Abstract
Background
Mangroves plants, an important wetland system in the intertidal shores, play a vital role in estuarine ecosystems. However, there is a lack of a very effective method for extracting protein from mangrove plants for proteomic analysis. Here, we evaluated the efficiency of three different protein extraction methods for proteomic analysis of total proteins obtained from mangrove plant Kandelia obovata leaves.
Results
The protein yield of the phenol-based (Phe-B) method (4.47 mg/g) was significantly higher than the yields of the traditional phenol (Phe) method (2.38 mg/g) and trichloroacetic acid-acetone (TCA-A) method (1.15 mg/g). The Phe-B method produced better two-dimensional electrophoresis (2-DE) protein patterns with high reproducibility regarding the number, abundance and coverage of protein spots. The 2-DE gels showed that 847, 650 and 213 unique protein spots were separated from the total K. obovata leaf proteins extracted by the Phe-B, Phe and TCA-A methods, respectively. Fourteen pairs of protein spots were randomly selected from 2-DE gels of Phe- and Phe-B- extracted proteins for identification by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/TOF-MS) technique, and the results of three pairs were consistent. Further, oxygen evolving enhancer protein and elongation factor Tu could be observed in the 2-DE gels of Phe and Phe-B methods, but could only be detected in the results of the Phe-B methods, showing that Phe-B method might be the optimized choice for proteomic analysis.
Conclusion
Our data provides an improved Phe-B method for protein extraction of K. obovata and other mangrove plant tissues which is rich in polysaccharides and polyphenols. This study might be expected to be used for proteomic analysis in other recalcitrant plants.
Similar content being viewed by others
Introduction
Proteomics has been developed as an important approach for studying plant functional genomics [1], which can be used to detect the post-transcriptional modification [2]. Two-dimensional gel electrophoresis (2-DE) was an efficient and powerful strategy to study complex gene expression at the protein level [3,4,5]. The quality of protein sample was the most crucial steps for optimal results in proteomic analysis. However, it can be problematic because of co-extraction and other non-protein components [6]. It was even worse in plant tissues due to the relative low content of proteins compared with other components, such as proteases and oxidative enzymes, cell walls and vacuoles, pigments, lipids, starches polysaccharides, organic acids, polyphenols, and other secondary metabolites [7,8,9]. The protein extraction methods used for processing plant samples may affect subsequent experimental results [10].
Mangroves plants, an important wetland vegetation in the intertidal shores, play a vital role in estuarine ecosystems [11, 12]. On account of high levels of interfering compounds, including tannins [13], which made it difficult to extract proteins from mangrove plants. How to obtain high quality proteins was crucially important for the proteomic study of mangrove plants. Many protocols have been applied to improve plant protein extraction for 2-DE, the method of trichloroacetic acid-acetone (TCA-A) precipitation was a classical strategy for most proteomic studies of in different plant tissues (leaves, roots, fruit, seeds and stems) [8, 14,15,16,17,18]. However, proteins could not be fully redissolved after TCA-A precipitation and some polymeric contaminants were often co-extracted by the TCA-A method [8, 19,20,21]. Furthermore, when the TCA-A method was used to extract leaf proteins for mangrove plant Kandelia candel, obvious vertical stripes and smearing were found in the gels of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels [22]. The quality of protein extraction will directly affect the proteomic analysis results. An alternative basic method was phenol (Phe) method. Phenol was observed as an effective agent for extracting proteins from aqueous solutions in some plants [8, 16]. Phe method has been applied to mangrove plants, such as Avicennia marina [23], Bruguiera gymnorhiza [24], Rhizophora stylosan [25], K. candel [26, 27] and K.obovata [28]. However, the profiles of SDS-PAGE gels in these studies exhibited a high background, indicating the presence of substantial interfering substance. It seemed that the Phe method was not powerful enough to remove interfering compounds for mangrove plants for proteomics research.
There is a lack of a very effective method for extracting protein from mangrove plants for proteomic analysis. Kandelia obovata, the most cold-resistant specie of mangrove plants, is widely distributed along the South China Coast [29]. With regard to low-abundance proteins in mangrove plants, we developed an improved protein extraction method (Phe-B) for proteomics studies on mangrove plant K. obovata in this study. Comparing with the traditional TCA-A and Phe menthods, the phenol-based (Phe-B) method has been successfully developed for extracting proteins from mangrove plant K. obovata. The protein yield and quality were also discussed.
Results and discussion
Quantitative comparison of protein yield
The protein yields of K. obovata leaves obtained by three extraction methods were summarized in Fig. 1. Quantitative comparison of protein extracts revealed that the yield amount of protein by the Phe-B method (4.47 ± 0.17 mg/g) was significantly greater than that by the Phe method (2.38 ± 0.15 mg/g) or TCA-A method (1.15 ± 0.10 mg/g).
SDS-PAGE evaluation of three methods with K. obovata leaves
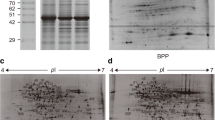
In Fig. 2A, the three protein extracts from K. obovata leaves were performed by SDS-PAGE gel. The Mr of proteins spanned from 6.5 kDa marker to more than 200 kDa. The proteins extracted by Phe-B method had higher quality according to the well-resolved bands distributed in a wide range of Mr (from 6.5 to 116 kD). Besides, proteins extracted by the Phe-B method also showed much less smearing than that by the TCA-A method or Phe method. These results revealed that the Phe-B method was much more effective for protein extraction of K. obovata leaves.
Representative SDS-PAGE and 2-DE gels of the total proteins extracted by three methods. (A) SDS-PAGE gel of three protein extracts and protein marker. The black arrow indicated the location of the Rubisco protein. (B) 2-DE gel of total proteins extracted by TCA-A method. (C) 2-DE gel of total proteins extracted by Phe method. (D) 2-DE gel of total proteins extracted by Phe-B method. In all the 2-DE gels, the isoelectric point ranged from 4 to 7. Fourteen pairs of protein spots were randomly selected from the Phe (C) and Phe-B (D) gels for MALDI TOF/TOF (indicated by arrows and numbers). Due to the failure to obtain a clear 2-DE map, no protein spots were selected from the TCA-A gel (B) for MS identification
There were many differences among the SDS-PAGE bands pattern of the three protein extracts. Particularly, there were much more polypeptides in protein extracts by the Phe-B or Phe methods than that by the TCA-A method. In addition, the obvious difference among the extracts was the relative abundance of Rubisco, as an arrow indicated in Fig. 2A. Protein extracted by Phe-B method contained the lower level of the Rubisco comparing with that by the TCA-A method or Phe method. Rubisco is the world’s most abundant protein in plants and shares more than 50% of the total leaf protein in some species [30]. It can reduce detectable protein spots during electrophoretic separation of leaf proteins. Therefore, several multistep techniques have been developed to remove Rubisco selectively as a pre-fractionation step [31, 32]. The results showed that the Phe-B method was a rapid and efficient method to reduce Rubisco amount. This further indicated that the Phe-B method was the optimal method to acquire high quality proteins from K. obovata leaves for proteomics analysis.
2-DE evaluation of three methods with K. obovata leaves
The proteins extracted by the Phe-B showed better solubility and less streaks than that by the Phe method or TCA-A method. In Fig. 2B–D, protein spots extracted by the Phe-B method (847 ± 93) were more than that by the Phe (650 ± 66) or TCA-A method (213 ± 49). Additionally, spot-to-spot comparison of 2-DE gels revealed that almost all the protein spots extracted by Phe method or TCA-A method were included in that by Phe-B method. These results indicated that the Phe-B method had improved the efficiency of protein extraction and had increased the solubility of proteins. This data further confirmed that the Phe-B method was effective to extract proteins from K. obovata leaves.
Compared theses three methods, the Phe-B method obtained the largest numbers of spots, while the TCA-A method got the lowest. Obvious differences were observed in the spot patterns among these extracts. The proteins extracted by Phe-B showed greater spot intensities than that by the Phe method or TCA-A method. The protein spots extracted by the Phe-B and Phe methods were highlighted by black frames in Fig. 2C, D. Based on different protein extraction methods, differential spot patterns reflected different degrees of proteolysis [16]. Table 1 gave the details about the identification of 14 protein spots that are extracted by Phe-B method. Fig. 2C (by Phe method) and Fig. 2D (by Phe-B method) showed the digging locations of these 14 protein spots, respectively. None of these 14 proteins spots were found in Fig. 2B (by TCA-A method). As for mass spectrum analysis, only spot nos. 3, 4, 6 were identified from the protein spots that are extracted both by the Phe (Fig. 2C) and Phe-B method (Fig. 2D). The other 11 spots were identified only in extracts by the Phe-B method (Fig. 2D). However, these missing protein spots, which were lost in the other two extracts, played very important roles in plants. The lost proteins contained aconitase (spot no. 2), adenosine kinase (spot no. 7), fructose-1, 6-bisphosphatase (spot no. 8), thioredoxin reductase (spot no. 10), oxygen evolving enhancer protein (spot no. 11) and ATP synthase (spot no. 13). Previous studies showed that these proteins are involved in the process of metabolism, photosynthesis and anti-stress physiology [33,34,35,36,37,38].
Several studies have involved in the protein extraction of mangrove plant [22,23,24,25,26,27,28, 39, 40], including Rhizophora stylosa, Bruguiera parviflora, Avicennia marina, Bruguiera gymnorhiza, Kandelia candel and K. obovata, but these protein extracts showed obvious smearing and streaking in 2-DE gels, and the protein quality were not good enough when applied to proteomics analysis in the studies. In this study, fewer smears and streaks, more spots and higher protein yields were obtained by the Phe-B method (as shown in Figs.1, 2). These results indicated that the Phe-B method was a very good method for the protein extraction of mangrove plants.
As is well-known that most interfering compounds would reduce solubility of proteins and prevent sample powder homogenizing thoroughly in aqueous buffer [41]. Thus, it is very important to remove interfering compounds before protein extraction. Here, the outstanding advantages of the Phe-B method were described as follows: (1) Pulverizing plant tissues with PVPP (0.1 g/g tissue) helps to increase the removal efficiency of phenolic compounds [42, 43]. (2) Briefly washing with 10% TCA/acetone can promote protein precipitation and contaminant removal (most lipids and lipid-like polymers) [44, 45]. Besides, this step also can help to reduce protein degradation and modifications during long-term exposure in low pH (TCA) [21]. (3) Washing with 80% methanol plus 0.1 M ammonium acetate helps to neutralize residual TCA and increase the pH to above 7. The alkaline environment facilitates the subsequent protein extraction by phenol [21]. (4) Phenol/SDS mixture helps to improve protein solubility. As an excellent solubilizing agent, SDS allowed the recovery of membrane-bound proteins [9, 43]. The cautions mentioned in the study were important for obtaining high quality proteins from mangrove plants.
As to comparative proteomics, a major objective is to maximize the number of polypeptides. The Phe-B method obtained the greatest numbers of proteins in comparison with the other two methods. Although the Phe-B method was somewhat complicated and time consuming, a greater number of proteins had been obtained. In addition, the Phe-B method might lose small amounts of proteins due to many steps, but the loss can be remedied by parallel experiments.
Conclusions
In comparing these three methods, the Phe-B method gave the greatest protein yield, the most protein spots, and the least of smearing and streaking. This is the first time for the well-resolved SDS-PAGE and 2-DE protein patterns of K. obovata leaves. The Phe-B method might be the optimal method for extracting proteins from mangrove plants. This study also provides a potential method for protein extraction from other recalcitrant plant tissues for proteomic analysis.
Methods
Plant materials and growth
Kandelia obovata propagules, collected from Dong-chong mangrove wetland (Shenzhen, China), were used in all the experiments. Propagules were surface sterilized and germinated in clean sands. Seedlings were grown in a greenhouse under a 14 h light/10 h dark cycle at 25/22 ℃, and were irrigated with 1/2 Hoagland solution once a week. After 3 months, two pairs of fully-expanded leaves were collected from seedlings. The leaves were washed with distilled water, and then dried with paper. All the harvested samples were immediately frozen in liquid nitrogen and stored at – 80 ℃ until protein extraction.
Protein extraction by TCA-A method
The TCA-A method was performed as described [15] with some modifications. Frozen plant leaves (2.0 g) were finely powdered in a pestle and mortar with liquid nitrogen. Three replicate leaf samples were used for protein extraction. The frozen powder was then transferred into a 50 mL centrifuge tube. Filled the tube with four volumes of ice-cold acetone containing 10% TCA and 2% 2-mercaptoethanol. The mixture was adequately homogenized using a vortex mixer and then was precipitated at – 20 ℃ overnight. Then centrifugated the tube at 12,000×g for 30 min at 4 ℃ to obtain the protein pellet. The protein pellets were washed with cold acetone, which containing 0.07% 2-mercaptoethanol and 0.1 mM phenylmethanesulfonyl fluoride (PMSF). Then centrifuged the protein pellets at 12,000×g for 10 min at 4 ℃ and discarded the supernatant. Repeated this step twice to obtain clean protein pellet. The process of protein extraction by TCA-A method was repeated at least three times.
Protein extraction by Phe method
Total protein extracts were prepared according to Wang et al. [26]. Frozen plant leaves (2.0 g) were finely powdered in liquid nitrogen, and then suspended completely in 15 mL cold acetone. Three duplicate leaf samples were used for protein extraction. After vibrated the mixture for 1 min, the suspension was centrifuged in a pre-cooling rotor at 12,000×g for 5 min at 4 ℃. The supernatant was discarded and the pellet was re-suspended with pre-cooling extraction buffer (100 mM Tris–HCl, pH 8.0, 50 mM L-ascorbic acid, 100 mM KCl, 50 mM disodium tetraborate decahydrate, 1% (v/v) Triton X-100, 2% (v/v) β-mercaptoethanol, and 1 mM PMSF). After adding an equal volume of ice-cold Tris-saturated phenol (pH 7.9), the homogenate was thoroughly mixed and centrifuged at 12,000×g for 10 min at 4 ℃. The upper phenolic phase was collected and then was transferred into a new 50 mL tube. The phenol phase was mixed with five volumes of methanol containing 0.1 M ammonium acetate. Then the mixture was precipitated at – 20 ℃ for overnight. After centrifuging the tube at 12,000×g for 20 min at 4 ℃, the supernatant was discarded. The protein pellets were washed with cold methanol for one time, and then were washed twice with acetone. During each wash step, mixed and centrifuged them well. The process of protein extraction by TCA-A method was repeated at least three times.
Protein extraction by the Phe-B method
Based on TCA-A precipitation and phenol extraction with SDS-containing buffer, the Phe-B method was proposed. Three repeated leaf samples were used for protein extraction. Frozen K. obovata leaves (2.0 g) were subjected to the follows: (1) tissue powder. After 10% PVPP of sample weight were added, frozen leaves were powdered with liquid nitrogen. (2) TCA/acetone washing. The powder was transferred into a 50 mL tube and then was filled with pre-cooling 10% TCA/acetone. The mixture was extensively homogenized and centrifuged at 12,000×g for 5 min at 4 ℃. The supernatant was carefully removed with pipetting. (3) Methanol washing. The pellets were washed by 80% methanol containing 0.1 M ammonium acetate to remove residual TCA (the pH value to above 7). The mixture was mixed well and centrifuged at 12000×g for 5 min at 4 ℃. Then discarded the supernatant. (4) Acetone washing. The pellet was washed and vibrated again with 80% acetone until the pellet was fully dispersed. Centrifuged the mixture at 12000×g for 5 min at 4 ℃ and discarded the supernatant. (5) Drying. The pellets were freeze-dried for at least 5 min to remove the residual acetone. (6) Protein extraction and precipitation. The protein particles were suspended with 0.4–0.8 mL/0.1 g starting material of 1:1 phenol (pH 7.9)/sodium dodecyl sulfate (SDS) buffer (30% sucrose, 2% SDS, 0.1 M Tris-HCl, pH 8.0, 5% 2-mercaptoethanol, 1 mM PMSF). The mixture was mixed and incubated for 5 min. Then centrifuged the mixture at 12,000×g for 10 min and transferred the upper phenol phase into a new 50 mL tube. The upper phenol phase was mixed with five–ten volumes of methanol containing 0.1 M ammonium acetate. The mixture was precipitated at – 20 ℃ for overnight. When a white pellet was visible, centrifuged the mixture at 12,000×g for 20 min at 4 ℃ and carefully discarded the supernatant (note: if no phase separation occurs, add more phenol (100 μL) into the mixture, then mixed and centrifuged the mixture again). (7) Washing and air-drying the pellet. The final protein pellet was washed once with 100% methanol and then was washed twice with 80% acetone. During each washing step, the sample should be mixed well and be centrifuged as above. Then discarded the supernatant. The process of protein extraction by Phe-B method was repeated at least three times.
Protein solubilization and concentration determinations
All the protein pellets by these three protocols were briefly lyophilized by Freeze Dry Systems (SCIENTZ-12N, NINGBO SCIENTZ Biotechnology CO, LTD). The samples were transferred into 1.5 mL microcentrifuge tubes respectively, and were re-dissolved directly in appropriate lysis buffer [7 M urea, 2 M thiourea, 4 % (v/v) CHAPS, 2 % (v/v) pharmalyte 4-7, and 50 mM dithiothreitol (DTT), 2 mM tri-butyl-phosphate (TBP), 0.1 mM PMSF]. Samples were incubated in water bath at 36 ℃ for 2 h, and then was centrifuged at 14,000×g for 30 min at 4 ℃. The supernatants were stored at – 80 ℃ for concentration analysis or downstream experiments. The concentrations of protein were quantified by Bradford [46].
Isoelectric focusing (IEF) and SDS-PAGE
IPG strips (17 cm pH 4-7, Bio-Rad ReadyStrip; Bio-Rad) were passively rehydrated at 17 ℃ for 14 h with 330 μL IEF buffer (7 M urea, 2 M thiourea, 4 % (v/v) 3-[(3-Cholanidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 2 % (v/v) pharmalyte 4-7, 50 mM DTT, 2 mM TBP, 0.1 mM PMSF, and 0.002% bromophenol blue) containing 2.0 mg of protein. Isoelectric focusing was performed with a Protean i12 IEF Cell (Bio-Rad) apparatus under the following program: 250 V for 30 min, 500 V for 30 min, 1000 V for 30 min, 8000 V for 5 h and 8000 V for a total of 40,000 Vh. After finished the IEF protocol, the focused strips should be equilibrated in reducing equilibration buffer (6 M urea, 20% w/v glycerol, 2% SDS, and 0.375 mM Tris-HCl, pH 8.8, 2% w/v DTT) as soon as possible. After that the strips were equilibrated in same solution that contained 2.5% w/v iodoacetamide instead of 2.0% DTT. The strips were then transferred to 12% SDS-PAGE gels for second dimension electrophoresis by the Bio-Rad PROTEAN xi 2-D Cell gel system (Bio-Rad, USA). There came to the following program: 50 V for 1 h, and then 200 V for 5.5 h for each strip. The SDS electrophoresis buffer (25 mM Tris-base solution (pH 8.3), 192 mM glycine and 0.1% SDS) was used as working solution.
Image acquisitions and analysis
The 2-DE gels were stained with Bio-Safe Coomassie Brilliant Blue R-250 (Amresco, USA) and then was scanned by GS-800 Calibrated Densitometer (Bio-Rad, USA). All the 2-DE gel separations were repeated three times. Image analysis was performed using PDQUEST software (Bio-Rad, USA). The automatic default spot analysis was used to edit the spot features with manual correction by combining semiautomatic method. The spots were quantified using the % volume criterion. Three repetitions were executed. Student’s t test was performed for statistical analysis.
MALDI-TOF/TOF-MS analysis
The selected protein spots were manually excised from stained 2-DE gels for mass spectrometric analysis. All the samples were digested by trypsin and then were analyzed by Ultraflex MALDI-TOF/TOF mass spectrometer (Bruker, Bremen, Germany). The analysis was performed under the control of FlexControl™ 3.3 software (Bruker) with external mass calibration. The mass spectrometer was set to perform data acquisition with a selected mass range of 800–3500 m/z. Internal calibration of the standard spectra was performed after every 10 consecutive spectra using Pepmix peptide calibration standards (Bruker Daltonics). The obtained spectrum was analyzed with FlexAnalysis™ 3.3 (Bruker) and Biotool™ 2.2 (Bruker). Peptide mass fingerprinting was searched using the program Mascot (Matrix Science, London, UK) against the NCBI database. Zero–two peptide cleavage sites were set on the MASCOT search engine. The mass tolerance was 100 ppm, and MS/MS tolerance was 0.6 Da. Protein scores more than 76 were considered statistically significant (p < 0.05) for peptide mass fingerprinting in MS/MS analysis.
Availability of data and materials
Not applicable.
Abbreviations
- TCA-A:
-
Trichloroacetic acid—acetone method
- Phe-B:
-
Phenol-based method
- Phe:
-
Phenol method
- PMSF:
-
Phenylmethanesulfonyl fluoride
- DTT:
-
Dithiothreitol
- TBP:
-
Tri-butyl phosphate
- MALDI-TOF/TOF-MS:
-
Matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry
References
Thiellement H, Bahrman N, Damerval C, Plomion C, Rossignol M, Santoni V, de Vienne D, Zivy M. Proteomics for genetic and physiological studies in plants. Electrophoresis. 1999;20:2013–26.
Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D. Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 2001;126:835–48.
Maximiano MR, Megias E, Santos IR, Santos LS, Ollero FJ, Megias M, Franco OL, Mehta A. Proteome responses of Rhizobium tropici CIAT 899 upon apigenin and salt stress induction. Appl Soil Ecol. 2021;159:103815.
Santos IR, Rios TB, Maximiano MR, Coutinho WM, De Lima LM, Silva LP, Olivera-Neto OB, Mehta A. Proteomic screening for the identification of proteins involved in resistance to Xanthomonas campestris pv. malvacearum in cotton. Physiol Mol Plant Pathol. 2021;113:101562.
Li HB, Li YL, Ke QB, Kwak SS, Zhang SQ, Deng XP. Physiological and differential proteomic analyses of imitation drought stress response in Sorghum bicolor root at the seedling stage. Int J Mol Sci. 2020;21:9174.
Gorg A, Obermaier C, Boguth G, Weiss W. Recent developments in two-dimensional gel electrophoresis with immobilized pH gradients: wide pH gradients up to pH 12, longer separation distances and simplified procedures. Electrophoresis. 1999;20:712–7.
Flemmig J, Rusch D, Czerwinska ME, Rauwald HW, Arnhold J. Components of a standardised olive leaf dry extract (Ph. Eur.) promote hypothiocyanite production by lactoperoxidase. Arch Biochem Biophys. 2014;549:17–25.
Isaacson T, Damasceno CM, Saravanan RS, He Y, Catalá C, Saladié M, Rose JK. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protoc. 2006;1:769–74.
Wang W, Scali M, Vignani R, Spadafora A, Sensi E, Mazzuca S, Cresti M. Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis. 2003;24:2369–75.
Joy AP, Ayre DC, Chute IC, Beauregard AP, Wajnberg G, Ghosh A, Lewis SM, Ouellette RJ, Barnett DA. Proteome profiling of extracellular vesicles captured with the affinity peptide Vn96: comparison of Laemmli and TRIzol protein-extraction methods. J Extracell Vesicles. 2018;7:1438727.
Donato DC, Kauffman JB, Murdiyarso D, Kurnianto S, Stidham M, Kanninen M. Mangroves among the most carbon-rich forests in the tropics. Nat Geosci. 2011;4:293–7.
zu Ermgassen PSE, Mukherjee N, Worthington TA, Acosta A, Araujo ARR, Beitl CM, Castellanos-Galindo GA, et al. Fishers who rely on mangroves: modelling and mapping the global intensity of mangrove-associated fisheries. Estuar Coast Shelf Sci. 2020;247:106975.
Yan ZZ, Tam NFY. Temporal changes of polyphenols and enzyme activities in seedlings of Kandelia obovata under lead and manganese stresses. Mar Pollut Bull. 2011;63:438–44.
Hassan M, Yusoff N, Aizat WM, Othman NW, Abd GI. Optimization method for proteomic analysis of the larva and adult tissues of Plutella xylostella (L.) (Lepidoptera: Plutellidae). Sains Malays. 2018;47:2975–83.
Lee TCH, Lai KKY, Kwok CZN, Xu SJL, Lee FWF. Comparison of five trizol-based protein preparation methods for 2-DE production from challenging marine dinoflagellate samples: a case study on two benthic Prorocentrum species. J Mar Sci Eng. 2020;8:363.
Saravanan RS, Rose JKC. A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics. 2004;4:2522–32.
Tan HS, Liddell S, Ong Abdullah M, Wong WC, Chin CF. Differential proteomic analysis of embryogenic lines in oil palm (Elaeis guineensis Jacq). J Proteom. 2016;143:334–45.
Tian XH, Xie JM, Yu JH. Study on signal induced expression of cold tolerance in edible lily in alpine environment. Appl Ecol Env Res. 2020;18:2687–701.
Jin X, Zhu LP, Tao C, Xie QL, Xu XY, Chang LL, Tan YH, Ding GH, Li HB, Wang X. An improved protein extraction method applied to cotton leaves is compatible with 2-DE and LC-MS. BMC Genom. 2019;20:285.
Wang YB, Wang XY, Yang RX, Niu LJ, Wang W. Comparison of protein extraction methods for 2DE-based proteomic analysis of duckweed Spirodela polyrhiza, a small aquatic model plant. Aquat Bot. 2020;163:103216.
Wang W, Vignani R, Scali M, Cresti M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis. 2006;27:2782–6.
Yu F, Zheng C, Shi M, Liu WC, Qiu JB, Ji DW, Huang L, Chen SB. Establishment and optimization of 2-DE technique system in leaf proteome of Kandelia candel. J Trop Subtrop Bot. 2011;19:519–23.
Shen ZJ, Chen J, Ghoto K, Hu WJ, Gao GF, Luo MR, Li Z, Simon M, Zhu XY, Zheng HL. Proteomic analysis on mangrove plant Avicennia marina leaves reveals nitric oxide enhances the salt tolerance by up-regulating photosynthetic and energy metabolic protein expression. Tree Physiol. 2018;38:1605–22.
Zhu Z, Chen J, Zheng HL. Physiological and proteomic characterization of salt tolerance in a mangrove plant, Bruguiera gymnorrhiza (L.) Lam. Tree Physiol. 2012;32:1378–88.
Fan JX, Deng YC, Huang X, Lin QF, Luo YH. Comparative proteomic analysis of salt-stress response proteins in Rhizophora stylosa roots. Chin J Biochem Mol Biol. 2009;25:72–7.
Wang LX, Liu X, Liang M, Tan FL, Liang WY, Chen YY, Lin YX, Huang L, Xing JH, Chen W. Proteomic analysis of salt-responsive proteins in the leaves of mangrove Kandelia candel during short-Term stress. Plos ONE. 2014;9:e83141.
Weng ZX, Wang LX, Tan FL, Huang L, Xing JH, Chen SP, Cheng CL, Chen W. Proteomic and physiological analyses reveal detoxification and antioxidation induced by Cd stress in Kandelia candel roots. Trees. 2013;27:583–95.
Liu YL, Shen ZJ, Simon M, Li H, Ma DN, Zhu XY, Zheng HL. Comparative proteomic analysis reveals the regulatory effects of H2S on salt tolerance of mangrove plant Kandelia obovata. Int J Mol Sci. 2020;21:118.
Ren H, Lu H, Shen W, Huang C, Guo Q, Li ZA, Jian S. Sonneratia apetala Buch. Ham in the mangrove ecosystems of China: an invasive species or restoration species? Ecol Eng. 2009;35:1243–8.
Ellis RJ. The most abundant protein in the world. Trends Biochem Sci. 1979;4:241–4.
Kim ST, Cho KS, Jang YS, Kang KY. Two-dimensional electrophoretic analysis of rice proteins by polyethylene glycol fractionation for protein arrays. Electrophoresis. 2001;22:2103–9.
Kim YJ, Lee HM, Wang Y, Wu J, Kim SG, Kang KY, Park KH, Kim YC, Choi IS, Agrawal GK, Rakwal R, Kim ST. Depletion of abundant plant RuBisco protein using the protamine sulfate precipitation method. Proteomics. 2013;13:2176–9.
Laus MN, Soccio M. First evidence of a protective effect of plant bioactive compounds against H2O2-induced aconitase damage in durum wheat mitochondria. Antioxidants. 2020;9:1256.
Hao J, Yang Z, Li J, Han LF, Zhang Y, Wang T. Discovery of natural adenosine monophosphate-activated protein kinase activators through virtual screening and activity verification studies. Mol Med Rep. 2021;23:203.
Daie J. Cytosolic fructose-1,6-bisphosphatase: a key enzyme in the sucrose biosynthetic pathway. Photosynth Res. 1993;38:5–14.
Jacquot JP, Eklund H, Rouhier N, Schuermann P. Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms. Trends Plant Sci. 2009;14:336–43.
Yang EJ, Oh YA, Lee ES, Park AR, Cho SK, Yoo YJ, Park OK. Oxygen-evolving enhancer protein 2 is phosphorylated by glycine-rich protein 3/wall-associated kinase 1 in Arabidopsis. Biochem Bioph Res Commun. 2003;305:862–8.
Seelert H, Dencher NA. ATP synthase superassemblies in animals and plants: two or more are better. BBA Bioenerg. 2011;807:1185–97.
Parida AK, Mittra B, Das AB, Das TK, Mohanty P. High salinity reduces the content of a highly abundant 23-kDa protein of the mangrove Bruguiera parviflora. Planta. 2005;221:135–40.
Tada Y, Kashimura T. Proteomic analysis of salt-responsive proteins in the mangrove plant, Bruguiera gymnorhiza. Plant Cell Physiol. 2009;50:439–46.
Islam N, Lonsdale M, Upadhyaya N, Higgins T, Hirano H, Akhurst R. Protein extraction from mature rice leaves for two-dimensional gel electrophoresis and its application in proteome analysis. Proteomics. 2004;4:1903–8.
Usuda H, Shimogawara K. Phosphate deficiency in maize. 6. Changes in the two-dimensional electrophoretic patterns of soluble-proteins from 2nd-leaf blades associated with induced senescence. Plant Cell Physiol. 1995;36:1149–55.
Yao Y, Yang YW, Liu JY. An efficient protein preparation for proteomic analysis of developing cotton fibers by 2-DE. Electrophoresis. 2006;27:4559–69.
Faurobert M, Mihr C, Bertin N, Pawlowski T, Negroni L, Sommerer N, Causse M. Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiol. 2007;143:1327–46.
Shaw MM, Riederer BM. Sample preparation for two-dimensional gel electrophoresis. Proteomics. 2003;3:1408–17.
Bradford MM. A rapid and sensitive method for quantitation of microgram quantities of proteinutilizing principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Acknowledgements
We greatly thank all of our colleagues in our laboratory for providing useful technical assistance.
Funding
This work was supported by the National Natural Science Foundation of China (No. 41706118, No. 41676086, No. U1901211, No. 41430966 and No. 41876126), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA23050200), the International Partnership Program of Chinese Academy of Sciences (No. 133244KYSB20180012), the National Key Research and Development Plan (No. 2017FY100700) and the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0305). The funding body was not involved in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
JF conceived, designed and undertook the experiments and wrote the manuscript. YSW designed, conceptualized and supervised the manuscript. HC conceived and supervised the research, revised and developed this manuscript. YBS participated in experiments as well as revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Fei, J., Wang, YS., Cheng, H. et al. An efficient protein extraction method applied to mangrove plant Kandelia obovata leaves for proteomic analysis. Plant Methods 17, 100 (2021). https://doi.org/10.1186/s13007-021-00800-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13007-021-00800-y