Abstract
Noncoding RNAs, including long noncoding RNAs (lncRNAs), microRNAs (miRNAs) and circular RNAs (circRNAs), are involved in regulating biological functions. In recent decades, miRNAs and lncRNAs have both inspired a wave of research, but the study of circRNA functions is still in its infancy. Studies have found that circRNAs actively participate in the occurrence and development of various diseases, which emphasizes the importance of circRNAs. Here, we review the features and classification of circRNAs and summarize their functions. Then, we briefly describe how to analyze circRNAs by bioinformatics procedures. In addition, the relationship between circRNAs and cancers is discussed with an emphasis on proving whether circRNAs can be potential biomarkers for the prognosis and diagnosis of cancer.
Similar content being viewed by others
Background
CircRNAs are a class of noncoding RNAs without 3′-poly(A) tails and 5′-caps that have closed circular structures formed by covalent bonding. They were first discovered in a plant virus by electron microscopy in 1976 [1], and then in 1979, Hsu et al. [2] provided evidence of the existence of circRNAs in several eukaryotic cells. In the following decades, although there were many discoveries about circRNAs, such as the first observation of circRNAs in humans [3] and the identification of four circRNAs expressed from the DCC (deleted in colorectal cancer) gene [4], the function of circRNAs had not come into great notice. Prior to 2013, circRNAs were regarded as a product of the incorrect splicing of pre-mRNA. Then, in the wake of the emergence of next-generation RNA sequencing (RNA-seq) and the development of bioinformatics, it was found that circRNAs are abundant and dynamically expressed in several eukaryotic cells, have high diversity and conservation, and play an irreplaceable role in the development of organisms [5]. Some publications have shown that circRNAs are closely related to cancers and can be involved in the regulation of tumorigenesis by adsorbing miRNAs, interacting with proteins, regulating the transcription of mRNA or translation into protein. However, more efforts are needed to explore the concrete functions of circRNAs in cancers.
The characteristics and categories of CircRNAs
Characteristics of circRNAs
In the human genome, RNAs that encode proteins make up only approximately 2% of the total sequence, and most of the remaining sequences are noncoding RNAs. Noncoding RNAs play an important role in regulating biological functions. CircRNAs, a member of the noncoding RNA family, possess some important features.
First, circRNAs have a special circular structure that protects them from degradation by RNase R or RNA exonucleases, which makes circRNAs more stable than their linear counterparts. In terms of stability, Jeck et al. [6] reported that the average half-life of circRNAs is approximately 5 times longer than that of mRNAs in most species.
Second, with the application of RNA sequencing technologies that are independent of poly(A) purification [7], several studies have reported that circRNAs are abundant in eukaryotic cells. More than 20,000 different circRNAs from eukaryotes have been identified [8], and the number will continue to grow. The study by Memczak et al. [9] identified 724 circRNAs in C. elegans, 1903 circRNAs in mice and 1950 circRNAs in humans. Evidence showed that circRNAs are widespread in human cancer genomes. There were 92,589 circRNAs identified across ~ 1000 cancer cell lines [10], and circRNAs were found in more than 800 tumor samples [11]. Additionally, some circRNAs have much higher expression than their liner mRNAs in human cells [5, 12].
Third, circRNAs exhibit sequence conservation among species to some extent [13]. For instance, it has been reported that 4522 out of 15,849 circRNAs in mice possess homologous sequences in humans [14]. This feature proves that circRNAs are not the outcome of aberrant splicing, as was previously thought.
Moreover, You et al. [15] observed that circRNAs originating from Rmst and Klhl2 are much more highly expressed in the mouse brain than in the liver or lung. Moreover, according to the sequence data, some nematode circRNAs are not expressed in 1- or 2-cell embryos but seem to exist in oocytes [9]. In general, these discoveries indicate that circRNAs are expressed in specific tissues and cells.
Biogenesis of circRNAs
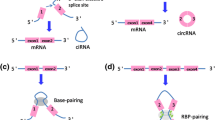
It is crucial to reveal how circRNAs are generated to establish their classification. Canonical splicing, with activation of spliceosomal machinery, is a process that removes the introns of precursor messenger RNAs (pre-mRNAs), after which exons are connected to form liner mRNAs [16] (Fig. 1a). However, unlike the canonical splicing of mRNAs, circRNAs originate from spliceosome-mediated nonsequential back-splicing of pre-mRNAs [17]. In the process of back-splicing, the upstream 5′ splice donor site is reversibly paired to the downstream 3′ splice acceptor site to generate a covalently closed structure [17, 18]. Recent studies have shown that canonical splicing signals and canonical splicing machinery are both indispensable for back-splicing [19, 20].
Biogenesis of CircRNAs. A: Canonical splicing. A process that removes the introns of pre-mRNAs, and then exons are connected to be a liner mRNA. B, C: Backing splicing. intron pairing and RNA binding proteins (RBPs) that can promote the circularization of back splicing to generate ecircRNAs or EIcicRNA. D: Intron splicing. Lariat introns that failure to debranch at the branch point site and trimming of the lariat tail can produce ciRNAs via intron splicing
Two main factors, intron pairing and RNA binding proteins (RBPs), can promote back-splicing circularization. On the one hand, intron pairing induces exons to approach each other by ALU elements [21] or nonrepetitive but complementary sequences [22] (Fig. 1b). For the formation of circHIPK3, long flanking introns that possess complementary ALU repeats are required [23]. On the other hand, some RBPs, such as MBL (MBNL1), QKI and FUS [24], can bind strongly and specifically to conserved sequences of flanking introns to promote exon circularization, and finally facilitating the formation of circRNAs [20] (Fig. 1c). In addition, the effect of double-stranded RNA (dsRNA) specific adenosine deaminase (ADAR) on the editing of adenosine to inosine [25] and the unwinding of dsRNA helix structure by ATP-dependent RNA helicase A (also known as DHX9) [26, 27] suppress the biogenesis of circRNAs.
Furthermore, circRNA generation can be facilitated by splicing factors (SFs) like ESRP1 [28] and the elongation velocity of RNA polymerase II [29].
Categories of circRNAs
CircRNAs are divided into three main classes based on their different structures: circRNA from exons (ecircRNA), circRNA with 3′,5′- or 2′,5′- phosphodiester bond (ciRNA) and circRNA from exon–intron junctions (EIciRNA).
EcircRNAs are the largest subclass of circRNAs, accounting for approximately 85% of circRNAs [9]. EcircRNAs include only exons that can be one or more, but most ecircRNAs span less than 5 exons [30]. Some researchers have found that ecircRNAs are mainly located in the cytoplasm [6]. The biogenesis of ecircRNAs consists of two mechanisms: direct back-splicing and exon skipping [5, 17, 31]. During the process of direct back-splicing, alternatively spliced RNA and a lariat intermediate that includes exons are first formed, after which canonical splicing removes the introns of the lariat to generate ecircRNA transcripts [12, 32, 33]. Comparatively speaking, exon skipping is the process of a downstream exon jumping over one or several exons to connect to an upstream exon, thus forming a circRNA after removal of the introns [5, 34, 35]. Recent reports have proven that direct back-splicing may be the main mechanism to form ecircRNAs [6]. EcircRNAs can interact with miRNA RBPs or can be translated into proteins to play a role in gene expression regulation [36].
CiRNAs, which with 3′,5′- or 2′,5′- phosphodiester bond and mainly exist in the nucleus, are involved in the regulation of their parental mRNA [37] and increase the expression of parental genes [36]. During conventional splicing intron lariats excised can sometimes escape debranching and retain a circular form with a 3′,5′- or 2′,5′-phosphodiester bond between the splice donor and the branch point [38] (Fig. 1d).
Composed of both exons and introns, EIciRNAs share some features with both ecircRNAs and ciRNAs [36]. Similar to ciRNAs, EIciRNAs are also predominantly located in the nucleus and regulate the expression of their parental gene in the nucleus [39, 40]. The biogenesis of EIciRNAs is similar to that of ecircRNAs; however, while ecircRNAs are generated when the introns in the lariat are completely removed, EIciRNAs are produced when the introns are retained.
Functions
Although circRNAs were discovered a long time ago, research on their functions is still in its infancy. The different distributions of circRNAs exhibit distinctive functions: circRNAs located in the cytoplasm mainly function as miRNA sponges and have the potential to be translated, while circRNAs scattered in the nucleus may regulate the transcription of parental genes.
Acting as miRNA sponges
As a pivotal regulator of gene expression [41], miRNA can bind to mRNA by complementary sequences in the 3′ UTR of target mRNA, thereby inhibiting mRNA translation or promoting mRNA degradation. Some scientific studies have reported that ecircRNAs distributed in the cytoplasm contain miRNA response elements (MREs) that can absorb miRNA and prevent them from binding to their target mRNAs [42] (Fig. 2a). circRNAs can bind to one miRNA with several binding sites or bind to multiple miRNAs. CDR1as, cerebellar degeneration-related protein 1 antisense, harbors over 70 conventional miR-7 binding sites; therefore, it can regulate the expression of target mRNAs by absorbing miR-7 [43].
Functions of CirxRNAs. A: Regulating transcription. CiRNAs which mainly located in the nucleus can bind to RNA polymerase II at the promoters of their host genes to enhance gene expression. B :Regulating alternative splicing. Circularization and splicing compete against each other, the formation of circRNAs impact the alternative splicing of pre-mRNAs, eventually giving rise to alteration of gene expression. C: Regulating transcription. EIciRNAs can interact with U1 snRNP and then the EIciRNAs-U1 snRNP complexes associate with RNA polymerase II (Pol II) at the promoters of their host genes to enhance gene expression. D: Acting as sponges. EcircRNAs can bind to miRNAs and proteins to regulate gene expression. E: Translating into proteins. Some ecircRNAs can be encoded to proteins, thereby influence their downstreams
circHIPK3 [44] can sponge to 9 miRNAs with 18 potential binding sites and circCCDC66 [45] was potentially targeted by 99 miRNAs. Another example of circRNA-miRNA interactions has been shown for circ-FOXP1. A recent report revealed that circ-FOXP1 controlling mesenchymal stem cell (MSC) identity and differentiation was enriched in MSCs. Circ-FOXP1 can act as a miRNA sponge targeting miR-17–3p and miR-127–5p and promotes the proliferation and differentiation of MSCs [46].
Translating into proteins
Although circRNAs are regarded as a class of noncoding RNAs, increasing evidence has demonstrated that some circRNAs are translatable [4] (Fig. 2b). Unlike mature mRNA translation, which normally requires a 5′ end 7-methylguanosine (m7G) cap structure and a 3′ poly(A) tail [47], circRNAs have different translation processes because they do not have these necessary structures. In 1998, it was confirmed that a circular mRNA containing a complete open reading frame (ORF) of green fluorescent protein (GFP) could successfully encode GFP in Escherichia coli [48]. A subsequent study showed that some circRNAs possessing internal ribosome entry sites (IRESs) can interact with ribosomes as the entry point, thus initiating translation [17, 41]. Moreover, surprisingly, it was found that N6-methyladenosine (m6A), the most common RNA modification, facilitates the protein-coding ability of circRNAs [49]. A recent study documented that FBXW7-185aa, a novel 21-kDa protein, was encoded by a spanning junction open reading frame in circ-FBXW7 driven by an internal ribosome entry site. Upregulation of FBXW7-185aa inhibited proliferation and cell cycle progression and reduced the half-life of c-Myc by antagonizing USP28-induced c-Myc stabilization in glioblastoma, while knockdown of FBXW7-185aa promoted malignant phenotypes in vitro and in vivo [50]. The protein-coding function provides a fresh viewpoint on the role of circRNAs in diseases.
Regulating transcription
Due to being predominantly located in the nucleus and containing a relatively low number of microRNA target sites, EIciRNAs and ciRNAs have been found to be involved in transcription regulation [4, 47]. Some studies have suggested that these circRNAs modulate the expression of their parental genes [51]. A recent study indicated that the intron of EIciRNAs has one putative U1 snRNP (small nuclear ribonucleoprotein) binding site [36]; therefore, EIciRNAs can interact with U1 snRNP, and then the EIciRNAs-U1 snRNP complexes associate with RNA polymerase II (Pol II) at the promoters of their host genes to enhance gene expression [40] (Fig. 2c). For ciRNAs, it has been reported that ci-ankrd52 and ci-sirt7 bind to the extended Pol II complex at the transcriptional start site and positively regulate Pol II-mediated transcription, suggesting that they may also have cis-regulatory effects on upstream genes [4, 37] (Fig. 2e). The interplay between circRNAs and the transcriptional machinery provides new perspectives for strategies regulating gene expression in cells [52].
Regulating alternative splicing
Circularization and splicing compete against each other, enabling ecircRNAs to function in alternative splicing [20] (Fig. 2d). When the formation of circRNA and liner mRNA involves the same exon, competition occurs, and the more exons are circularized, the less the same exons will appear in the processed mRNA [4, 53]. Therefore, the formation of circRNAs impacts the alternative splicing of pre-mRNAs, eventually resulting in altered gene expression. This is best illustrated by the example of circMbl, which is circularized from the second exon of the splicing factor muscleblind (MBL) gene, whereby modulation of MBL levels strongly affects the biosynthesis of circMbl [20].
Binding to proteins
In addition to interacting with miRNAs, circRNAs can bind to proteins to regulate gene expression and ultimately influence the development of some diseases (Fig. 2a). For example, circ-Foxo3 can interact with the stress-related proteins FAK and HIF-1α in the cytoplasm and then block these proteins from entering the nucleus, which ultimately promotes cardiac senescence [54]. In contrast, Yang et al. revealed that circ-Amotl1 can interact with Stat3 and facilitate Stat3 nuclear translocation. By binding to the Stat3 and Dnmt3a promoters, circ-Amotl1 promotes Dnmt3a transcription, and the increased Dnmt3a can methylate the miR-17 gene promoter to inhibit the expression of miR-17-5p but increase fibronectin expression, which leads to accelerated wound repair [55].
Although circRNAs have different functions, they mainly adsorb miRNAs and then affect downstream targets involved in the development of cancers; the other functions account for only a small portion (Fig. 3). Table 1 shows the proportion of circRNAs with different functions in different tumors. Therefore, efforts need to be made to explore other functions of circRNAs in tumors.
CircRNAs are closely related to cancers
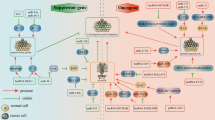
It has been reported that circRNAs play an important role in cancer progression by modulating many of the hallmarks of cancer, such as circRNAs being involved in the regulation of sustained proliferative signaling [56, 57], evasion of growth suppressors [58], impairment of differentiation signals, contribution to tumor metastasis and invasion [59], and induction of angiogenesis [60]. In addition, increasing reports have shown that circRNAs can serve as tumor promoters or suppressors to regulate signaling pathways of cancer, including the Wnt/β-catenin signaling [61], MAPK/ERK and PTEN/PIK3/AKT pathways [62]. Meanwhile, some important molecules such as p53 [63] and K-Ras [64] participate in the regulation of circRNAs.
Recently, the relationship between circRNAs and tumor microenvironment (TME) has raised some concern due to its influence on tumor immunity, angiogenesis, metastasis, and hypoxia. CircRNAs can modulate TME by mediating tumor immune surveillance, regulating immune escape via a circRNA-miRNA-PD-1/PD-L1 axis, and regulating the cytotoxicity of natural killer cells, thus promote or inhibit the immune system and angiogenesis, improve the permeability of endothelial cells, and remodel the extracellular matrix (ECM) [65].
Herein, we will discuss the relationship between circRNAs and cancers (Fig. 4) and evaluate whether circRNAs can be biomarkers of the diagnosis and prognosis of cancer (see Table 2).
CircRNAs and hepatocellular cancer
Due to the aggressive nature and recurrence rate of hepatocellular cancer (HCC), the prognosis of HCC remains poor. Therefore, effective biomarkers are necessary to improve early diagnosis and prognosis analysis [66]. There is increasing evidence that circRNAs are related to the development and invasion of hepatoma, although the mechanism is not entirely clear. Liang et al. [67] reported that circβ-catenin was upregulated in liver cancer tissues compared to adjacent normal tissues. Circβ-catenin encodes a novel β-catenin isoform that promotes liver cancer growth and migration in vitro and attenuates tumorigenesis and metastasis in vivo by activating the Wnt pathway. A study by Song et al. [68] showed that circ0003998 was markedly upregulated in portal vein tumor thrombus (PVTT) metastasis and HCC tissues. Its expression was related to the aggressive characteristics of HCC patients. Circ0003998 promoted mesenchymal transition (EMT) in HCC by acting as a competing endogenous RNA (ceRNA) to sponge miR-143-3p to relieve the repressive effect on FOSL2 (EMT-related stimulator). Furthermore, circ0003998 could bind with PCBP1 to increase the expression level of the EMT-related gene CD44v6. In addition, Wang et al. [69] found that circHIAT1 was obviously downregulated in HCC samples and cell lines. The low expression of circHIAT1 was positively correlated with the poor overall survival of HCC patients. The overexpression of circHIAT1 suppressed HCC progression in vitro and in vivo by regulating the miR-3171/PTEN axis. Zheng et al. [70] found that hsa_circ_0079929 expression was lower in HCC tissues. The inhibitory effect of hsa_circ_0079299 was partially mediated by the PI3K/AKT/mTOR signaling pathway. Circ-0051443 was also strongly downregulated in plasma exosomes and tissues, and exosomal circ-0051443 could inhibit HCC progression by functioning as a sponge of miR-331-3p to promote BAK1 expression [71]. Circ-0051443 spread from normal cells to HCC cells through exosomes and inhibited malignant biological behaviors by promoting cell apoptosis and suppressing the cell cycle, indicating that exosomal circ-0051443 can serve as a predictor and potential therapeutic target for HCC.
CircRNAs and breast cancer
CircRNAs are abnormally expressed in breast cancer. Chen et al. [72] performed high-throughput circular RNA microarray assays in 3 pairs of triple-negative breast cancer (TNBC) patient samples and identified TNBC-related circRNAs, of which 173 circRNAs were obviously upregulated, whereas 77 circRNAs were significantly downregulated. They found that circEPSTI1, derived from the EPSTI1 gene, was the most upregulated circRNA among the candidates. Due to the crucial role of the EPSTI1 gene in cancer invasion and metastasis, the authors focused their attention on circEPSTI1. They demonstrated that circEPSTI1 was highly expressed in most TNBC cell lines and tissues, and knockdown of circEPSTI1 reduced cell proliferation and induced apoptosis. The effects of circEPSTI1 could be explained by its interaction with miR-4753 and miR-6809, which then affected downstream BCL11A expression. In addition, high expression of circEPSTI1 was positively correlated with tumor size, lymph node infiltration and TNM stage, and TNBC patients with increased circEPSTI1 and BCL11A levels were significantly associated with reduced disease-free survival (DFS) and overall survival (OS), suggesting that circEPSTI1 is an independent prognostic marker of TNBC patient survival. Hsa_circ_0025202 was found to be downregulated in breast cancer cells and tissues and inversely correlated with lymphatic metastasis and histological grade, which reduced cell proliferation, colony formation, and migration and enhanced cell apoptosis and sensitized cells to tamoxifen (TAM) treatment [73]. In addition, hsa_circ_0025202 could absorb miR-182-5p to regulate the expression and activity of FOXO3a and then affect tumor inhibition and tamoxifen sensitization effects, indicating that hsa_circ_0025202 could be a therapeutic target in patients with HR-positive BC, especially in those receiving TAM therapy. In contrast to its host gene SMARCA5, circSMARCA5 is reduced in breast cancer tissues [74]. The enforced expression of circSMARCA5 could induce drug sensitivity in breast cancer cell lines in vitro and in vivo. There is a competitive relationship between the expression of circSMARCA5 and SMARCA5. circSMARCA5 can interact with its parental locus to form an R loop, which results in the suppression SMARCA5 exon 15 transcription. The expression of circSMARCA5 leads to the downregulation of SMARCA5 and the production of a truncated nonfunctional protein, while circSMARCA5 overexpression was sufficient to increase the sensitivity to cytotoxic drugs. The above evidence shows that circSMARCA5 may be used as a therapeutic target for patients with resistant breast cancer.
CircRNAs and gastric cancer
Gastric cancer (GC) is the fifth most common cancer in the world with high morbidity and mortality [75]. With the deepening of research on circular RNAs, the relationship between circular RNAs and the diagnosis and prognosis of gastric cancer has received increasing attention. Ding et al. [76] used an online dataset to analyze overexpressed circRNAs in GC tissues compared to adjacent tissues and found that circ-DONSON was the most upregulated circRNA among the candidates. Circ-DONSON was proven to be highly expressed in GC tissues and cells. Additionally, the authors found that circ-DONSON expression was positively correlated with TNM stage and lymphoid metastasis. Higher expression of circ-DONSON in GC patients was correlated with a lower overall survival rate and disease-free survival rate, indicating that circ-DONSON might be a prognostic marker. In contrast to the miRNA sponge mechanism, circ-DONSON might regulate transcription due to its nuclear location. Ding et al. found that circ-DONSON could recruit the NURF complex to the SOX4 promoter and initiate its transcription, thereby regulating GC cell malignant behaviors. Zhang et al. [77] found that circNRIP1, upregulated in GC tissues, acted as a miRNA-149-5p sponge to promote GC progression via the AKT/mTOR pathway, and the expression level of circNRIP1 significantly correlated with GC tumor size and lymphatic invasion. Moreover, a new report showed that circPSMC3 was significantly downregulated in gastric cancer plasma, tissues and cells and associated with poor prognosis, whereby patients with lower circPSMC3 expression showed a reduced overall survival time [78]. CircPSMC3 could act as a ceRNA by sponging miR-296-5p to suppress the proliferation and metastasis of gastric cancer. CircLMTK2 expression was also significantly reduced in GC [79]. A lower level of circLMTK2 expression was correlated with decreased overall survival in GC patients, and a multivariate Cox hazards analysis showed that high circLMTK2 expression was an independent factor for OS, suggesting that circLMTK2 could be used as a prognostic factor in GC cases. Huang et al. [80] found that circAKT3 was upregulated in cisplatin (CDDP)-resistant GC tissues and cells compared with CDDP-sensitive samples, and the upregulation of circAKT3 in GC patients receiving CDDP therapy was significantly associated with aggressive characteristics and was an independent risk factor of disease-free survival. These results suggest that circRNAs may be novel biomarkers of the diagnosis and prognosis of gastric cancer and can be therapeutic targets.
CircRNAs and lung cancer
Lung cancer is the leading cause of cancer-related death worldwide, and most cases present with advanced local infiltration and/or distant metastasis at diagnosis [81]. Despite ongoing efforts to improve the response to treatment and the discovery of lung cancer treatments that have shown survival benefits, the overall 5-year survival rate for advanced lung cancer remains below 15%, underscoring the need for early diagnosis and timely treatment [82]. Recent studies have shown that circRNAs may be involved in the development of lung cancer, which is beneficial to the diagnosis, prognosis and even treatment of lung cancer. Recently, Jin et al. [83] reported that the expression of circARHGAP10 was significantly increased in both non-small cell lung cancer tissues and cell lines, and a high level of circARHGAP10 expression was correlated with a poor prognosis. Knockdown of circARHGAP10 suppressed proliferation, metastasis and glycometabolism by targeting the miR-150-5p/GLUT1 axis in NSCLC. Thus, circARHGAP10 may be a potential target for NSCLC treatment. Similarly, circTP63, which was upregulated in lung squamous cell carcinoma (LUSC), was shown to be a sponge of miR-873-3p, thus abolishing the suppressive effect of miR-873-3p and increasing the level of FOXM1, which upregulated CENPA and CENPB, thereby facilitating cell cycle progression [84]. The upregulation of circTP63 was correlated with larger tumor size and higher TNM stage in LUSC patients, suggesting that circTP63 is associated with the prognosis of LUSC. More recently, Qin et al. [85] determined that circ-UBR5 was markedly reduced in NSCLC tissues and correlated with tumor differentiation. Surprisingly, circ-UBR5, a novel snRNA, could play a role in spliceosome-mediated RNA splicing regulation by binding to the splicing regulatory factors QKI and NOVA1 and U1 snRNA in the nucleus.
CircRNAs and glioma
Glioma is the most common intracranial primary cancer with extraordinarily high morbidity and mortality worldwide [86, 87]. Although there are common methods for clinical treatment, such as surgery, radiotherapy, and chemotherapy, the long-term effects and postoperative outcomes for patients with glioma are still unsatisfactory [88, 89]. Recently, circSCAF11 was found to be significantly upregulated in glioma tissues and cell lines, and ectopic overexpression of circSCAF11 was closely correlated with the poor clinical outcome of glioma patients [90]. Mechanistic experiments further indicated that circSCAF11 accelerated glioma tumorigenesis through the miR-421/SP1/VEGFA axis, providing a potential target for circRNAs and glioma treatment. CircRNAs can not only act as a miRNA sponge but can also absorb some proteins to influence the progression of tumors. For instance, circCPA4, which is highly expressed in glioma tissues and positively related to poor outcome of glioma, could act as a sponge for let-7 to regulate the expression of CPA4 and glioma progression [91]. Therefore, circCPA4 could be a novel prognostic biomarker and target for glioma treatment. Furthermore, circRNAs have the ability to encode proteins involved in the development of glioma. Zhang et al. [92] found that circ-SHPRH used overlapping genetic codes to generate a 'UGA' stop codon, which resulted in the translation of the 17 kDa SHPRH-146aa, and the expression of circ-SHPRH and SHPRH-146aa was low in glioma. The overexpression of SHPRH-146aa could protect full-length SHPRH from degradation by the ubiquitin proteasome, and then stabilized SHPRH subsequently ubiquitinating proliferating cell nuclear antigen (PCNA) as an E3 ligase, leading to inhibited cell proliferation and tumorigenicity. These results suggest that circRNAs are involved in the development of glioma in different ways.
CircRNAs and colorectal cancer
Xu et al. [93] performed secondary sequencing to profile circRNA expression in colorectal cancer (CRC) tissues and matched normal tissues. They found that in CRC tissues, 92 circRNAs were significantly upregulated, and 21 circRNAs were downregulated. In particular, the expression of circRNA_0001178 and circRNA_0000826 was increased in CRC tissues and showed potential diagnostic value with AUCs of 0.945 and 0.816, respectively. CircHIPK3 was also significantly upregulated in CRC tissues and cell lines, and ectopic expression of circHIPK3 effectively reversed the miR-7-induced attenuation of the malignant phenotype of CRC cells by increasing the expression level of miR-7 targeting proto-oncogenes (FAK, IGF1R, EGFR, YY1) [94]. Increased expression of circHIPK3 was significantly associated with tumor T status, lymph node metastasis, distant metastasis, and advanced clinical stage, and it was an independent prognostic factor of poor overall survival in CRC. These results indicate that circHIPK3 upregulation is an early event in CRC development and has a vital role in CRC progression. Li et al. [95] demonstrated that circITGA7 and its linear host gene ITGA7 were both significantly downregulated in colorectal cancer tissues and cell lines, and their decreased expression was correlated with CRC progression. CircITGA7 inhibited colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating the transcription of its host gene ITGA7. Furthermore, the noncoding effects of circCCDC66 were demonstrated in a previous publication. CircCCDC66 expression was elevated in polyps, and all stages of colorectal cancer tissues and patients with a higher level of circCCDC66 had a poor prognosis, as evidenced by lower overall survival rates [45]. CircCCDC66 could exert its function by regulating various oncogenes, thus modulating multiple pathological processes, including cell proliferation, migration, invasion, and anchorage-independent growth.
Taken together, these findings highlight novel oncogenic and tumor suppressor functions of circRNAs in colon cancer progression and metastasis.
CircRNAs and other tumors
The abnormal expression of circRNAs was also observed in other cancers, such as bladder cancer, osteosarcoma, esophageal squamous cell carcinoma and leukemia. CircSLC8A1 was downregulated in bladder cancer tissues and cell lines, and it could function as a tumor suppressor through interaction with miR-130b and miR-494 to inhibit bladder cancer progression by regulating PTEN [96]. CircLRP6 was significantly increased in osteosarcoma, and high expression of circLRP6 resulted in shorter disease-free survival and overall survival. CircLRP6 acted as an oncogene by binding to LSD1 and EZH2 to inhibit the expression of KLF2 and APC [97]. In addition, circRNA microarray analysis revealed that 698 circRNAs were differentially expressed in acute myeloid leukemia (AML) patients, with 282 circRNAs found to be upregulated and 416 to be downregulated [98].
Fan et al. [99] identified ESCC-related circRNAs and found that the expression of 1045 circRNAs was obviously upregulated, whereas that of 1032 circRNAs was significantly downregulated by using a circular RNA microarray analysis in 3 pairs of esophageal squamous cell carcinoma (ESCC) frozen tumor and nontumor tissues. Cao et al. [100] revealed that circRNA_100876 expression was significantly higher in ESCC tissues and cell lines, and its high expression was strongly correlated with tumor invasion depth, lymph node metastasis and vascular invasion, indicating that circRNA_100876 expression was linked to the clinical progression of ESCC and might represent a promising prognostic biomarker.
These findings indicate that circRNAs regulate multiple signaling pathways through multiple modes of action and are closely linked to tumors.
Bioinformatic analysis of circRNAs
Although circRNAs have been studied for a long time, they are not easy to detect due to the previously inadequate technology. With the emergence of next-generation RNA sequencing, an increasing number of circRNAs have been identified. Herein, we summarize the research procedures to study circRNAs.
First, a cDNA library is constructed. The ribosomal rRNA in total RNA is first removed using a biotinylated specific probe. After purification, the RNA is fragmented with temperature and ionic environment. Subsequently, dNTPs are added to synthesize a cDNA strand, and then DNA polymerase I and RNase H are used to synthesize double-stranded cDNA. During cDNA double strand synthesis, the RNA template is removed, and dTTP is replaced by dUTP. The ligation product of the double-stranded cDNA product followed by the addition of the "A" base and the linker is amplified, and the final cDNA library is obtained after purification. Finally, the constructed sequencing library is sequenced.
Second, the raw data generated by sequencing are analyzed. In the first step, the low-quality, joint contamination, and unknown N content of the base are filtered out. The filtered data are called clean reads. In the second step, clean reads are mapped to the reference genome, and circRNAs are predicted by two software programs, CIRI and find_circ. In the third step, quantitative and differential expression analysis of circRNAs is performed after combining the results of the two software programs. The functional analysis of circRNAs mainly focuses on regulation of gene expression through interactions with multiple miRNAs, which can be analyzed by some databases like analysis of common targets (ACT) [101] and CircNet [102].
Then, experiments such as reverse transcription-PCR (RT-PCR), droplet digital PCR, northern blotting, and fluorescence in situ hybridization are conducted to verify the circRNAs [103]. To study the function of circRNAs, gene overexpression and knockdown are used to manipulate circRNA expression. For mechanistic studies and with reference to some databases, such as CircNet, circBase, Circ2Traits and circRNA Disease (see Table 3), researchers perform bioinformatic prediction, RNA immunoprecipitation, luciferase reporter assay, and RNA pull-down combined with mass spectrometry to reveal circRNA-miRNA and circRNA-protein interactions.
Conclusion
In this review, we briefly summarized the characteristics, biogenesis, classification, and mechanisms of circRNAs and their relationship with cancer. CircRNAs are stable, highly conserved and cell- and tissue-specific molecules in the RNA interaction network, indicating that they are not erroneous or random byproducts but are derived from a strictly controlled biological process. Currently, research on circRNAs mainly focuses on their expression pattern in cancer and their biological role in tumorigenesis. CircRNAs can function as miRNA sponges, which is by far the most studied mechanism. In addition, circRNAs can regulate gene expression at the transcriptional and posttranscriptional levels and can be translated into proteins. Among mechanistic studies, miRNA sponges are the most studied; thus, more attention should be paid to the exploration of gene regulation and translation mechanisms of circRNAs in the future.
It has been reported that the close relationship between the level of circRNAs and clinical features, such as tumor size, TNM stage and lymphatic metastasis, and circRNAs could be reflected in extracellular fluid through exosomes, and the steady expression of circRNAs in plasma [104] indicates that circRNAs can be biomarkers of the diagnosis and prognosis of cancer.
Due to the steady development of RNA technology, we expect that the field of circRNAs will have considerable development in the future. The exact location, transport, degradation of circRNAs in living cells, and the complete circRNAs interactome will all make progress in this field.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- lncRNAs:
-
Long noncoding RNAs
- miRNAs:
-
MicroRNAs
- circRNAs:
-
Circular RNAs
- DCC:
-
Deleted in colorectal cancer
- RNA-seq:
-
RNA sequencing
- pre-mRNAs:
-
Precursor messenger RNAs
- RBPs:
-
RNA binding proteins
- ADAR:
-
Adenosine deaminase
- SFs:
-
Splicing factors
- ecircRNA:
-
CircRNA from exons
- ciRNA:
-
CircRNA from introns
- EIciRNA:
-
CircRNA from exon–intron junctions
- MREs:
-
MiRNA response elements
- MSC:
-
Mesenchymal stem cell
- m7G:
-
7-methylguanosine
- ORF:
-
Open reading frame
- GFP:
-
Green fluorescent protein
- IRESs:
-
Internal ribosome entry sites
- m6A:
-
N6-methyladenosine
- snRNP:
-
Small nuclear ribonucleoprotein
- Pol II:
-
Polymerase II
- MBL:
-
Muscleblind
- TME:
-
Tumor microenvironment
- ECM:
-
Extracellular matrix
- HCC:
-
Hepatocellular cancer
- ceRNA:
-
Competing endogenous RNA
- PVTT:
-
Portal vein tumor thrombus
- EMT:
-
Mesenchymal transition
- TNBC:
-
Triple-negative breast cancer
- DFS:
-
Disease-free survival
- OS:
-
Overall survival
- TAM:
-
Tamoxifen
- GC:
-
Gastric cancer
- CDDP:
-
Cisplatin
- LUSC:
-
Lung squamous cell carcinoma
- PCNA:
-
Proliferating cell nuclear antigen
- CRC:
-
Colorectal cancer
- AML:
-
Acute myeloid leukemia
- ESCC:
-
Esophageal squamous cell carcinoma
- ACT:
-
Analysis of common targets
References
Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–6.
Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–40.
Kos A, Dijkema R, Arnberg AC, et al. The hepatitis delta (δ) virus possesses a circular RNA. Nature. 1986;323(6088):558–60.
Qian L, Yu S, Chen Z, et al. The emerging role of circRNAs and their clinical significance in human cancers. Biochimica et Biophysica Acta (BBA) Rev Cancer. 2018;1870(2):247–60.
Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–57.
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61.
Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–92.
Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–70.
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Ruan H, Xiang Y, Ko J, et al. Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med. 2019;11(1):55.
Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–81.
Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7(2):e30733.
Chen B, Huang S. Circular RNA: AN emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50.
Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–85.
You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–10.
Chen L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–8.
Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172–9.
Zhang X, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26(9):1277–87.
Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–11.
Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with Pre-mRNA splicing. Mol Cell. 2014;56(1):55–66.
Chen L, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2014;7(21):3294–301.
Zhang X, Wang H, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–47.
Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(1):11215.
Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8(1):14741.
Kristensen LS, Andersen MS, Stagsted L, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91.
Lin Y, Yu Y, Lin H, et al. Oxaliplatin-induced DHX9 phosphorylation promotes oncogenic circular RNA CCDC66 expression and development of chemoresistance. Cancers. 2020;12(3):697.
Aktaş T, Ilık AO, Maticzka D, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544(7648):115–9.
Yu CY, Li TC, Wu YY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8(1):1149.
Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2016;14(8):1035–45.
Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409.
Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51.
Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A. 1996;93(13):6536–41.
Qian Y, Lu Y, Rui C, et al. Potential significance of circular RNA in human placental tissue for patients with preeclampsia. Cell Physiol Biochem. 2016;39(4):1380–90.
Kelly S, Greenman C, Cook PR, et al. Exon skipping is correlated with exon circularization. J Mol Biol. 2015;427(15):2414–7.
Cocquerelle C, Mascrez B, Hetuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7(1):155–60.
Floris G, Zhang L, Follesa P, et al. Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol. 2017;54(7):5156–65.
Zhang Y, Zhang X, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806.
Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–90.
Quan G, Li J. Circular RNAs: biogenesis, expression and their potential roles in reproduction. J Ovarian Res. 2018;11(1):9.
Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–64.
Xin Z, Ma Q, Ren S, et al. The understanding of circular RNAs as special triggers in carcinogenesis. Briefings Functional Genomics. 2017;16(2):w1.
Thomas LF, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30(16):2243–6.
Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8.
Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
Hsiao K, Lin Y, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Can Res. 2017;77(9):2339–50.
Cherubini A, Barilani M, Rossi RL, et al. FOXP1 circular RNA sustains mesenchymal stem cell identity via microRNA inhibition. Nucleic Acids Res. 2019;47(10):5325–40.
Li X, Yang L, Chen L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–42.
Perriman R, Ares JM. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA. 1998;4(9):1047–54.
Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–41.
Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. JNCI. 2018;110(3):304–15.
Lei K, Bai H, Wei Z, et al. The mechanism and function of circular RNAs in human diseases. Exp Cell Res. 2018;368(2):147–58.
Meng X, Li X, Zhang P, et al. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2017;18(4):w45.
Khan MAF, Reckman YJ, Aufiero S, et al. RBM20 regulates circular RNA production from the titin gene. Circ Res. 2016;119(9):996–1003.
Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):w1.
Yang Z, Awan FM, Du WW, et al. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther. 2017;25(9):2062–74.
Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–64.
Liang G, Liu Z, Tan L, et al. HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res. 2017;37(8):4337–43.
Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–37.
Huang X, Huang Z, Xu Y, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7(1):5428.
Bach D, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–29.
Wan L, Zhang L, Fan K, et al. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed Res Int. 2016;2016:1–11.
Pan H, Li T, Jiang Y, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119(1):440–6.
Chen S, Thorne RF, Zhang XD, et al. Non-coding RNAs, guardians of the p53 galaxy. Seminars in Cancer Biology. 2020.
Chen J, Sun Y, Ou Z, et al. Androgen receptor-regulated circFNTA activatesKRAS signaling to promote bladder cancer invasion. EMBO Rep. 2020;21(4):e48467.
Zhang Q, Wang W, Zhou Q, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):14.
Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20(1):84.
Song L, Qiao G, Yu J, et al. Hsa_circ_0003998 promotes epithelial to mesenchymal transition of hepatocellular carcinoma by sponging miR-143-3p and PCBP1. J Exp Clin Cancer Res. 2020;39(1):114.
Wang Z, Zhao Y, Wang Y, et al. Circular RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by regulating miR-3171/PTEN axis. Biomed Pharmacother. 2019;116:108932.
Zheng H, Chen T, Li C, et al. A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma. Cancer Manag Res. 2019;11:443–54.
Chen W, Quan Y, Fan S, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–28.
Chen B, Wei W, Huang X, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8(14):4003–15.
Sang Y, Chen B, Song X, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27(9):1638–52.
Xu X, Zhang J, Tian Y, et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer. 2020;19(1):128.
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86.
Ding L, Zhao Y, Dang S, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18(1):45.
Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20.
Rong D, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2019;18(1):25.
He J, Chen J, Ma B, et al. CircLMTK2 acts as a novel tumor suppressor in gastric cancer. Biosci Rep. 2019;39(5):8.
Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(1):71.
Williams CD, Gajra A, Ganti AK, et al. Use and impact of adjuvant chemotherapy in patients with resected non-small cell lung cancer. Cancer. 2014;120(13):1939–47.
Lei B, Tian Z, Fan W, et al. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301.
Jin M, Shi C, Yang C, et al. Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in NSCLC through regulation of the miR-150-5p/GLUT-1 Axis. Mol Therapy Nucleic Acids. 2019;18:219–31.
Cheng Z, Yu C, Cui S, et al. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10(1):3200.
Qin M, Wei G, Sun X. Circ-UBR5: An exonic circular RNA and novel small nuclear RNA involved in RNA splicing. Biochem Biophys Res Commun. 2018;503(2):1027–34.
Chen T, Da Fonseca C, Schönthal A. Intranasal perillyl alcohol for glioma therapy: molecular mechanisms and clinical development. Int J Mol Sci. 2018;19(12):3905.
Wank M, Schilling D, Schmid T, et al. Human glioma migration and infiltration properties as a target for personalized radiation medicine. Cancers. 2018;10(11):456.
Dworkin M, Mehan W, Niemierko A, et al. Increase of pseudoprogression and other treatment related effects in low-grade glioma patients treated with proton radiation and temozolomide. J Neurooncol. 2019;142(1):69–77.
Noorlag L, De Vos FY, Kok A, et al. Treatment of malignant gliomas with ketogenic or caloric restricted diets: a systematic review of preclinical and early clinical studies. Clin Nutr. 2019;38(5):1986–94.
Meng Q, Li S, Liu Y, et al. Circular RNA circSCAF11 accelerates the glioma tumorigenesis through the miR-421/SP1/VEGFA Axis. Mol Therapy Nucleic Acids. 2019;17:669–77.
Peng H, Qin C, Zhang C, et al. circCPA4 acts as a prognostic factor and regulates the proliferation and metastasis of glioma. J Cell Mol Med. 2019;23(10):6658–65.
Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–14.
Xu H, Wang C, Song H, et al. RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18(1):8.
Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417.
Li X, Wang J, Zhang C, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host geneITGA7. J Pathol. 2018;246(2):166–79.
Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):111.
Zheng S, Qian Z, Jiang F, et al. CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am J Transl Res. 2019;11(7):4126–38.
Chen H, Liu T, Liu J, et al. Circ-ANAPC7 is upregulated in acute myeloid leukemia and appears to target the MiR-181 family. Cell Physiol Biochem. 2018;47(5):1998–2007.
Fan L, Cao Q, Liu J, et al. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol Cancer. 2019;18(1):16.
Cao S, Chen G, Yan L, et al. Contribution of dysregulated circRNA_100876 to proliferation and metastasis of esophageal squamous cell carcinoma. Onco Targets Ther. 2018;11:7385–94.
Lin YC, Lee YC, Chang KL, et al. Analysis of common targets for circular RNAs. BMC Bioinformatics. 2019;20(1):372.
Liu YC, Li JR, Sun CH, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44(D1):D209–15.
Zhang Y, Liang W, Zhang P, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. 2017;36(1):152.
Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–4.
Wei J, Wei W, Xu H, et al. Circular RNA hsa_circRNA_102958 may serve as a diagnostic marker for gastric cancer. Cancer Biomark. 2020;27(2):139–45.
Xie Y, Shao Y, Sun W, et al. Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomark Med. 2018;12(1):11–20.
Tian M, Chen R, Li T, et al. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. 2018;32(3):e22281.
Chen S, Li T, Zhao Q, et al. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–71.
Tang W, Fu K, Sun H, et al. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17(1):137.
Sun H, Tang W, Rong D, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21(2):299–306.
Wu L, Liu D, Yang Y. Enhanced expression of circular RNA circ-DCAF6 predicts adverse prognosis and promotes cell progression via sponging miR-1231 and miR-1256 in gastric cancer. Exp Mol Pathol. 2019;110:104273.
Ouyang Y, Li Y, Huang Y, et al. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J Cell Physiol. 2019;234(7):10458–69.
Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151.
Shao Y, Chen L, Lu R, et al. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39(4):1393390539.
Shao Y, Tao X, Lu R, et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res. 2020;26(3):1475–82.
Li XN, Wang ZJ, Ye CX, et al. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother. 2019;112:108611.
Tian J, Xi X, Wang J, et al. CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag Res. 2019;11:5413–23.
Li X, Wang Z, Ye C, et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):325.
Zhuo F, Lin H, Chen Z, et al. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–93.
Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–5.
Yuan Y, Liu W, Zhang Y, et al. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503(2):870–5.
Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. 2018;102:639–44.
Zhang S, Zeng X, Ding T, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8(1):2878.
Huang MS, Liu JY, Xia XB, et al. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front Oncol. 2019;9:508.
Gu X, Wang G, Shen H, et al. Hsa_circ_0033155: a potential novel biomarker for non-small cell lung cancer. Exp Ther Med. 2018;16(4):3220–6.
Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci. 2018;22(10):3053–60.
Li S, Sun X, Miao S, et al. hsa_circ_0000729, a potential prognostic biomarker in lung adenocarcinoma. Thorac Cancer. 2018;9(8):924–30.
Liu W, Ma W, Yuan Y, et al. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–51.
Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–6.
Zhang C, Zhang C, Lin J, et al. Circular RNA Hsa_Circ_0091579 serves as a diagnostic and prognostic marker for hepatocellular carcinoma. Cell Physiol Biochem. 2018;51(1):290–300.
Chen D, Zhang C, Lin J, et al. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res. 2018;10:1275–83.
Kou P, Zhang C, Lin J, et al. Circular RNA hsa_circ_0078602 may have potential as a prognostic biomarker for patients with hepatocellular carcinoma. Oncol Lett. 2019;17(2):2091–8.
He R, Liu P, Xie X, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):145.
Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res. 2019;11:1033–41.
Tang H, Huang X, Wang J, et al. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer. 2019;18(1):23.
Wang ST, Liu LB, Li XM, et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/beta-catenin pathway. Neoplasma. 2019;66(2):232–9.
Liang Y, Song X, Li Y, et al. circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR-548p/PBLD axis in breast cancer. Oncogene. 2019;38(42):6850–66.
Li M, Wang Y, Liu Y, et al. Low expression of hsa_circ_0018069 in human bladder cancer and its clinical significance. Biomed Res Int. 2019;2019:9681863.
Chen X, Chen RX, Wei WS, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24(24):6319–30.
Lin G, Sheng H, Xie H, et al. circLPAR1 is a novel biomarker of prognosis for muscle-invasive bladder cancer with invasion and metastasis by miR-762. Oncol Lett. 2019;17(3):3537–47.
Li Y, Wan B, Liu L, et al. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2019;508(4):991–6.
Chi BJ, Zhao DM, Liu L, et al. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma. 2019;66(2):197–202.
Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14(3):321–30.
Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–62.
Wu Z, Shi W, Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/beta-catenin pathway by regulating miR-1205/APC2 axis. Biochem Biophys Res Commun. 2018;502(4):465–71.
Kun-Peng Z, Chun-Lin Z, Jian-Ping H, et al. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int J Biol Sci. 2018;14(11):1513–20.
Shi Y, Guo Z, Fang N, et al. hsa_circ_0006168 sponges miR-100 and regulates mTOR to promote the proliferation, migration and invasion of esophageal squamous cell carcinoma. Biomed Pharmacother. 2019;117:109151.
Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576.
Li W, Zhong C, Jiao J, et al. Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular rna profile and bioinformatics analysis. Int J Mol Sci. 2017;18(3):597.
Xia L, Wu L, Bao J, et al. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/beta-catenin pathway. Biochem Biophys Res Commun. 2018;503(1):385–90.
Li F, Ma K, Sun M, et al. Identification of the tumor-suppressive function of circular RNA ITCH in glioma cells through sponging miR-214 and promoting linear ITCH expression. Am J Transl Res. 2018;10(5):1373–86.
Shuai M, Hong J, Huang D, et al. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett. 2018;16(5):6495–501.
Sun S, Li B, Wang Y, et al. Clinical significance of the decreased expression of hsa_circ_001242 in oral squamous cell carcinoma. Dis Markers. 2018;2018:6514795.
Jiang XM, Li ZL, Li JL, et al. A novel prognostic biomarker for cholangiocarcinoma: circRNA Cdr1as. Eur Rev Med Pharmacol Sci. 2018;22(2):365–71.
Yang F, Liu DY, Guo JT, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–54.
Lan X, Cao J, Xu J, et al. Decreased expression of hsa_circ_0137287 predicts aggressive clinicopathologic characteristics in papillary thyroid carcinoma. J Clin Lab Anal. 2018;32(8):e22573.
Liu J, Wang D, Long Z, et al. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p /519-5p family and modulating the expression of CBX8. Cell Physiol Biochem. 2018;48(1):173–84.
Acknowledgements
Not applicable.
Funding
None.
Author information
Authors and Affiliations
Contributions
DC conceived, designed and writing of the manuscript. JW participated in revising the manuscript. ZD and XL critically reviewed and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have reviewed the manuscript and agree to publish it in its current form.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Cheng, D., Wang, J., Dong, Z. et al. Cancer-related circular RNA: diverse biological functions. Cancer Cell Int 21, 11 (2021). https://doi.org/10.1186/s12935-020-01703-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-020-01703-z