics, with poor scientific methodology, or without an accessible abstract.
At the end of the first screening, we selected n = 76 articles that were eligible for full-text reading. After reading the full text, we ultimately selected n = 48 articles that satisfy the criteria. A PRISMA [16] flowchart of the selection and screening method is provided in Fig. 1. Reference lists from the selected papers were also screened.
Results
The included articles [3, 4, 12, 17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62] mainly focus on genetic research [17, 21,22,23, 25, 27, 29,30,31, 37,38,39,40,41,42,43,44,45,46,47,48,49,50, 52, 54,55,56, 58, 60, 61], epidemiological studies [12, 19, 20, 24, 32, 33, 35, 36, 53], MRI analysis [26, 51, 59], and histological histochemical analysis [18, 24, 28, 57] in ICTEV patients. Two previous reviews [3, 4, 62] reporting a significant analysis of the current evidence and research prospective were also included since they are part of the present evidence of the topic. The main findings of the included articles except for the two reviews were summarized (Table 1.)
ICTEV has historically been linked to several risk factors: oligohydramnios, smoking, parental age, parental education, parity, maternal anxiety or depression, alcohol use, and season of birth. A previous epidemiological study based on the difference in prevalence in different communities suggested that environmental factors have a role in pathogenesis. In 2014, a twin study done in Denmark surveyed 34,485 twins and found evidence of a role of environmental factors. The authors concluded that the presence of a genetic role in the development of the disease was not enough to explain the results. Therefore, they reported strong evidence of the presence of environmental factors to explain the statistical analysis [32]. Another study from 2013 [20] was conducted in a rural area of Turkey and showed that parental consanguineous marriage was associated with a higher risk of ICTEV. Even though the sample investigated was small, this result may support an etiology based on multiple genes and environmental factors.
Even though the role of environmental factors has been confirmed by several studies, all the proposed factors except for smoking were not significantly associated with ICTEV, which was linked to DNA oxidative damage caused by tobacco smoking [33,34,35,36]. A meta-analysis examined 172 reports containing cumulative data for 173,687 individuals and 11,674,332 unaffected controls published from 1959 to 2010. The analysis looked at the effects of smoking during pregnancy and showed that 15,673 individuals with CTEV and maternal smoking had an OR of 1.28 (95% CI 1.10–1.47) [34].
Genes involved in the metabolism of smoke-derived products may also contribute to the development of birth defects. Therefore, N-acetylation genes including NAT1, NAT2, and other related genes were screened for association analysis. Polymorphisms in the NAT2 gene cause decreased acetylation activity and have been associated with TEV. This suggests a deficit in the biotransformation of aromatic amines and the accumulation of DNA adducts, leading to a potential toxic effect and the development of TEV [27].
Hecht et al. [27] examined the variants of the NAT2 gene in 56 ICTEV multiplex families, 57 trios with a positive family history, and 160 simplex individuals. They reported a slight decrease in the expected number of homozygotes for the normal NAT2 allele in the Hispanic simplex trios. Significantly, a slow NAT2 acetylator phenotype was detected among the ICTEV patients, suggesting that slow acetylation may be a risk factor for ICTEV.
Genetic factors
Genetics has a crucial role in the development of ICTEV, even though no major gene candidate has been identified [3]. There is evidence of a family history of TEV in 24–50% of cases [22]. Results from twin studies showed concordance in monozygotic twins (32%) compared to dizygotic twins (2.9%), and a frequency of recurrence in 10–20% of families supports a role for genes in ICTEV [19, 32, 63]. There is also reported a unique case of bilateral ICTEV in preterm triplets, which provides even further support for a genetic etiology [19]. Many different families of genes were identified to play a role in the disease and a prospective role in the development of personalized conservative and surgical approaches [64]. Several families of genes and pathways were identified and investigated using mainly the candidate gene approach.
Homeobox family genes
The homeobox genes represent a family of transcription factors that play a central role in the morphogenesis processes of embryonic development. In particular, this family determines the correct genesis of the axial skeleton and limbs, which is why they were proposed as candidate genes for ICTEV pathogenesis [65]. Several candidate gene studies found a locus of genetic susceptibility associated with ICTEV in the HOX domain and the caspase domain [25, 37].
In 2009 and 2016, two large studies showed that ICTEV was associated with alteration in the regulator domain of HoxA and HoxD [38, 39]. Higher activity of the promotor was also reported as a result of promotor variation [40]. Based on the emerging evidence, we can assume that perturbation to the HOXA, HOXC, and HOXD clusters of genes may play a role in the etiology and pathogenesis of ICTEV [66].
Caspases pathway genes
Cysteine-dependent aspartate-directed proteases (caspases) are part of a family of cysteine proteases that play essential roles in apoptosis, necrosis, and inflammation processes. This family was investigated since caspase activity seems to be related to correct limb development, and related genes were first associated with ICTEV in 2005 by Heck et al. [50].
A CASP10 gene variant was found in simplex ICTEV in white and Hispanic trios. In 2007, Ester et al. [58] researched other alterations in three caspase genes. They genotyped SNPs of three different genes (Casp8, Casp10, and CFLAR) to investigate their association with ICTEV. One SNP in each of the genes was associated with the disease. Several haplotypes constructed from these SNPs displayed altered transmission, suggesting that genetic variation in apoptotic genes may play a role in the development of ICTEV.
Collagen family genes
The collagen family genes were also linked to ICTEV. The focus of related genetic research has been on the COL9A1 and COL1A1 genes. COL9A1 encodes for one of the three alpha chains of type IX collagen, a component of the hyaline cartilage, while COL1A1 encodes for pro-alpha 1 chains of type I collagen, a component of most connective tissue that is abundant in bone and tendons. In 2008, COL1A1 was investigated in healthy and ICTEV patients. The study reported a higher expression of COL1A1 in patients with ICTEV than in healthy patients. A − 161(T → C) heterozygous mutation and a + 274(C → G) homozygous mutation were also identified in the COL1A1 gene in patients with ICTEV, suggesting that COL1A1 variants could be linked to the onset of ICTEV [37].
Based on previous studies, Wang et al. [30] investigated genes that regulate COL91A1 expression (SOX9) in 2012. They reported no mutations of the gene but a higher expression of SOX9 in the muscular cells of ICTEV patients. COL9A1 polymorphism seems to modulate the gene expression and influence the protein function. Three studies reported a role of these polymorphisms in ICTEV in the populations examined [41, 42, 67].
GLI3 gene
The GLI3 gene encodes for a C2H2-type zinc finger protein of the GLI family. In 2005, a study showed how a mutation of this gene was associated with the occurrence of ICTEV [43]. In 2009, another study [31] reported how HoxD13 directly interacts with the promoter of GLI3. They observed that GLI3 mRNA and protein expression levels were increased in ICTEV-model rats. This may mean that HOXD13 is a transcription factor of GLI3. Low expression of HOXD13 might lead to increased GLI3 expression level during limb formation, which likely plays a key role in ICTEV pathogenesis.
T-box family
The T-box family comprises transcription factors that play a crucial role in embryogenesis and morphogenesis. Like other genes with a similar role, they are candidates for possible genetics inducers of ICTEV. TBX3 and TBX4 are the main family members studied. The TBX3 protein is a transcriptional factor of the T-box family. A 2014 study reported that mutations in this gene affect limb development were proven to have transmission disequilibrium in ICTEV patients, suggesting susceptibility to ICTEV [65].
PITX1-TBX4 pathway
TBX4 protein is a transcriptional factor that is mainly expressed in the hindlimb and is thus associated with ICTEV pathogenesis [68]. It was further studied in association with another transcriptional factor, PITX1, which is part of the same pathway. The PITX1-TBX4 pathway is responsible for early limb development. Numerous studies report that mutations in the genes encoding the transcription factors PITX1 and TBX4 lead to a reduction in lower-limb musculature and classic clubfoot phenotypes in both humans and mice [17, 44,45,46]. Studies support a role of the PITX1-TBX4 developmental pathway in TEV etiology.
Gurnett et al. [17] researched these pathway alterations in a five-generation family with asymmetric ICTEV segregating as an autosomal dominant condition. A single missense mutation (E130K) located in a highly conserved domain of the PITX1 gene has been identified. Another study showed that PITX1 downregulation causes a clubfoot-like phenotype in mice, thus providing evidence of the involvement of PITX in ICTEV pathogenesis [46].
TBX4 microdeletions and microduplications have been reported in patients affected by ICTEV, suggesting that chromosome 17q23.1q23.2 microduplication is a relatively common cause of familial isolated clubfoot [47]. However, in 2012, Lu et al. [44] examined the possible correlation between the hindfoot-specific gene TBX4 and ICTEV. They concluded that the microduplication is a rare cause of familial isolated clubfoot and can be segregated as an autosomal dominant phenotype. Significant variations were not present in the two known TBX4 hindlimb enhancers sequenced in 95 patients from simplex families.
A recent study conducted in 2017 reported that the PITX1-TBX4 pathway can be associated with HOXC alteration in vertical talus. They identified a HOXC13 deletion that segregated with clubfoot in a three-generation family [21]. Deletions of part of the HOXC gene cluster were later identified in two of five families with autosomal dominant isolated congenital vertical talus, suggesting that it is a possible cause of familial vertical talus [39]. Interestingly, HOXD10 mutations were previously identified in two families with vertical talus [48, 49], which strongly supports a role of homeobox gene mutations in the etiology of isolated vertical talus. However, because mutations in the PITX1-TBX4-HOXC pathway are infrequent in patients with clubfoot, other genetic mechanisms remain to be discovered and investigated [64].
Troponin and tropomyosin genes
The troponin (Tn) family is a protein complex involved in striated muscle contraction and has three subunits: Tn-I, Tn-T, and Tn-C. The Tn-I subunit inhibits actomyosin ATPase, while the Tn-T subunit binds tropomyosin and Tn-C. The Tn-C subunit binds calcium and overcomes the inhibitory action of the troponin complex on actin filaments.
A 2011 study analyzed 15 genes encoding proteins that control myofiber contractility in a cohort of both non-Hispanic white (NHW) and Hispanic families. They reported an association between ICTEV patients and multiple SNPs of two genes regulating troponin activity, TNNC2 and TPM1, suggesting a possible role in the etiology [56].
TPM1 is a member of the tropomyosin family, which comprises actin-binding proteins involved in the contraction of both striated and smooth muscles and the cytoskeleton of non-muscular cells. The associations of multiple SNPs in the TPM1 gene with ICTEV suggest a potential role of genes that encode contractile proteins of skeletal myofibers in the etiology of ICTEV [23]. ICTEV patients present a clinically evident alteration of the calf muscle at birth, which usually resituates after treatment [26, 69, 70]. This suggests the involvement of genes that play a role in muscle morphogenesis.
Distal arthrogryposis is a cause of syndromic TEV that is characterized by variations in genes that encode for components of the muscle contractile complex (MYH3, TPM2, TNNT3, TNNI2, and MYH8), resulting in muscle contractures. The similar phenotype suggests that these genes could be candidate genes. However, one study found that the development of the disease was different in ICTEV and in DA, even though it suggested a potential role of many regulatory candidate genes that could cause developmental defects in the hypaxial musculature that is invariably observed in clubfoot [24].
In contrast to other studies, Gurnett et al. [29] investigated 39 patients in 2009 to find mutations in the TNNT3, MYH3, and TPM2 genes in patients with ICTEV. The results showed an absence of correlation of these mutations in ICTEV patients. Recent evidence showed an absence of significant histological and cytological alteration of muscles after treatment [24]. Another work proposed an innovative 3D RM study of the muscle morphology to show how intramuscular fat distribution plays an important role in the morphology of the leg [51]. The potential of using MRI has also been suggested to better understand the clinical severity of an affected patient [71].
CAND2 and Wnt7a
In 2009, a study investigated two candidate genes, CAND2 and Wnt7a, and tested their role in the pathogenesis of ICTEV. They genotyped the CAND2 gene in 256 clubfoot patients and 75 control patients, while Wnt7a was screened using 56 clubfoot patients and 50 control patients. The study reported a polymorphism in each gene. However, the association results indicated that CAND2 and WNT7a are not major genes involved in the etiology of ICTEV [22].
In 2009, Poon et al. [18] showed that foot tissues were related to higher beta catenin levels. This was probably related to the Wnt signaling pathway and the synthesis of type III collagen. In particular, a higher amount of type III collagen was reported in studies analyzing the extracellular matrix of ICTEV tissues [28, 57]. More research is needed to understand the interactions of these growth factors with other proteins and their role in ICTEV etiology.
Dysplasia sulfate transporter gene
The dysplasia sulfate transporter (DTDST) gene was suggested to cause ICTEV and investigated by Bonafé et al. [61]. They tested whether R279 W mutations are responsible for the occurrence, but alterations in the coding region were not identified in 10 probands with ICTEV and a positive family history. The authors concluded that the R279 W mutation is no more frequent in this population of ICTEV probands than in controls.
Methylenetetrahydrofolate reductase gene
In 2006, Sharp et al. found that children who carry the 677T variant of the methylenetetrahydrofolate reductase gene (MTHFR) have a lower risk of ICTEV [60]. Another study later used whole genome sequencing to investigate the variants of MTHFR and the annexin A3 gene (ANXA3). They reported an MTHFR variant that is different from the variant associated with clubfoot in the study by Sharp et al. [52]. Bioinformatic analysis showed that the protein-binding region could be altered by this mutation (a sequence shift: the wild type is 264, while the mutant type is 267). Despite sharing some similar symptoms, these findings imply that the variant was associated with another genetic disease and not ICTEV. Furthermore, specific CNV profiles were identified in association with the diseased samples, thus further demonstrating the complexity of this multigenerational disorder [52].
Discussion
The etiology of ICTEV remains unknown as stated in recent reviews [3, 4, 72]. Many theories have been developed, but no one has clarified the major roles in the pathogenesis of idiopathic clubfoot. Recent studies have focused on the interaction between genetics and environmental factors, showing a multifactorial identity of the disease. Today, this remains the most validated theory.
A recent paper [73] reported a genetic analysis on a spontaneous autosomal recessive mouse model of peroneal muscular atrophy (PMA). It was used to understand the underlying developmental causes of ICTEV. The PMA mutation was mapped, and several candidate genes were identified, of which LIMK1 was upregulated in mutant mice. Collison et al. also reported that in chickens, LIMK1 upregulation can cause sciatic nerve defects and a TEV phenotype [73]. Further studies should be conducted using these models.
The years of research using the candidate gene approach has provided us more knowledge on the possible pathways involved in ICTEV pathogenesis, but it has failed to find a major gene causing the disease. The literature illustrates the great heterogeneity of the genetic causes of ICTEV. The candidate approach has probably not recognized the real amount of various causative variants and has likely underestimated the phenotypical and genotypical variants. The reported studies were also done using also different technical approaches, such as genome-wide association analysis (GWAS), linkage analysis, the technique of copy number variation, and whole exome sequencing. These next-generation genetic analyses should lead future studies on ICTEV etiology. Collaborative multicenter studies involving large populations might be a necessary step to shed light on the etiology of this complex disease. ICTEV inheritance is most often considered complex, with more than 75% of all cases reporting no family history [17, 53]. Thus, a large-scale GWAS study might reveal interesting results.
The filamin B (FLNB) gene encodes a member of the filamin family. The encoded protein interacts with glycoprotein Ib alpha as part of the process of repairing vascular injuries. The platelet glycoprotein Ib complex includes glycoprotein Ib alpha and binds the actin cytoskeleton. In 2016, Yang et al. performed WES sequencing and Sanger sequencing to identify and validate disease-causing mutations in a three-generation pedigree and 53 sporadic patients with ICTEV, respectively. A c.4717G>T (p.D1573Y) mutation in the FLNB gene, which co-segregated with ICETV, was identified in the pedigree. Two additional novel missense mutations in the same gene, c.1897A>G (p.M633V) and c.2195A>G (p.Y732C), were identified in the 53 sporadic patients, thus providing evidence of the involvement of the FLNB gene in ICTEV [54].
In 2014, Zhang et al. performed a GWAS study of the DNA of 396 isolated clubfoot patients and 1000 controls of European descent. The DNA was genotyped for > 600,000 single nucleotide polymorphisms (SNPs) to identify novel genes for ICTEV. The variants selected were then replicated with an independent cohort of 370 isolated clubfoot cases and 363 controls of European descent. The study found a strong association with the disease for an intergenic SNP on chromosome 12q24.31 between NCOR2 and ZNF664 (rs7969148, OR = 0.58, p = 1.25 × 10–5), which was significant on replication (combined OR = 0.63, p = 1.90 × 10–7). However, additional suggestive SNPs (Hox Genes, PITX1, TBX4, FOXN3, SORCS1, and MMP7/TMEM123) in the identified pathways were not significant in the replication phase [55]. With the aid of a new animal model, next-generation studies may have the potential to identify genes underlying the phenotype and elucidate the inheritance pattern and penetrance of the disorder [3].
Conclusions
The available literature on the etiology of ICTEV presents major limitations in terms of great heterogeneity and lack of high-profile studies. Although many studies have focused on the genetic background of the disease, there is a lack of consensus on one or multiple targets. Recent evidence shows a major role of both genetic and environmental factors. Thus far, smoking is the major environmental factor supported by recent evidence. The etiology of ICTEV is probably multifactorial and associated with multiple gene alterations, and large multi-center studies are required to investigate them. Further large international collaborative studies using next-generation sequencing technology in ICTEV patients are strongly encouraged.
Abbreviations
- DTDST:
-
Dysplasia sulphate transporter
- FLNB:
-
Filamin B
- GWAS:
-
Genome wide association analysis
- ICTEV:
-
Idiopathic congenital talipes equinovarus
- MTHFR:
-
Methylenetetrahydrofolate reductase
- PMA:
-
Peroneal muscular atrophy
- PRISMA:
-
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
References
Lovell ME, Morcuende JA. Neuromuscular disease as the cause of late clubfoot relapses: report of 4 cases. Iowa Orthop J. 2007;27:82–4.
Hester TW, Parkinson LC, Robson J, Misra S, Sangha H, Martin JE. A hypothesis and model of reduced fetal movement as a common pathogenetic mechanism in clubfoot. Med Hypotheses. 2009;73:986–8.
Basit S, Khoshhal KI. Genetics of clubfoot; recent progress and future perspectives. Eur J Med Genet. 2018;61(2):107–13.
Yong BC, Xun FX, Zhao LJ, Deng HW, Xu HW. A systematic review of association studies of common variants associated with idiopathic congenital talipes equinovarus (ICTEV) in humans in the past 30 years. Springerplus. 2016;5(1):896.
Smythe T, Kuper H, Macleod D, Foster A, Lavy C. Birth prevalence of congenital talipes equinovarus in low- and middle-income countries: a systematic review and meta-analysis. Trop Med Int Heal. 2017;22(3):269–85.
Chapman C, Stott NS, Port RV, Nicol RO. Genetics of club foot in Maori and Pacific people. J Med Genet. 2000;37(9):680–3.
Zionts LE, Jew MH, Ebramzadeh E, Sangiorgio SN. The influence of sex and laterality on clubfoot severity. J Pediatr Orthop. 2017;37(2):e129–33.
Pavone V, Bianca S, Grosso G, Pavone P, Mistretta A, Longo MR, et al. Congenital talipes equinovarus: an epidemiological study in Sicily. Acta Orthop. 2012;83(3):294–8.
Wijayasinghe SR, Abeysekera WYM, Dharmaratne TSS. Descriptive epidemiology of congenital clubfoot deformity in Sri Lanka. J Coll Physicians Surg Pak. 2018;28(2):166–8.
McConnell L, Cosma D, Vasilescu D, Morcuende J. Descriptive epidemiology of clubfoot in Romania: a clinic-based study. Eur Rev Med Pharmacol Sci. 2016;20(2):220–4.
Palma M, Cook T, Segura J, Pecho A, Morcuende JA. Descriptive epidemiology of clubfoot in Peru: a clinic-based study. Iowa Orthop J. 2013;33:167–71.
Kruse LM, Dobbs MB, Gurnett CA. Polygenic threshold model with sex dimorphism in clubfoot inheritance: the Carter effect. J Bone Joint Surg Am. 2008;90(12):2688–94.
Shabtai L. Worldwide spread of the Ponseti method for clubfoot. World J Orthop. 2014;5(5):585.
Pavone V, Testa G, Costarella L, Pavone P, Sessa G. Congenital idiopathic talipes equinovarus: an evaluation in infants treated by the Ponseti method. Eur Rev Med Pharmacol Sci. 2013;17(19):2675–9.
Hosseinzadeh P, Kelly DM, Zionts LE. Management of the relapsed clubfoot following treatment using the Ponseti method. J Am Acad Orthop Surg. 2017;25(3):195–203.
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred Reporting Items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097.
Gurnett CA, Alaee F, Kruse LM, Desruisseau DM, Hecht JT, Wise CA, et al. Asymmetric lower-limb malformations in individuals with homeobox PITX1 gene mutation. Am J Hum Genet. 2008;83(5):616–22.
Poon R, Li C, Alman BA. Beta-catenin mediates soft tissue contracture in clubfoot. Clin Orthop Relat Res. 2009;467(5):1180–5.
Pagnotta G, Boccanera F, Rizzo G, Agostino R, Gougoulias N, Maffulli N. Bilateral clubfoot in three homozygous preterm triplets. J Foot Ankle Surg. 2011;50(6):718–20.
Sahin O, Yildirim C, Akgun RC, Haberal B, Yazici AC, Tuncay IC. Consanguineous marriage and increased risk of idiopathic congenital talipes equinovarus: a case-control study in a rural area. J Pediatr Orthop. 2013;33(3):333–8.
Alvarado DM, Buchan JG, Frick SL, Herzenberg JE, Dobbs MB, Gurnett CA. Copy number analysis of 413 isolated talipes equinovarus patients suggests role for transcriptional regulators of early limb development. Eur J Hum Genet. 2013;21(4):373–80.
Shyy W, Dietz F, Dobbs MB, Sheffield VC, Morcuende JA. Evaluation of CAND2 and WNT7a as candidate genes for congenital idiopathic clubfoot. Clin Orthop Relat Res. 2009;467(5):1201–5.
Shyy W, Wang K, Sheffield VC, Morcuende JA. Evaluation of embryonic and perinatal myosin gene mutations and the etiology of congenital idiopathic clubfoot. J Pediatr Orthop. 2010;30(3):231–4.
Herceg MB, Weiner DS, Agamanolis DP, Hawk D. Histologic and histochemical analysis of muscle specimens in idiopathic talipes equinovarus. J Pediatr Orthop. 2006;26(1):91–3.
Wang LL, Fu WN, Li-Ling J, Li ZG, Li LY, Sun KL. HOXD13 may play a role in idiopathic congenital clubfoot by regulating the expression of FHL1. Cytogenet Genome Res. 2008;121(3–4):189–95.
Ippolito E, De Maio F, Mancini F, Bellini D, Orefice A. Leg muscle atrophy in idiopathic congenital clubfoot: is it primitive or acquired? J Child Orthop. 2009;3(3):171–8.
Hecht JT, Ester A, Scott A, Wise CA, Iovannisci DM, Lammer EJ, et al. NAT2 variation and idiopathic talipes equinovarus (clubfoot). Am J Med Genet Part A. 2007;143A(19):2285–91.
Ošt'ádal M, Eckhardt A, Herget J, Mikšík I, Dungl P, Chomiak J, et al. Proteomic analysis of the extracellular matrix in idiopathic pes equinovarus. Mol Cell Biochem. 2015;501(1–2):133–9.
Gurnett CA, Alaee F, Desruisseau D, Boehm S, Dobbs MB. Skeletal muscle contractile gene (TNNT3, MYH3, TPM2) mutations not found in vertical talus or clubfoot. Clin Orthop Relat Res. 2009;467(5):1195–200.
Wang Z, Yan N, Liu L, Cao D, Gao M, Lin C, Jin C. SOX9 overexpression plays a potential role in idiopathic congenital talipes equinovarus. Mol Med Rep. 2013;7(3):821–5.
Cao D, Jin C, Ren M, Lin C, Zhang X, Zhao N. The expression of Gli3, regulated by HOXD13, may play a role in idiopathic congenital talipes equinovarus. BMC Musculoskelet Disord. 2009;10(1):142.
Engell V, Nielsen J, Damborg F, Kyvik KO, Thomsen K, Pedersen NW. Heritability of clubfoot: a twin study. J Child Orthop. 2014;8(1):37–41.
Parker SE, Mai CT, Strickland MJ, Olney RS, Rickard R, Marengo L, et al. Multistate study of the epidemiology of clubfoot. Birth Defects Res A Clin Mol Teratol. 2009;85(11):897–904.
Hackshaw A, Rodeck C, Boniface S. Maternal smoking in pregnancy and birth defects: a systematic review based on 173 687 malformed cases and 11.7 million controls. Hum Reprod Update. 2011;17(5):589–604.
Dickinson KC, Meyer RE, Kotch J. Maternal smoking and the risk for clubfoot in infants. Birth Defects Res A Clin Mol Teratol. 2008;82(2):86–91.
Honein MA, Paulozzi LJ, Moore CA. Family history, maternal smoking, and clubfoot: an indication of a gene-environment interaction. Am J Epidemiol. 2000;152(7):658–65.
Wang L, Jin C, Liu L, Zhang X, Ji S, Sun K. Analysis of association between 5’ HOXD gene and idiopathic congenital talipes equinovarus. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2005;22(6):653–6.
Ester AR, Weymouth KS, Burt A, Wise CA, Scott A, Gurnett CA. Altered transmission of HOX and apoptotic SNPs identify a potential common pathway for clubfoot. Am J Med Genet A. 2009;149A(12):2745–52.
Alvarado DM, McCall K, Hecht JT, Dobbs MB, Gurnett CA. Deletions of 5’ HOXC genes are associated with lower extremity malformations, including clubfoot and vertical talus. J Med Genet. 2016;53(4):250–5.
Weymouth KS, Blanton SH, Powell T, Patel CV, Savill SA, Hecht JT. Functional assessment of clubfoot associated HOXA9, TPM1, and TPM2 variants suggests a potential gene regulation mechanism. Clin Orthop Relat Res. 2016;474(7):1726–35.
Liu LY, Jin CL, Jiang L, Lin CK. Expression of COL9A1 gene and its polymorphism in children with idiopathic congenital talipes equinovarus. Zhongguo Dang Dai Er Ke Za Zhi. 2011;13(6):478–81.
Zhao XL, Wang YJ, Wu YL, Han WH. Role of COL9A1 genetic polymorphisms in development of congenital talipes equinovarus in a Chinese population. Genet Mol Res. 2016;15(4).
Zhang X, Jin CL, Liu LY, Zhao N, Zhang LJ, Ji SJ. Association and mutation analysis of GLI3 gene in idiopathic congenital talipes equinovarus. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23(5):551–4.
Lu W, Bacino CA, Richards BS, Alvarez C, VanderMeer JE, Vella M, et al. Studies of TBX4 and chromosome 17q23.1q23.2: an uncommon cause of nonsyndromic clubfoot. Am J Med Genet A. 2012;158(7):1620–7.
Peterson JF, Ghaloul-Gonzalez L, Madan-Khetarpal S, Hartman J, Surti U, Rajkovic A, et al. Familial microduplication of 17q23.1-q23.2 involving TBX4 is associated with congenital clubfoot and reduced penetrance in females. Am J Med Genet A. 2014;164(2):364–9.
Alvarado DM, McCall K, Aferol H, Silva MJ, Garbow JR, Spees WM, et al. Pitx1 haploinsufficiency causes clubfoot in humans and a clubfoot-like phenotype in mice. Hum Mol Genet. 2011;20(20):3943–52.
Alvarado DM, Aferol H, McCall K, Huang JB, Techy M, Buchan J, et al. Familial isolated clubfoot is associated with recurrent chromosome 17q23.1q23.2 microduplications containing TBX4. Am J Hum Genet. 2010;87:154–60.
Dobbs MB, Gurnett CA, Pierce B, Exner GU, Robarge J, Morcuende JA, et al. HOXD10 M319K mutation in a family with isolated congenital vertical talus. J Orthop Res. 2006;24(3):448–53.
Shrimpton AE, Levinsohn EM, Yozawitz JM, Packard DS Jr, Cady RB, Middleton FA, et al. A HOX gene mutation in a family with isolated congenital vertical talus and Charcot-Marie-Tooth disease. Am J Hum Genet. 2004;75(1):92–6.
Heck AL, Bray MS, Scott A, Blanton SH, Hecht JT. Variation in CASP10 gene is associated with idiopathic talipes equinovarus. J Pediatr Orthop. 2005;25(5):598–602.
Duce SL, D’Alessandro M, Du Y, Jagpal B, Gilbert FJ, Crichton L, et al. 3D MRI analysis of the lower legs of treated idiopathic congenital talipes equinovarus (clubfoot). PLoS One. 2013;8(1):e54100.
Zhang Z, Kong Z, Zhu M, Lu W, Ni L, Bai Y. Whole genome sequencing identifies ANXA3 and MTHFR mutations in a large family with an unknown equinus deformity associated genetic disorder. Mol Biol Rep. 2016;43(10):1147–55.
Lochmiller C, Johnston D, Scott A, Risman M, Hecht JT. Genetic epidemiology study of idiopathic talipes equinovarus. Am J Med Genet. 1998;79(2):90–6.
Yang H, Zheng Z, Cai H, Li H, Ye X, Zhang X. Three novel missense mutations in the filamin B gene are associated with isolated congenital talipes equinovarus. Hum Genet. 2016;135(10):1181–9.
Zhang TX, Haller G, Lin P, Alvarado DM, Hecht JT, Blanton SH, et al. Genome-wide association study identifies new disease loci for isolated clubfoot. J Med Genet. 2014;51(5):334–9.
Weymouth KS, Blanton SH, Bamshad MJ, Beck AE, Alvarez C, Richards S. Variants in genes that encode muscle contractile proteins influence risk for isolated clubfoot. Am J Med Genet A. 2011;155(9):2170–9.
Gilbert JA, Roach HI, Clarke NMP. Histological abnormalities of the calcaneum in congenital talipes equinovarus. J Orthop Sci. 2001;6(6):519–26.
Ester AR, Tyerman G, Wise CA, Blanton SH, Hecht JT. Apoptotic gene analysis in idiopathic talipes equinovarus (clubfoot). Clin Orthop Relat Res. 2007;462(462):32–7.
Ippolito E, Dragoni M, Antonicoli M, Farsetti P, Simonetti G, Masala S. An MRI volumetric study for leg muscles in congenital clubfoot. J Child Orthop. 2012;6(5):433–8.
Sharp L, Miedzybrodzka Z, Cardy AH, Inglis J, Madrigal L, Barker S. The C677T polymorphism in the methylenetetrahydrofolate reductase gene (MTHFR), maternal use of folic acid supplements, and risk of isolated clubfoot: a case-parent-triad analysis. Am J Epidemiol. 2006;164(9):852–61.
Bonafe L, et al. DTDST mutations are not a frequent cause of idiopathic talipes equinovarus (club foot). J Med Genet. 2002;39(4):e20.
Chesney D, Barker S, Miedzybrodzka Z, Haites N, Maffulli N. Epidemiology and genetic theories in the etiology of congenital talipes equinovarus. Bull Hosp Jt Dis. 1999;58(1):59–64.
Barker S, Chesney D, Miedzybrodzka Z, Maffulli N. Genetics and epidemiology of idiopathic congenital talipes equinovarus. J Pediatr Orthop. 2003;23(2):265–72.
Dobbs MB, Gurnett CA. The 2017 ABJS Nicolas Andry award: advancing personalized medicine for clubfoot through translational research. Clin Orthop Relat Res. 2017;475(6):1716–25.
Raines AM, Magella B, Adam M, Potter SS. Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev Biol. 2015;15(1):28.
Dobbs MB, Gurnett CA. Genetics of clubfoot. J Pediatr Orthop B. 2012;21(1):7–9.
Brachvogel B, Zaucke F, Dave K, Norris EL, Stermann J, Dayakli M, et al. Comparative proteomic analysis of normal and collagen IX null mouse cartilage reveals altered extracellular matrix composition and novel components of the collagen IX interactome. J Biol Chem. 2013;288(19):13481–92.
Menke DB, Guenther C, Kingsley DM. Dual hindlimb control elements in the Tbx4 gene and region-specific control of bone size in vertebrate limbs. Development. 2008;135(15):2543–53.
Irani RN, Sherman MS. The pathological anatomy of idiopathic clubfoot. Clin Orthop Relat Res. 2012;84:14–20.
Isaacs H, Handelsman JE, Badenhorst M, Pickering A. The muscles in club foot--a histological histochemical and electron microscopic study. J Bone Joint Surg Br. 1977;59–B(4):465–72.
Moon DK, Gurnett CA, Aferol H, Siegel MJ, Commean PK, Dobbs MB. Soft-tissue abnormalities associated with treatment-resistant and treatment-responsive clubfoot. J Bone Jt Surg. 2014;96(15):1249–56.
Cardy A, Barker S, Chesney D, Sharp L, Maffulli N, Miedzybrodzka Z. Pedigree analysis and epidemiological features of idiopathic congenital talipes equinovarus in the United Kingdom: a case-control study. BMC Musculoskelet Disord. 2007;8(1):62.
Collinson JM, Lindström NO, Neves C, Wallace K, Meharg C, Charles RH, et al. The developmental and genetic basis of ‘clubfoot’ in the peroneal muscular atrophy mutant mouse. Development. 2018;145(3):dev160093.
Availability of data and materials
All the data will be available upon motivated request to the corresponding author of the present paper.
Author information
Authors and Affiliations
Contributions
The design of the study was made by VP, EC, and GT. EC wrote the manuscript. EC searched the literatures and analyzed and collected the data. VP and GT assisted in the critical revision of the manuscript for important intellectual content. Editing was made by VP, GT, and EC. VP provided financial funding. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Pavone, V., Chisari, E., Vescio, A. et al. The etiology of idiopathic congenital talipes equinovarus: a systematic review. J Orthop Surg Res 13, 206 (2018). https://doi.org/10.1186/s13018-018-0913-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-018-0913-z