Abstract
In recent years, with the development of RNA sequencing technology and bioinformatics methods, the epigenetic modification of RNA based on N6-methyladenosine (m6A) has gradually become a research hotspot in the field of bioscience. m6A is the most abundant internal modification in eukaryotic messenger RNAs (mRNAs). m6A methylation modification can dynamically and reversibly regulate RNA transport, localization, translation and degradation through the interaction of methyltransferase, demethylase and reading protein. m6A methylation can regulate the expression of proto-oncogenes and tumor suppressor genes at the epigenetic modification level to affect tumor occurrence and metastasis. The morbidity and mortality of esophageal cancer (EC) are still high worldwide. Esophageal squamous cell carcinoma (ESCC) is the most common tissue subtype of EC. This article reviews the related concepts, biological functions and recent advances of m6A methylation in ESCC, and looks forward to the prospect of m6A methylation as a new diagnostic biomarker and potential therapeutic target for ESCC.
Similar content being viewed by others
Introduction
Esophageal cancer (EC) is one of the most invasive malignant tumors of digestive tract in the world, and its morbidity and mortality are still high in China [1]. 90% of the histopathological types of esophageal cancer are esophageal squamous cell carcinoma (ESCC) [2]. EC is caused by a variety of causes, among which genetic and epigenetic modifications play a key role in the occurrence and development of ESCC [3, 4]. In recent years, although surgical resection, combined radiotherapy and chemotherapy have improved the prognosis of patients with ESCC, the 5-year overall survival rate is still very low [5], between 20 and 30% [6]. Therefore, there is an urgent need to find new diagnostic biomarkers and potential therapeutic targets for ESCC patients.
In recent years, with the continuous development of tumor epigenetics, N6-methyladenine (m6A) has not only initiated a new era of post-transcriptional gene regulation in eukaryotes, but also rapidly become a research hotspot in the field of RNA methylation modification. As a reversible RNA methylation modification, m6A methylation is dynamically regulated by a variety of regulatory factors [7]. The imbalance of m6A methylation regulators changes the biological functions of cell proliferation, migration and invasion, and finally leads to the occurrence and development of tumor.
This article reviews the related concepts and biological functions of m6A methylation and the research progress in ESCC. In addition, it also emphasizes the prospect of m6A methylation as a new diagnostic biomarker and potential therapeutic target for ESCC.
Related concepts and biological functions of m6A methylation
m6A refers to the methylation at the N6 position of adenosine, which is mainly concentrated in the 3'untranslated region near the mRNA Terminator. m6A modification mainly occurs in the RRm6ACH sequence [8]. As one of the most common and abundant internal modifications in mammals and eukaryotes [9], m6A RNA modification involves almost all aspects of RNA metabolism [10]. Its regulation process is mainly dynamically and reversibly regulated by a variety of regulatory factors. The methylated proteins involved in m6A methylation are mainly methyltransferases, demethylases and RNA binding proteins [11,12,13]. They can add, remove or give priority to recognize m6A sites, which play a key regulatory role in the expression of the whole genome and have a great impact on normal physiological function or pathological status [14].
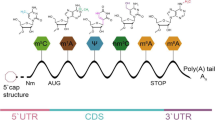
m6A is involved in many cellular RNA processes, including transcription, splicing, nuclear transport, translation and degradation [12, 15]. By affecting the stability and half-life of mRNA, m6A regulates gene expression and regulates important biological functions such as mammalian reproductive function, circadian rhythm, adipogenesis and human lifespan [16]. Its interference may affect gene table regulation and cell biological function [17], and indirectly affect the stability and half-life of mRNA, leading to tumors and many diseases. The basic mechanism of m6A methylation in RNA is shown in Fig. 1.
Basic mechanism of m6A methylation in RNA. Basic mechanism of m6A. The m6A methylation is catalyzed by the writer complex including METTL3, METTL14, WTAP, VIRMA, RBM15, ZC3H13 and CBLL1. The m6A modification is removed by demethylase FTO or ALKBH5. Reader proteins recognize m6A and determine target RNA nature
m6A modified writing gene (writers)
The written genes are various methyltransferases that promote m6A RNA methylation modification. At present, the components identified by m6A methyltransferase include methyltransferase like 3 (METTL3), METTL14, Wilms' tumor 1 associated protein (WTAP), and Virilizer like m6A methyltransferase associated protein (VIRMA/KIAA1429), RNA binding motif protein 15 (RBM15), zinc finger CCCH domain-containing protein 13 (ZC3H13), Casitas B-lineage lymphoma-transforming sequence-like protein 1, CBLL1/HAKAI).
Methyltransferase mainly forms stable complexes through core proteins such as METTL3, METTL14 and WTAP. m6A methylation occurs on the bases of mRNA [18], in which METTL3 is a subunit with catalytic activity, METTL14 is responsible for recognizing the substrate, WATP is mainly responsible for assisting METTL3 and METTL14 to target to nuclear spots, and WTAP has independent methylation sites, which can make some m6A sites specific methylation. VIRMA/KIAA1429, another component of methyltransferase complex, is a protein involved in alternative splicing and interacts with WTAP.
m6A modified eraser gene (erasers)
The erase gene can remove the m6A modification in the RNA molecule by encoding m6A demethylase, which is the key to the reversible process of m6A modification [19]. m6A demethylase has been identified as AlkB homolog 5 (ALKBH5) [20] and fat mass and obesity associated (FTO) [21]. Although they have similar functions, but the action process is different.
ALKBH5 can catalyze the m6A modification demethylation of RNA in vitro [20], which can significantly affect the output of mRNA and RNA metabolism as well as the assembly of mRNA processing factors in nuclear spots. FTO has the function of demethylation of m6A on single-stranded DNA and RNA, and they are highly expressed in fat, brain and hypothalamus [22], which is vital to metabolism.
m6A modified reading protein (readers)
The protein that selectively binds to the post-transcriptional product of m6A is called m6A reading protein. At present, the research on m6A modified reading protein is mainly focused on the YT521-B homology domain family (YTHDF), YT521-B homology domain containing (YTHDC), heterogeneous nuclear ribonucleoprotein binding protein (HNRNP), and insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2), eukaryotic translation initiation factor (eIF3) and ELAV like RNA binding protein 1 (ELAVL1/HuR).
YTHDC1 may affect the splicing of mRNA and its output from the nucleus [15]. YTHDF2, YTHDF3 and YTHDC2, such as YTHDF2, which promote the degradation of mRNA, can selectively bind to methylated mRNA, participate in the storage and degradation of mRNA, affect the decay process of mRNA, and may have a positive effect on human lifespan [23]. It is IGF2BP1/2/3 that maintains the stability of mRNA [24], and it is YTHDF1, YTHDF3, YTHDC2 and IGF2BP1/2/3, that promote the translation of target mRNA. Among them, YTHDF3 can assist YTHDF1 to jointly promote the translation of related mRNA [25].
The role and significance of m6A methylation in ESCC
Studies have shown that m6A modified mRNA is maladjusted in many cancers, and its role in cancer has been gradually confirmed in vivo and in vitro, not only non-coding "writing genes", "erasing genes" and "reading genes", but also other protein factors, including oncogenes, transcription factors and signal transduction factors. the overexpression or consumption of these m6A-related factors may change the m6A modification in the tumor and interfere with the progression of cancer. The role of m6A regulatory factor in ESCC is shown in Table 1. Therefore, to clarify the molecular mechanism of these changes of m6A modified RNA and identifying the abnormal expression of m6A regulatory factors in clinical biopsy specimens. It is of great significance for early diagnosis of tumors, prediction of tumor prognosis and provision of new approaches of tumor treatment.
m6A modification related protein was downregulated in ESCC.
ALKBH5
Previous studies [26] have found that, the expression of ALKBH5 in EC tissue decreased. Functional analysis showed that ALKBH5 could inhibit the proliferation, migration and invasion of EC cells. However, a recent study found that ALKBH5 promotes the proliferation and migration of ESCC [27]. These results contradict previous findings. The reason may be that this study did not compare the differential expression patterns between ESCC and normal esophageal tissue, but only detected the expression of ALKBH5 in ESCC. The results of the cancer genome atlas (TCGA) database through Gene Expression Profiling Interactive Analysis (GEPIA) online tool show that the overall survival time of patients with high expression of ALKBH5 is longer than that of patients with low expression, indicating that ALKBH5 plays a tumor inhibitory role in EC [28]. ALKBH5 regulates cell proliferation, migration, invasion, tumor progression, metastasis, tumorigenesis and chemotherapy resistance might by regulating m6A methylation.
YTHDC2
Yang et al. [29] through the study of the database, it was observed that the expression of YTHDC2 was downregulated in esophageal cancer. In the proliferation experiment, it was found that the low expression of YTHDC2 significantly promoted the growth of cells, suggesting that YTHDC2 may play a role as a tumor suppressor in ESCC. In the further enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, it was observed that several pathological pathways related to ESCC, including p53 signal pathway, NF-kappaB signal pathway and JAK-STAT signal pathway, were significantly rich in downregulated genes, thus promoting the proliferation of ESCC cells.
m6A modification related protein expression upregulated in ESCC
METTL3
METTL3 is the core catalytic component of methyltransferase complex. Xia et al. [30] detected the expression of METTL3 in 207 patients with ESCC. The results of open data set and immunohistochemistry showed that the expression of METTL3 in tumor tissues was up-regulated compared with normal tissues adjacent to cancer, and the higher the expression level of METTL3 was, the worse the survival time was. In addition, it was also found that the expression level of METTL3 was an independent predictor of disease-free survival and overall survival in patients with ESCC. Another study [31] showed that small interfer RNA (siRNA) gene knockout of METTL3 inhibited the survival, colony formation, migration and invasion of EC cells, induced apoptosis, and significantly inhibited the malignant phenotype of ESCC cells by down-regulating PI3K/AKT signal pathway. In conclusion, METTL3 is a good predictor of ESCC and can be used as a potential biomarker for the prognosis of ESCC.
FTO
A study [32] through immunohistochemistry and data mining of 80 pairs of ESCC tissues, it was found that the expression of FTO in ESCC tissues was higher than that in adjacent normal tissues. The survival curve shows that the high expression of FTO has the trend of poor prognosis. A large number of evidence in the study confirmed that the downregulated expression of FTO can significantly inhibit the proliferation and migration of ESCC. In addition, FTO promotes cell proliferation and migration in ESCC by upregulating matrix metallopeptidase 13 (MMP13). Therefore, FTO acts as a tumor promoter in the progression of ESCC.
HNRNPA2B1
Guo et al. [33] found that the expression of m6A and its regulatory factor HNRNPA2B1 was significantly increased in ESCC tissues, and the high expression of HNRNPA2B1 was positively correlated with tumor diameter and lymph node metastasis of ESCC. In addition, functional studies have shown that HNRNPA2B1 gene knockout inhibits the proliferation, migration and invasion of ESCC. In terms of mechanism, HNRNPA2B1 promotes the progress of ESCC by up-regulating the expression of fatty acid synthase ATP citrate lyase (ACLY) and aminocyclopropane- 1-carboxylate (ACC1), indicating that HNRNPA2B1, as a carcinogenic factor, promotes the progression of ESCC by accelerating fatty acid synthesis, and may become a prognostic biomarker and therapeutic target of ESCC.
In addition, ALKBH5 and HNRNPA2B1 are effective indicators for predicting Overall Survival (OS) in patients with ESCC. High expression of HNRNPA2B1 and low expression of ALKBH5 are risk factors for ESCC survival. The combination of these two factors shows better predictive ability than using these two factors alone.
HNRNPC
Studies have shown that HNRNPC is overexpressed in ESCC tissues, and its expression is negatively correlated with the overall survival of patients with ESCC [34]. The double gene prognostic markers composed of ALKBH5 and HNRNPC have been proved to be a good predictor of survival outcome in ESCC.
m6A methylation as a therapeutic strategy for ESCC
More and more studies have found that microRNAs [35, 36] and circular RNAs [37] can be used as potential biomarkers for prognosis, diagnosis and treatment of EC. However, due to the high morbidity and mortality of EC, it is necessary to find new anticancer drugs, such as curcumin [38], which can be potentially used in chemotherapy and chemoprevention of EC by regulating miRNAs. Allicin [39] can achieve its anticancer effect by inhibiting cell growth and inducing apoptosis.
As described in Table 1, m6A plays different regulatory roles in ESCC through different biological functions. In addition, a study [40] constructed and verified prognostic markers of ESCC based on m6A RNA methylation regulators, which may be a promising tool for predicting patient survival and provide important information for ESCC to make diagnosis and treatment strategies.
In recent years, many studies have revealed that m6A plays an important role in the formation of many kinds of tumors, such as breast cancer [41, 42], ovarian cancer [43, 44], cervical cancer [45], acute myeloid leukemia [46,47,48], glioblastomas [49,50,51], non-small cell lung cancer [52,53,54,55], hepatocellular cancer [56,57,58,59], gastric cancer [60], colorectal cancer [61], pancreatic cancer [62], etc. Furthermore, it is found that m6A modification plays a key role in tumor radiotherapy, chemotherapy and drug therapy. Table 2 summarizes the mechanism and drug resistance of m6A regulatory factors in related tumors, which may provide clinical reference value and significance for the treatment of esophageal cancer in the future.
Although many scholars have reported the study of m6A modification in tumor therapy, because the mechanism of tumor formation is very complex, accurate m6A targeting therapy needs to be explored. By changing the m6A level of some specific genes corresponding to mRNA in cells, it affects the expression of a series of downstream oncogenes or transcription factors. It is possible that regulating the level of m6A in tumor cells will become the entry point of tumor radiotherapy, chemotherapy and drug therapy [63, 64]. There is still a long way to go in the treatment of tumor in the future.
Conclusion
In summary, with the development of biological techniques such as high-throughput sequencing, the role of m6A methylation in ESCC has been gradually revealed, at present, it has been found that there are abnormal expressions of METTL3, ALKBH5, FTO, YTHDC2, HNRNPA2B1 and HNRNPC in ESCC, mainly by affecting the stability of mRNA, regulating cancer cell proliferation and affecting tumor cell metastasis and invasion.
The discovery of m6A opens a new way for the study of epigenetics and tumor-related diseases, but the study of m6A modification is still in its infancy and there are still many challenges. the aim is to further study the role of epigenetic network in the occurrence and development of ESCC and to strengthen the evaluation of the safety and effectiveness of m6A-related regulatory factors and pathways as new targets for tumor therapy. To further explore the correlation between m6A and drug sensitivity and long-term prognosis of patients with ESCC, and to realize the application of m6A from basic research to clinical drug development as soon as possible.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- m6A:
-
N6-methyladenosine
- mRNAs:
-
Messenger RNAs
- EC:
-
Esophageal cancer
- ESCC:
-
Esophageal squamous cell carcinoma
- METTL3:
-
Methyltransferase like 3
- WTAP:
-
Wilms' tumor 1 associated protein
- VIRMA:
-
Virilizer like m6A methyltransferase
- RBM15:
-
RNA binding motif protein 15
- ZC3H13:
-
Zinc finger CCCH domain-containing protein 13
- CBLL1:
-
Casitas B-lineage lymphoma-transforming sequence-like protein 1
- ALKBH5:
-
AlkB homolog 5
- FTO:
-
Fat mass and obesity associated
- YTHDF:
-
YT521-B homology domain family
- YTHDC:
-
YT521-B homology domain containing
- HNRNP:
-
Heterogeneous nuclear ribonucleoprotein binding protein
- IGF2BP2:
-
Insulin like growth factor 2 mRNA binding protein 2
- eIF3:
-
Eukaryotic translation initiation factor
- ELAVL1:
-
ELAV like RNA binding protein 1
- TCGA:
-
The cancer genome atlas
- GEPIA:
-
Gene Expression Profiling Interactive Analysis
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- SiRNA:
-
Small interfer RNA
- MMP13:
-
Upregulating matrix metallopeptidase 13
- ACLY:
-
ATP citrate lyase
- ACC1:
-
Aminocyclopropane- 1-carboxylate
- OS:
-
Overall survival
References
Abnet CC, et al. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. 2018;154:360–73.
Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2015;65(1):5–29.
Cheung WY, et al. Genetic variations in esophageal cancer risk and prognosis. Gastroenterol Clin North Am. 2009;38:75–91.
Hassanabad AF, Chehade R, Breadner D, Raphael J. Esophageal carcinoma: towards targeted therapies. Cell Oncol. 2020;43(2):195–209.
van Rossum PSN, Mohammad NH, Vleggaar FP, van Hillegersberg R. Treatment for unresectable or metastatic oesophageal cancer: current evidence and trends. Nat Rev Gastroenterol Hepatol. 2018;15:235–49.
Abbas G, Krasna M. Overview of esophageal cancer. Ann Car-diothorac Surg. 2017;6(2):131.
Zhao Y, Shi Y, Shen H, Xie W. m6A-binding proteins: the emerging crucial performers in epigenetics. Hematol Oncol. 2020;13:35.
Kane SE, Beemon K. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: Implications for RNA processing. Mol Cell Biol. 1985;5(9):2298–306.
Zhang C, Fu J, Zhou Y. A review in research progress concerning m6A methylation and immunoregulation. Front Immunol. 2019;10:922.
Bokar JA, et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3(11):1233–47.
Zhang S. Mechanism of N(6)-methyladenosine modification and its emerging role in cancer. Pharmacol Ther. 2018;189:173–83.
Liu N, Dai Q, Zheng G, et al. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4.
Meyer KD, Jaffrey SR. Rethinking m(6)A readers, writers, and erasers. Annu Rev Cell Dev Biol. 2017;33:319–42.
Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18(1):103.
Wang X, Lu Z, Gomez A, et al. N-6-methyladenosine -dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–20.
Parashar NC, Parashar G, Nayyar H, et al. N6-adenine DNA methylation demystified in eukaryotic genome: From biology to pathology. Biochimie. 2018;144:56–62.
Edupuganti RR, Geiger S, Lindeboom RGH, et al. N- 6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017;24(10):870.
Wang Y, Li Y, Toth JI, et al. N6- methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–8.
Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–200.
Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29.
Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity- associated FTO. Nat Chem Biol. 2011;7(12):885–7.
Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–72.
Cardelli M, Marchegiani F, Cavallone L, et al. A polymorphism of the YTHDF2 gene (1p35) located in an Alu-rich genomic domain is associated with human longevity. J Gerontol A Biol Sci Med Sci. 2006;61(6):547–56.
Huang H, Weng H, Sun W, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–95.
Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–28.
Xiao Y, Xue J, Zhang L, et al. Misalignment fault diagnosis for wind turbines based on information fusion. Entropy (Basel). 2021;23(2):243.
Nagaki Y, Motoyama S, Yamaguchi T, et al. m(6)A demethylase ALKBH5 promotes proliferation of esophageal squamous cell carcinoma associated with poor prognosis. Genes Cells. 2020;25(8):547–61.
Xue J, Xiao P, Yu X, Zhang X. A positive feedback loop between AlkB homolog 5 and miR-193a-3p promotes growth and metastasis in esophageal squamous cell carcinoma. Hum Cell. 2021;34(2):502–14.
Yang N, Ying P, Tian J, et al. Genetic variants in m6A modification genes are associated with esophageal squamous-cell carcinoma in the Chinese population. Carcinogenesis. 2020;41(6):761–8.
Xia TL, Li X, Wang X, et al. Upregulation of METTL3 expression predicts poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Manag Res. 2020;12:5729–37.
Hu W, Liu W, Liang H, et al. Silencing of methyltransferase-like 3 inhibits oesophageal squamous cell carcinoma. Exp Ther Med. 2020;20(6):138.
Liu S, Huang M, Chen Z, et al. FTO promotes cell proliferation and migration in esophageal squamous cell carcinoma through up-regulation of MMP13. Exp Cell Res. 2020;389(1):111894.
Guo H, Wang B, Xu K, et al. m6A reader HNRNPA2B1 promotes esophageal cancer progression via up-regulation of ACLY and ACC1. Front Oncol. 2020;10:553045.
Xu LC, Pan JX, Pan HD. Construction and validation of an m6A RNA methylation regulators-based prognostic signature for esophageal cancer. Cancer Manag Res. 2020;12:5385–94.
Pourhanifeh MH, Vosough M, Mahjoubin-Tehran M, et al. Autophagy-related microRNAs: possible regulatory roles and therapeutic potential in and gastrointestinal cancers. Pharmacol Res. 2020;161:105133.
Jamali L, Tofigh R, Tutunchi S, et al. Circulating microRNAs as diagnostic and therapeutic biomarkers in gastric and esophageal cancers. J Cell Physiol. 2018;233(11):8538–50.
Naeli P, Pourhanifeh MH, Karimzadeh MR, et al. Circular RNAs and gastrointestinal cancers: epigenetic regulators with a prognostic and therapeutic role. Crit Rev Oncol Hematol. 2020;145:102854.
Hesari A, Azizian M, Sheikhi A, et al. Chemopreventive and therapeutic potential of curcumin in esophageal cancer: current and future status. Int J Cancer. 2019;144(6):1215–26.
Sarvizadeh M, Hasanpour O, Naderi Ghale-Noie Z, et al. Allicin and digestive system cancers: from chemical structure to its therapeutic opportunities. Front Oncol. 2021;11:650256.
Xu LC, Pan JX, Pan HD. Construction and validation of an m6A RNA methylation regulators-based prognostic signature for esophageal cancer. Cancer Manag Res. 2020;6(12):5385–94.
Liu X, Gonzalez G, Dai X, et al. Adenylate kinase 4 modulates the resistance of breast cancer cells to tamoxifen through an m6A-based epitranscriptomic mechanism. Mol Ther. 2020;28(12):2593–604.
Zhang CZ, Samanta D, Lu HQ, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. PNAS. 2016;113(14):E2047–56.
Hao L, Wang JM, Liu BQ, et al. m6A-YTHDF1-mediated TRIM29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim Biophys Acta Mol Cell Res. 2021;1868:118878.
Fukumoto T, Zhu H, Nacarelli T, et al. N(6)-methylation of adenosine of FZD10 mRNA contributes to PARP inhibitor resistance. Cancer Res. 2019;79:2812–20.
Wang XL, Li ZH, Kong BH, et al. Reduced m6A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget. 2017;8(58):98918–30.
Barbieri I, Tzelepis K, Pandolfini L, et al. Promoter- bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;552(7683):126–31.
Vu LP, Pickering BF, Cheng YM, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23(11):1369–76.
Bansal H, Yihua Q, Iyer SP, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. 2014;28(5):1171–4.
Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–33.
Cui Q, Shi HL, Ye P, et al. m6A RNA methylation regulates the Self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–34.
Zhang SC, Zhao BS, Zhou AD, et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591-606.e6.
Jin D, Guo J, Wu Y, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. Hematol Oncol. 2019;12:135.
Liu S, Li Q, Li G, et al. The mechanism of m(6)A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by beta-elemene. Cell Death Dis. 2020;11:969.
Ding N, You A, Tian W, et al. Chidamide increases the sensitivity of Non-small Cell Lung Cancer to Crizotinib by decreasing c-MET mRNA methylation. Int J Biol Sci. 2020;16:2595–611.
Shi Y, Fan S, Wu M, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10:4892.
Chen MN, Wei L, Law CT, et al. RNA N6-methyladenosine methyl-transferase- like 3 promotes liver cancer progression through YTH-DF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–70.
Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, et al. RNA m(6) A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39:e103181.
Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methylade-nosine-dependent primary Micro RNA processing. Hepatology. 2017;65(2):529–43.
Yang Z, Li J, Feng GX, et al. MicroRNA-145 modulates N6-methy-ladenosine levels by targeting the 3’- untranslated mRNA region of the N6-methyladenosine binding YTH domain family 2 protein. J Biol Chem. 2017;292(9):3614–23.
Sun Y, Li S, Yu W, et al. N(6)-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis. 2020;11:836.
Nishizawa Y, Konno M, Asai A, et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9(7):7476–86.
Taketo K, Konno M, Asai A, et al. The epitranscriptome m6A writer METTL3 promotes chemo and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52:621–9.
Wang S, Sun C, Li J, et al. Roles of RNA methylation by means of N6-methyladenosine (m6A) in human cancers. Cancer Lett. 2017;408:112–20.
Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8:284–96.
Acknowledgements
None.
Funding
This work was supported by Shaanxi Province Key Research and Development Program (2021SF-129), Medical Project of Xi'an Science and Technology Bureau [2019114713YX002SF035(1) and 2019114713YX002SF035(6)], and Scientific research plan projects of Shaanxi Education Department (19JK0765).
Author information
Authors and Affiliations
Contributions
XZ and NL conceived, designed and writing of the manuscript. LW and YW performed the literature retrieval, data analysis and interpretation. ML and YZ participated in revising the manuscript. MC, MZ and LZ critically reviewed and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have reviewed the manuscript and agree to publish it in its current form.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, X., Lu, N., Wang, L. et al. Recent advances of m6A methylation modification in esophageal squamous cell carcinoma. Cancer Cell Int 21, 421 (2021). https://doi.org/10.1186/s12935-021-02132-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-021-02132-2