Abstract
Background
The burden and characteristics of respiratory viral infections in children hospitalized for acute respiratory tract infections (ARTIs) during the post-COVID-19 pandemic era are unclear. We analyzed the epidemiological and clinical characteristics of pediatric patients hospitalized with common respiratory virus infections before and after relaxation of non-pharmaceutical interventions in Hangzhou, China and evaluated the diagnostic value of the six-panel respiratory pathogen detection system.
Methods
Six types of respiratory viruses were detected in respiratory samples from children with suspected ARTIs by multiplex real-time quantitative polymerase chain reaction (RT-qPCR). Changes in virus detection rates and epidemiological and clinical characteristics, obtained from electronic health records, were analyzed. Binary logistic regression was used to identify respiratory tract infections risk factors. Multiplex RT-qPCR and targeted next-generation sequencing results were compared in random samples.
Results
Among the 11,056 pediatric samples, 3228 tested positive for one or more of six common respiratory pathogens. RSV and PIV-3 detection rates differed significantly across age groups (both P < 0.001), and were more common in younger children. PIV-1 was more common in infants, toddlers, and preschoolers than in school-age children (P < 0.001). FluB was predominantly detected in school-age children (P < 0.001). RSV-, ADV-, and PIV-1-positivity rates were higher in 2022 than in 2023. Seasonal viral patterns differed across years. RSV (OR 9.156. 95% CI 5.905–14.195) and PIV-3 (OR 1.683, 95% CI 1.133–2.501) were risk factors for lower respiratory tract infections. RSV-positivity was associated with severe pneumonia (P = 0.044). PIV-3 (OR 0.391, 95% CI 0.170–0.899), summer season (OR 1.982, 95% CI 1.117–3.519), and younger age (OR 0.938, 95% CI 0.893–0.986) influenced pneumonia severity. Multiplex RT-qPCR showed good diagnostic performance.
Conclusion
After changes in COVID-19 prevention and control strategies, six common respiratory viruses in children were prevalent in 2022–2023, with different seasonal epidemic characteristics and age proclivities. RSV and PIV-3 cause lower, and FluA, FluB, and ADV more typically cause upper respiratory tract infections. Infancy and summer season influence severe pneumonia risk. Multiplex RT-qPCR is valuable for accurate and timely detection of respiratory viruses in children, which facilitates management, treatment, and prevention of ARTIs.
Similar content being viewed by others
Background
Acute respiratory tract infections (ARTIs), particularly among children, are a major public health concern worldwide [1]. ARTIs are caused by various pathogens, including bacteria, mycoplasmas, Chlamydia, fungi, parasites, and viruses. Among these pathogens, viruses are the leading cause of ARTIs in children [2]. Common viral pathogens associated with ARTIs in children include influenza virus (Flu), respiratory syncytial virus (RSV), rhinovirus, parainfluenza virus (PIV), and adenovirus (ADV).
The immature immune and respiratory systems of children, particularly those of infants and preschool-aged children, render them susceptible to the harmful effects of viral respiratory infections. Infections occurring early in life can hinder lung development and increase the risk of asthma [3, 4]. The diagnosis of virus-induced ARTIs is challenging because of similar symptoms and changes on clinical images observed across different viral type infections. In addition, the same viral pathogen can manifest with various clinical presentations [5]. Consequently, accurate and timely detection of these viruses in children with respiratory infections is essential for a precise diagnosis and to facilitate optimal clinical management strategies, while reducing the excessive use of antibiotics [6].
Traditional diagnostic methods, such as viral culture and serological tests, are time-consuming and have limitations in terms of sensitivity and specificity. Therefore, accurate and rapid diagnostic methods are needed to identify specific viral pathogens that cause ARTIs in children. In recent years, advances in molecular detection and gene sequencing techniques have significantly improved our ability to identify respiratory pathogens. According to the Chinese Center for Disease Control and Prevention, ARTIs in children are commonly caused by Flu, RSV, rhinovirus, PIV, and ADV [7]. Multiple viral pathogens can be detected in a single test in clinical samples by using multiplex real-time quantitative polymerase chain reaction (RT-qPCR), a high-throughput technique that provides rapid and accurate results.
Furthermore, after the emergence of COVID-19, the government implemented non-pharmacological interventions (NPIs), such as wearing masks and social distancing measures, to reduce the transmission of SARS-CoV-2 [8, 9]. In late 2022, the government stopped implementing NPIs. Since then, there have been limited reports emerging on the incidence, age distribution, seasonal patterns, geographical variations, and pathogens associated with certain infectious diseases, in comparison to the conditions before the onset of the COVID-19 pandemic.[10]. Therefore, the epidemiological and clinical characteristics of pediatric patients hospitalized with common respiratory virus infections before and after relaxation of NPIs remain to be explored.
This study evaluated the clinical applications and epidemiological characteristics of common pediatric respiratory pathogens during the post-pandemic era, detected by using multiplex assays, to determine the prevalence, distribution, and clinical manifestations of respiratory viruses in different pediatric age groups. Furthermore, we evaluated the utility of multiplex RT-qPCR and targeted next-generation sequencing (tNGS) for diagnosing and characterizing respiratory viral infections, to provide a basis for improving diagnostic strategies, guiding clinical management, and implementing preventive measures.
Methods
Study population
We enrolled 11,056 children with suspected ARTIs who were hospitalized at Hangzhou Children's Hospital from March 2022 to February 2024. Their median age was 4 years (range, 31 days–17 years). The cohort comprised 5,862 boys (53%) and 5,194 girls (47%).
The inclusion criteria were age under 17 years and presence of at least two of the following symptoms: fever (> 38.0 °C), sore throat, cough, hoarseness, runny nose, nasal congestion/purulent pharyngeal exudate, rapid breathing, or wet rales. Patients with autoimmune diseases, immunodeficiency, repeated the detection within 2 weeks, or a primary diagnosis unrelated to respiratory diseases were excluded from the study.
This observational study was conducted in compliance with the Declaration of Helsinki and approved by the Hangzhou Children's Hospital Clinical Research Ethics Committee (Protocol Number: No. 2021-IRB-74). Patient privacy was protected through encryption of all identifiable information in the electronic database. Informed consent was not required as no interventions were involved, and no patient-identifiable information was included in this study.
Specimen collection and testing
Nasopharyngeal swabs were collected from all hospitalized patients by trained medical personnel following standardized protocols. Nasal swabs were mixed with 1 mL extraction buffer containing surfactant. The samples were stored at a temperature of 2–8 °C for a maximum of 8 h before further processing. Nucleic acids were extracted from each specimen using a viral nucleic acid extraction kit (TIANLONG, Xi’an, China). A multiplex RT-qPCR assay with Taqman probe (X-ABT, Beijing, China) was used with an ABI7500 system (Thermo Fisher Scientific, Waltham, MA, USA) to detect six respiratory viruses: RSV, FluA, FluB, PIV-1, PIV-3, and ADV. The multiplex RT-qPCR was carried out at 45 °C for 10 min, followed by 5 min at 95 °C, and then 45 cycles of 15 s at 95 °C and 45 s at 60 °C. The results were interpreted according to the manufacturer’s instructions provided with the assay kit.
tNGS
Total nucleic acids were either extracted manually or automatedly using a MagPure Viral DNA/RNA Kit (IVD5412, Magen Biotechnology, Guangzhou, China) on the KingFisher™ Flex Purification Platform (Thermo Fisher Scientific). Nuclease-free water (Invitrogen, Waltham, MA, USA) used in the extraction process and also served as non-template-control to monitor for possible contamination.
Reverse transcription, multiplex PCR preamplification, and library preparation for tNGS were performed using the URP50TM Respiratory Pathogen Microorganism Multiplex Testing Kit (KingCreate, Guangzhou, China). Nucleic acid concentration was measured in all samples using an Equalbit DNA HS Assay Kit (Vazyme Biotech Co., Ltd, Nanjing, China) with an Invitrogen™ Qubit™ 3.0/4.0 (Thermo Fisher Scientific) to ensure a library concentration of ≥ 0.5 ng/μL; if this was not achieved, nucleic acid was re-extracted or libraries were re-constructed. Qualified libraries were used as inputs for sequencing, which was performed on the KM MiniSeq Dx-CN platform (KingCreate).
Data collection
Clinical data, including age, sex, nasopharyngeal swab collection date, clinical diagnosis results, and multiplex RT-qPCR test results for the six respiratory viruses were extracted from the laboratory information system of Hangzhou Children's Hospital. ARTIs in children were grouped into four categories based on age: infants (< 1 year), toddlers (1–3 years), preschoolers (3–7 years), and school-aged children (7–17 years). ARTIs in children were grouped into four categories based on the season in Hangzhou: spring (March to May), summer (June to August), autumn (September to November), and winter (December to February). ARTIs were classified as upper respiratory tract infections (URTIs) or lower respiratory tract infections (LRTIs), based on the inferior border of the cricoid cartilage. URTIs included pharyngitis, rhinitis, tonsillitis, and influenza without lower respiratory tract symptoms, whereas LRTIs included bronchitis, bronchiolitis, and pneumonia.
Statistical analysis
Statistical analyses were performed using the SPSS software (version 26.0; IBM SPSS, Inc., Armonk, NY, USA). Count (percentage) data were used for analysis. The 95% confidence interval (CI) for the detection rate was calculated based on the approximate normal distribution method. The chi-square test was used to analyze the positivity rates for the six common respiratory pathogens in different sexes, ages, seasons, and clinical diagnoses. The chi-square trend test was conducted to compare multiple groups. Multivariate analysis was performed using binary logistic regression analysis, and P < 0.05 was considered statistically significant. Sensitivity, specificity, positive-predictive value (PPV), and negative-predictive value (NPV) were calculated to analyze the diagnostic performance of multiplex RT-qPCR.
Results
Overall findings of the six-panel respiratory pathogen detection assay
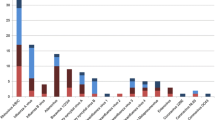
Among the 11,056 collected samples from pediatric patients, 3228 cases tested positive for at least one of the six common respiratory pathogens, with an overall detection rate of 29.20% (95% confidence interval [CI]: 28.35–30.06%). Among the 3228 positive cases, 3,078 (95.35%; 95% CI 94.57–96.05%) were single-pathogen positive, while 150 cases (4.65%; 95% CI 3.95–5.43%) were mixed-pathogen positive. The detection rates from highest to lowest of each of the six pathogens was in the order RSV, FluA, PIV-3, FluB, ADV, and PIV-1, with detection rates of 13.58% (95% CI 12.95–14.23%), 4.79% (95% CI 4.40–5.20%), 4.52% (95% CI 4.14–4.92%), 3.12% (95% CI 2.80–3.46%), 2.67% (95% CI 2.38–2.99%), and 1.90% (95% CI 1.65–2.17%), respectively. Among the 150 patients with mixed-pathogen infections, 148 showed dual positivity, whereas two showed triple positivity. The top-three combinations of mixed infections were RSV + PIV-3 (32.67%, 49/150), RSV + ADV (20.67%, 31/150), and RSV + FluB (9.33%, 14/150) (Table 1 and Additional file 1).
Age and sex characteristics of six respiratory pathogen infections in children
The detection rates of six common respiratory pathogens varied among different age groups in children, with the highest detection rate of 51.50% (95% CI 48.63–54.36%) in the infant group, followed by 42.01% (95% CI 40.01–44.03%) in the toddler group, 26.49% (95% CI 25.21–27.80%) in the preschool group, and 14.05% (95% CI 12.82–15.35%) in the school-age group. The differences were statistically significant (χ2 = 821.261, P < 0.001).
In the univariate chi-square test, the highest RSV detection rate was found in the infant group at 31.39% (95% CI 28.78–34.10%), followed by 24.67% (95% CI 22.94–26.46%) in the toddler group, 10.92% (95% CI 10.03–11.86%) in the preschool group, and 1.55% (95% CI 1.14–2.06%) in the school-age group. The differences in positive detection rates among the age groups were statistically significant (χ2 = 965.212, P < 0.001).
The PIV-3 detection rate showed a similar pattern to RSV, with higher detection rates in the infant and toddler groups (11.71%, 95% CI 9.95–13.66%; 8.52%, 95% CI 7.42–9.72%), and a significant decrease in detection rate in children above 3 years old. The differences in detection rates among the four age groups were statistically significant (χ2 = 354.712, P < 0.001). PIV-1 was relatively more common in the infant, toddler, and preschool groups, and less common in the school-age group (χ2 = 50.492, P < 0.001).
FluB showed a higher detection rate in school-age children and a lower detection rate in infants and toddlers, with statistically significant differences among the age groups (χ2 = 19.262, P < 0.001). FluA and ADV showed no statistically significant differences in detection rates among different age groups in children (χ2 = 2.521, P = 0.472; χ2 = 4.241, P = 0.237) (Table 1).
The overall detection rate in boys was 30.33% (95% CI 29.15–31.52%), while that in girls was 27.92% (95% CI 26.70–29.16%). The difference in overall detection rates between the sexes was statistically significant (χ2 = 7.648, P = 0.006). However, no significant difference in the detection rates of individual pathogens were found between boys and girls in univariate chi-square analysis.
Comparison of detection rates of six respiratory pathogens in children during 2022–2023
In 2022, of a total of 1625 cases, 593 cases were positive for at least one of the six common respiratory pathogens, with a detection rate of 36.49% (95% CI 34.14–38.88%). In 2023, of a total of 9,431 cases, 2,635 cases were positive for at least one of the six common respiratory pathogens, with a detection rate of 27.94% (95% CI 27.04–28.86%). The overall detection rate in 2022 was statistically significantly higher than that in 2023 (χ2 = 48.634, P < 0.001). Further analysis of the detection rates of individual pathogens between these two years showed that the detection rates of RSV, ADV, and PIV1 were significantly higher in 2022 than in 2023 (χ2 = 31.772, P < 0.001; χ2 = 25.237, P < 0.001; χ2 = 151.887, P < 0.001). The detection rate of influenza B virus was statistically significantly higher in 2023 (3.66%) than in 2022 (0.00%, 0/1625; χ2 = 60.156, P < 0.001). There were no statistically significant differences in the detection rates of influenza A virus and parainfluenza virus type 3 between these two years (Table 2).
Seasonal characteristics of six respiratory pathogen infections in children
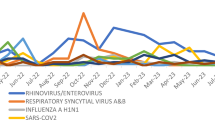
In the analysis of the seasonal prevalence characteristics of various pathogens in 2022 and 2023, we found that RSV had a high prevalence during the autumn and winter seasons of 2022, with an extremely low prevalence during the summer season, displaying a typical seasonal pattern. However, in 2023, RSV showed a year-round prevalence, with the highest detection rate in the summer season at 16.18% (95% CI 14.76–17.68%), followed by 14.61% (95% CI 13.25–16.05%) in winter, 13.09% (95% CI 10.97–15.45%) in spring, and 8.97% (95% CI 8.04–9.97%) in autumn. The seasonal prevalence pattern of RSV differed significantly between 2022 and 2023 (χ2 = 192.807, P < 0.001).
FluA showed a normal prevalence in the winter of 2022, with a relatively low prevalence in other seasons. However, in 2023, FluA had a high prevalence in the spring, at 13.20% (95% CI 11.08–15.56%), followed by 6.77% (95% CI 5.82–7.82%) in the winter, and the lowest prevalence, 0.28% (95% CI 0.11–0.57%), in the summer. The seasonal prevalence pattern of FluA differed significantly between 2022 and 2023 (χ2 = 34.277, P < 0.001).
FluB was not detected in 2022, but in 2023, a sudden outbreak of this virus occurred during the winter season, with a prevalence of 13.42% (95% CI 12.11–14.81%), but with an extremely low prevalence in the other three seasons. The seasonal prevalence of FluB differed significantly between 2022 and 2023 (χ2 = 654.487, P < 0.001).
PIV-3 was prevalent during the summer and autumn seasons of 2022, but in 2023, its prevalence was 10.08% (95% CI 8.93–11.32%) during only the summer season, with a low prevalence in the other three seasons. The seasonal prevalence pattern of PIV-3 differed significantly between 2022 and 2023 (χ2 = 109.484, P < 0.001).
PIV-1 was prevalent during the autumn seasons of 2022 and 2023: 9.47% (95% CI 7.59–11.63%) and 2.13% (95% CI 1.68–2.67%), respectively. The seasonal prevalence pattern of PIV-1 differed significantly between these years (χ2 = 19.009, P < 0.001).
ADV had a relatively high prevalence during the autumn and winter seasons of 2022, but in 2023, it had a high prevalence during the winter season, at 4.71% (95% CI 3.92–5.61%), whereas its prevalence was low in the other three seasons. The seasonal prevalence pattern of ADV differed significantly between the two years (χ2 = 42.837, P < 0.001) (Fig. 1).
Further analysis was conducted in this study to investigate the distribution characteristics of seasons of six common pediatric respiratory pathogens during the year 2023. Results revealed significant differences in the seasonal detection rates of these respiratory pathogens throughout the year (p < 0.001).The distribution of six pathogens in the four seasonal groups in 2023 was shown in Table 2.
Multivariate regression analysis of the impact of factors influencing six-panel respiratory pathogen infections
Using multivariate logistic regression analysis, we examined the relationship between each pathogen and age, sex and seasons. Pearson’s correlation coefficients express the linear relation between variables. The data showed no correlation between any two pathogens, with coefficients consistently ≤ 0.1 (Additional file 2). For RSV, the odds ratio (OR) for age was 0.622 (95% CI 0.601–0.644), indicating that a younger age was associated with a higher risk of RSV infection. For PIV-3, the OR for age was 0.622 (95% CI 0.587–0.660), also suggesting that younger age is associated with a higher risk of PIV-3 infection. For PIV-1, the OR for age was 0.782 (95% CI 0.725–0.842), indicating that a younger age was associated with a higher risk of PIV-1 infection. For FluA and ADV, after multivariate logistic regression analysis, the ORs for age were 0.961 (95% CI 0.931–0.991) and 0.953 (95% CI 0.911–0.997), respectively, suggesting that younger children were only slightly more at risk of FluA and ADV infections, although the differences were still statistically significant (Wald = 6.311, P = 0.012; Wald = 4.461, P = 0.035, respectively). However, in the case of FluB, no significant correlation with age was found by logistic regression analysis, indicating that age does not notably affect the risk of FluB infection. In multivariate logistic regression analysis, sex group exhibited statistically non-significant association (Table 3).
The results of multiple logistic regression analysis showed that the OR for RSV infection in the summer and winter seasons were 1.486 (95% CI 1.175–1.880) and 2.046 (95% CI 1.607–2.606), respectively. Thus, RSV exhibited a bimodal seasonal pattern, with peaks in the summer and winter of 2023. For FluA virus, the ORs for seasonal infection were 0.021 (95% CI 0.010–0.045) in summer, 0.319 (95% CI 0.246–0.413) in autumn, and 0.622 (95% CI 0.481–0.804) in winter. Compared with the peak of infections in the spring season, the other three seasons were considered low-risk fact periods. For FluB, the OR for winter infection was 73.930 (95% CI 18.336–298.072), indicating a high risk of outbreaks. For human PIV-3, the ORs for the summer and autumn seasons were 6.166 (95% CI 3.812–9.973) and 2.449 (95% CI 1.480–4.051), respectively, with the highest risk of PIV-3 infection occurring in the summer season. Human PIV-1 predominantly circulated during autumn and winter seasons, with ORs for autunm and winter seasons were 26.690 (95% CI 3.690–193.051) and 14.230 (95% CI 1.914–105.824). ADV had a high prevalence in the winter season, with an OR of 3.466 (95% CI 1.999–6.011) (Table 3).
Diagnostic value of the six-panel respiratory pathogen detection assay for LRTIs and severe pneumonia
In total, 9431 children were enrolled as study participants for 2023. As shown in Additional File 3, the positivity rate for the six pathogens in patients with URTIs (32.62%, 95% CI 29.27–36.10%) was significantly higher than that observed in patients with LRTIs (27.53%, 95% CI 26.59–28.48%; P < 0.001). We designated one as the outcome for LRTIs and 0 as the outcome for URTIs, considering them as the dependent variables. Additionally, we considered the six pathogens, age, sex, and season of onset as independent variables for conducting binary logistic regression analysis. As shown in Table 4, RSV (OR 9.156. 95% CI 5.905–14.195, P < 0.001) and PIV-3 (OR 1.683, 95% CI 1.133–2.501, P = 0.010) were identified as significant risk factors for LRTIs, while FluA (OR 0.347, 95% CI 0.263–0.456, P < 0.001), FluB (OR 0.421, 95% CI 0.286–0.619, P < 0.001), and ADV (OR 0.249, 95% CI 0.171–0.361, P < 0.001) were more likely to cause URTIs. In 2023, compared to spring, the likelihood of LRTIs was higher during summer (OR 3.996, 95% CI 3.199–4.993, P < 0.001), autumn (OR 9.181, 95% CI 7.179–11.741, P < 0.001), and winter (OR 5.385, 95% CI 4.223–6.867, P < 0.001).
An older age was found to be associated with a higher risk of developing LRTIs (OR 1.296, 95% CI 1.253–1.340, P < 0.001). Girls were more likely to experience such infections (OR 1.254, 95% CI 1.063–1.479, P = 0.007) (Table 4). Notably, PIV-1 did not show any differences in its association with LRTIs or URTIs.
We enrolled 8,680 pediatric patients clinically diagnosed with LRTIs, such as pneumonia, in 2023 and categorized them into two groups based on the severity of pneumonia: severe pneumonia and non-severe pneumonia. The overall positivity rate for the six pathogens in the severe pneumonia group was 25.23% (56/222), whereas that in the non-severe pneumonia group was 27.60% (2334/8458). The positivity rate for RSV in the severe pneumonia group was higher than that in the non-severe pneumonia group (χ2 = 4.051, P = 0.044). Single-factor analysis revealed that RSV infection was significantly associated with the development of severe pneumonia (OR 1.447, 95% CI 1.025–2.042) (Additional File 3).
Binary logistic regression analysis was conducted to determine the factors associated with severe pneumonia. In this study, patients with severe pneumonia were defined as 1, and those with non-severe pneumonia, as 0. The independent variables included the six pathogens, along with age, sex, and season. PIV-3 infection was associated with non-severe pneumonia (OR 0.391, 95% CI 0.170–0.899, P = 0.027). The incidence of severe pneumonia was influenced by both season and age, with a higher occurrence observed during summer (OR 1.982, 95% CI 1.117–3.519, P = 0.019). Additionally, younger age groups exhibited a higher risk for this condition (OR 0.938, 95% CI 0.893–0.986, P = 0.012) (Table 5). After adjusting for seasonal and age-related factors, multivariate regression analysis revealed no statistically significant difference in RSV infection between non-severe and severe pneumonia.
Evaluation of the diagnostic performance of the six-panel respiratory pathogen panel using tNGS
We randomly selected 138 patients from 8680 clinical cases diagnosed with LRTI in 2023. Among them, 77 patients tested positive in the respiratory panel test (including 12 cases of mixed infections), while 61 tested negative. These cases were verified using tNGS of respiratory pathogens. The agreement between the multiplex respiratory panel test and tNGS overall and for each of the viruses are shown in Table 6.
Discussion
In this large-sample real-world cohort study, we found that, among the 11,056 pediatric specimens tested, the total positivity rate for the six different respiratory viruses was 29.20% (95% CI 28.35–30.06%). In descending order, the positivity rates were 13.58% for RSV, 4.79% for FluA, 4.52% for PIV-3, 3.12% for FluB, 2.67% for ADV, and 1.90% for PIV-1. RSV and PIV-3 detection rates differed significantly across age groups (both P < 0.001), and were more common in younger children. PIV-1 was more common in infants, toddlers, and preschoolers than in school-age children (P < 0.001). FluB was predominantly detected in school-age children (P < 0.001). RSV-, ADV-, and PIV-1-positivity rates were higher in 2022 than in 2023. Seasonal viral patterns differed across years. RSV (OR 9.156. 95% CI 5.905–14.195) and PIV-3 (OR 1.683, 95% CI 1.133–2.501) were risk factors for lower respiratory tract infections. RSV-positivity was associated with severe pneumonia (P = 0.044). PIV-3 (OR 0.391, 95% CI 0.170–0.899), summer season (OR 1.982, 95% CI 1.117–3.519), and younger age (OR 0.938, 95% CI 0.893–0.986) influenced pneumonia severity. Multiplex RT-qPCR showed good diagnostic performance.
In a previous study, 30.67% of children with ARTIs in Shanghai were positive for at least one respiratory virus [11], similar to our findings. The positivity rate for respiratory viruses in boys was higher than that in girls, suggesting that boys may be more susceptible to viral infections than girls, possibly because their higher activity levels increase their risk of infection. This was consistent with the findings of previous studies [12, 13].
The incidence of respiratory viral infections decreased with age. The positivity rate was highest among the infant and toddler groups, which could be attributed to the immaturity of the immune system in children under 3 years old. RSV is a prominent pathogen responsible for respiratory infections in children under the age of 5 years [14]. This study demonstrated the extensive prevalence of RSV infections among children aged ≤ 3 years, with a subsequent decline in infection rates as children grow older [15]. Our study showed that the positivity rate for PIV-3 was third after that for RSV and FluA. The age distribution of PIV-3 infections closely resembled that of RSV infection, with infants predominantly being affected. Different PIV subtypes have different epidemiological characteristics. The incidence rate of PIV-1 was lower than that of PIV-3, with the highest positivity rates observed among preschool-aged children, which was consistent with previous research findings [16, 17]. Our study provided novel evidence of the epidemiological characteristics of these 6 respiratory viruses in this region of China following NPI relaxation.
Autumn and winter are typically characterized by a higher incidence of respiratory pathogens, primarily due to the diminished local resistance of the children's respiratory mucosa caused by low temperatures [18]. Additionally, the enhanced stability of viruses in colder environments facilitates their invasion of the respiratory tract [19]. However, the findings of this study indicated that the seasonal prevalence of RSV in 2022 closely aligned with that of pre-COVID-19 outbreak reports and epidemic patterns [20]. RSV infections were infrequent during summer months and peaked during winter. In contrast, an atypical pattern emerged in 2023, characterized by a significantly higher infection rate, of up to 16.18%, during summer followed by a sustained high prevalence throughout the year. Following the relaxation of NPIs, Germany and the United States, among other countries, experienced an off-season RSV epidemic [21, 22]; moreover, New Zealand and Australia observed a more than five-fold increase in RSV incidence as compared to pre-epidemic levels [23]. Additionally, based on 2 years of monitoring data, PIV-3 was found to be mostly prevalent during the summer season [17]. Both PIV-3 and RSV are highly prevalent among children under the age of 3 years. Given the seasonal outbreak pattern observed for RSV in 2023, our data suggest that is important to conduct differential diagnostic tests to distinguish between RSV and PIV-3. The highest prevalence of PIV-1 was observed during autumn. The FluA epidemic reached its peak in the spring of 2023. Based on influenza surveillance data from China, a wave of influenza epidemics dominated by subtype A (H1N1) emerged between mid-February and late April 2023 [24, 25]. This analysis attributed the occurrence of this winter-like outbreak to the relaxation of NPIs in late 2022, resulting in a significant number of individuals being infected with COVID-19 from December 2022 to January 2023, which subsequently delayed the onset of the FluA virus infection from February to April 2023. Furthermore, an outbreak of FluB occurred during the winter of 2023, indicating the need for close monitoring of changes in virus epidemiology.
Univariate analysis revealed that RSV and PIV-1 exhibited a predisposition towards LRTIs, while multivariate analysis identified RSV and PIV-3 as the primary influential factors for such infections. PIV infection can lead to both URTIs and LRTIs, including common cold, pertussis, bronchitis, bronchiolitis, and pneumonia [26]. PIV-1 is the leading cause of laryngitis, while PIV-3 exhibits a predilection for inducing LRTIs, such as bronchiolitis and pneumonia, particularly in the pediatric population [26, 27]. The likelihood of developing URTIs increases when individuals are infected with FluA, FluB, and ADV. The findings of a study conducted in Belgium indicated that children who contract an influenza virus infection exhibit a comparatively lower risk of developing associated complications than those infected with other respiratory viruses [28]. According to a hospital-based surveillance study, the risk of severe ADV infection in Vietnamese children was consistently low [29]. In addition, risk factors such as young age, season, and sex were significantly associated with LRTIs.
Our study indicated that RSV is more commonly found in patients with severe pneumonia than in those with non-severe pneumonia, and that RSV is a high-risk factor for the development of severe pneumonia. These findings were consistent with those of previous studies [30,31,32]. The pathogenesis of RSV infection involves a complex interplay among multiple pathogenic factors, immune system responses, and environmental influences that exacerbate the disease in children [33]. Binary logistic regression analysis suggested that PIV-3 was associated with non-severe pneumonia as the predominant manifestation of LRTIs. A case–control study conducted in seven countries across Africa and Asia revealed that approximately 60% of children requiring hospitalization for severe pneumonia were infected with viruses, of which PIV accounted for approximately 5%. Moreover, the proportion of PIV-3 cases varied according to the severity of pneumonia. Notably, the prevalence of PIV-3 was lower in patients with severe pneumonia than in those without severe pneumonia [33]. However, no comparison has been made with individuals suffering from non-severe pneumonia. Further research is required to address this issue. Additionally, young age and the summer season were identified as significant risk factors for severe pneumonia.
The six-panel respiratory pathogen detection assay is a testing kit that has been approved for listing by the National Medical Products Administration in China However, further validation is needed to assess its methodological performance. We conducted comparative verification experiments using the tNSG methodology.tNGS technology can detect more than 100 common respiratory infectious pathogens, including viruses, bacteria, and Mycoplasma pneumoniae. Additionally, it can be used to identify specific viral genotypes. A total of 138 patients were randomly selected for tNGS in this study. The results demonstrated a high level of concordance between tNGS and multiplex RT-qPCR, with 133 cases exhibiting identical detection outcomes for all six pathogens. The overall concordance rate was 96.38%, indicating that multiplex RT-qPCR is a valuable diagnostic tool that is fast and accurate, and covers target genes commonly found in six respiratory viruses.
This study had several limitations. First, it was a retrospective study conducted at a single center, which may restrict the generalizability of the findings. Future studies should consider recruiting participants from multiple centers to enhance the external validity of the results. Second, although the study focused on six common respiratory viruses, it did not include several other common respiratory infectious pathogens, such as SARS-CoV-2, rhinovirus, and Mycoplasma pneumoniae. Third, our data provided only a snapshot of 2022–2023, emphasizing the importance of evaluating long-term trends in positive rates in large samples across different age groups.
Conclusion
This study conducted a comparative analysis of the prevalence and clinical diagnostic value of six common respiratory pathogens in children in the Hangzhou area before and after the adjustment for epidemic prevention and control strategies in 2022–2023, with a large sample size. The overall detection rate of the six common respiratory pathogens in children was 29.20% (95% CI 28.35–30.06%), and the prevalence of each pathogen was determined. The six most common respiratory pathogens in children had unique characteristics in terms of age and season of infection. RSV, PIV-3, and PIV-1 were more common in infants and young children, with a higher probability of infection at younger ages. A slight negative correlation between FluA and ADV infection and age was found, with a slightly higher infection rate in the younger age groups. The prevalence rates and seasonal characteristics of the six pathogens differed significantly across 2022 and 2023. RSV showed a seasonal anomaly with a peak in the summer of 2023, and seasonal peaks in both summer and winter. PIV-3 and PIV-1 showed relatively stable peaks in summer and autumn, respectively. Therefore, the seasonal characteristics of FluA, FluB, and ADV should constantly be monitored. RSV and PIV-3 are high-risk factors for LRTIs, whereas FluA, FluB, and ADV are more likely to cause URTIs. Young age and summer season were high risk factors for severe pneumonia, whereas PIV-3 was a low-risk factor for severe pneumonia. The six-panel respiratory pathogen assay yields a high sensitivity and specificity, making it particularly suitable for the clinical diagnosis of respiratory tract infections in young children.
Availability of data and materials
No datasets were generated or analysed during the current study.
Abbreviations
- ADV:
-
Adenovirus
- ARTIs:
-
Acute respiratory tract infections
- CI:
-
Confidence interval
- Flu:
-
Influenza virus
- LRTI:
-
Lower respiratory tract infection
- NPIs:
-
Non-pharmacological interventions
- NPV:
-
Negative-predictive value
- OR:
-
Odds ratio
- PIV:
-
Parainfluenza virus
- PPV:
-
Positive-predictive value
- RSV:
-
Respiratory syncytial virus
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
- tNGS:
-
Targeted next-generation sequencing
- URTI:
-
Upper respiratory tract infection
References
Global Burden of Disease Child and Adolescent Health Collaboration; Kassebaum N, Kyu HH, Zoeckler L, Olsen HE, Thomas K, et al. Child and adolescent health from 1990 to 2015: findings from the Global Burden of Diseases, Injuries, and Risk Factors 2015 Study. JAMA Pediatr. 2017;171:573–92.
Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128.
Makrinioti H, Hasegawa K, Lakoumentas J, Xepapadaki P, Tsolia M, Castro-Rodriguez JA, et al. The role of respiratory syncytial virus- and rhinovirus-induced bronchiolitis in recurrent wheeze and asthma-A systematic review and meta-analysis. Pediatr Allergy Immunol. 2022;33:e13741.
van Meel ER, Mensink-Bout SM, den Dekker HT, Ahluwalia TS, Annesi-Maesano I, Arshad SH, et al. Early-life respiratory tract infections and the risk of school-age lower lung function and asthma: a meta-analysis of 150 000 European children. Eur Respir J. 2022;60:2102395.
Van den Bruel A, Haj-Hassan T, Thompson M, Buntinx F, Mant D; European Research Network on Recognising Serious Infection investigators. Diagnostic value of clinical features at presentation to identify serious infection in children in developed countries: a systematic review. Lancet. 2010;375:834–45.
van Houten CB, Cohen A, Engelhard D, Hays JP, Karlsson R, Moore E, et al. Antibiotic misuse in respiratory tract infections in children and adults-a prospective, multicentre study (TAILORED Treatment). Eur J Clin Microbiol Infect Dis. 2019;38:505–14.
Li ZJ, Zhang HY, Ren LL, Lu QB, Ren X, Zhang CH, et al. Etiological and epidemiological features of acute respiratory infections in China. Nat Commun. 2021;12:5026.
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–33.
Spinelli MA, Glidden DV, Gennatas ED, Bielecki M, Beyrer C, Rutherford G, et al. Importance of non-pharmaceutical interventions in lowering the viral inoculum to reduce susceptibility to infection by SARS-CoV-2 and potentially disease severity. Lancet Infect Dis. 2021;21:e296–301.
Li M, Cong B, Wei X, Wang Y, Kang L, Gong C, et al. Characterising the changes in RSV epidemiology in Beijing, China during 2015–2023: results from a prospective, multi-centre, hospital-based surveillance and serology study. Lancet Reg Health West Pac. 2024;45:101050.
Zhang L, Wang Y, Xu H, Hao L, Zhao B, Ye C, et al. Prevalence of respiratory viruses in children with acute respiratory infections in Shanghai, China, from 2013 to 2022. Influenza Other Respir Viruses. 2024;18:e13310.
Duan Y, He J, Cui Y, Li W, Jiang Y. Characteristics and forecasting of respiratory viral epidemics among children in west China. Medicine (Baltimore). 2021;100:e25498.
Gao Z-X, Wang Y, Yan L-Y, Liu T, Peng L-W. Epidemiological characteristics of respiratory viruses in children during the COVID-19 epidemic in Chengdu, China. Microbiol Spectr. 2024;12:e0261423.
Li Y, Wang X, Blau DM, Caballero MT, Feikin DR, Gill CJ, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet. 2022;399:2047–64.
Wan L, Li L, Zhang H, Liu C, Li R, Wu X, et al. The changing pattern of common respiratory viruses among children from 2018 to 2021 in Wuhan. China Arch Virol. 2023;168:291.
Li L, Jia R, Zhang Y, Sun H, Ma J. Changes of parainfluenza virus infection in children before and after the COVID-19 pandemic in Henan. China J Infect. 2023;86:504–7.
Gao Y, Ma Y, Feng D, Zhang F, Wang B, Liu X, et al. Epidemiological characteristics of human parainfluenza viruses infections—China, 2019–2023. China CDC Wkly. 2024;6:235–41.
Liang J, Wang Z, Liu Y, Zeng L, Li Z, Liang J, et al. Epidemiology and co-infection patterns in patients with respiratory tract infections in southern China between 2018 and 2020. J Infect. 2021;83:e6-8.
Sauni R, Verbeek JH, Uitti J, Jauhiainen M, Kreiss K, Sigsgaard T. Remediating buildings damaged by dampness and mould for preventing or reducing respiratory tract symptoms, infections and asthma. Cochrane Database Syst Rev. 2015;2015:CD007897.
Zhang Y, Yuan L, Zhang Y, Zhang X, Zheng M, Kyaw MH. Burden of respiratory syncytial virus infections in China: systematic review and meta-analysis. J Glob Health. 2015;5:020417.
Abu-Raya B, Viñeta Paramo M, Reicherz F, Lavoie PM. Why has the epidemiology of RSV changed during the COVID-19 pandemic? EClinicalMedicine. 2023;6(61):102089.
Billard MN, Bont L. RSV immunisation: lessons from the COVID-19 pandemic. Lancet Child Adolesc Health. 2023;7:147–9.
Hatter L, Eathorne A, Hills T, Bruce P, Beasley R. Respiratory syncytial virus: paying the immunity debt with interest. Lancet Child Adolesc Health. 2021;5:e44–5.
Du Z, Shao Z, Zhang X, Chen R, Chen T, Bai Y, et al. Nowcasting and forecasting seasonal influenza epidemics—China, 2022–2023. China CDC Wkly. 2023;5:1100–6.
Wei M, Li S, Lu X, Hu K, Li Z, Li M. Changing respiratory pathogens infection patterns after COVID-19 pandemic in Shanghai. China J Med Virol. 2024;96:e29616.
Branche AR, Falsey AR. Parainfluenza virus infection. Semin Respir Crit Care Med. 2016;37:538–54.
Howard LM, Edwards KM, Zhu Y, Williams DJ, Self WH, Jain S, et al. Parainfluenza virus types 1–3 infections among children and adults hospitalized with community-acquired pneumonia. Clin Infect Dis. 2021;73:e4433–43.
Fischer N, Moreels S, Dauby N, Reynders M, Petit E, Gérard M, et al. Influenza versus other respiratory viruses—assessing severity among hospitalised children, Belgium, 2011 to 2020. Euro Surveill. 2023;28:2300056.
Althouse BM, Flasche S, Toizumi M, Nguyen HT, Vo HM, Le MN, et al. Differences in clinical severity of respiratory viral infections in hospitalized children. Sci Rep. 2021;11:5163.
Sun YP, Zheng XY, Zhang HX, Zhou XM, Lin XZ, Zheng ZZ, et al. Epidemiology of respiratory pathogens among children hospitalized for pneumonia in Xiamen: a retrospective study. Infect Dis Ther. 2021;10:1567–78.
Li Y, Zhai Y, Lin Y, Lu C, He Z, Wu S, et al. Epidemiology of respiratory syncytial virus in hospitalized children with community-acquired pneumonia in Guangzhou: a 10-year study. J Thorac Dis. 2023;15:967–76.
Pneumonia Etiology Research for Child Health (PERCH) Study Group. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet. 2019;394:757–79.
Rossi GA, Colin AA. Respiratory syncytial virus-Host interaction in the pathogenesis of bronchiolitis and its impact on respiratory morbidity in later life. Pediatr Allergy Immunol. 2017;28:320–31.
Acknowledgements
Not applicable.
Funding
This study was supported by the Zhejiang Provincial Health and Medical Science and Technology Plan Project (2022KY1011) and Hangzhou Science and Technology Bureau Biomedical and Health Industry Development Support Science and Technology Special Project (2022WJC126).
Author information
Authors and Affiliations
Contributions
Yidong Wu and Lizhong Du conceived and designed the study. Yidong Wu, Jun Zhou, Ting Shu, and Wei Li performed the experiments. Yidong Wu wrote the original draft. Lizhong Du and Shiqiang Shang supervised the study and revised the manuscript. All the authors have personally reviewed and given final approval of the submitted version of this manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study adhered to the tenets of the Declaration of Helsinki and was approved by the Hangzhou Children's Hospital Clinical Research Ethics Committee (Protocol Number: No. 2021-IRB-74). Informed consent was not required due to the nature of the study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, Y., Zhou, J., Shu, T. et al. Epidemiological study of post-pandemic pediatric common respiratory pathogens using multiplex detection. Virol J 21, 168 (2024). https://doi.org/10.1186/s12985-024-02441-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12985-024-02441-8