Abstract
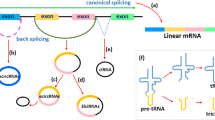
Circular RNAs (circRNAs) are a class of long noncoding RNAs that are characterized by the presence of covalently linked ends and have been found in all life kingdoms. Exciting studies in regulatory roles of circRNAs are emerging. Here, we summarize classification, characteristics, biogenesis, and regulatory functions of circRNAs. CircRNAs are found to be preferentially expressed along neural genes and in neural tissues. We thus highlight the association of circRNA dysregulation with neurodegenerative diseases such as Alzheimer’s disease. Investigation of regulatory role of circRNAs will shed novel light in gene expression mechanisms during development and under disease conditions and may identify circRNAs as new biomarkers for aging and neurodegenerative disorders.





Similar content being viewed by others
References
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733. doi:10.1371/journal.pone.0030733
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9:e1003777. doi:10.1371/journal.pgen.1003777
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461. doi:10.1038/nbt.2890
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:409. doi:10.1186/s13059-014-0409-z
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806. doi:10.1016/j.molcel.2013.08.017
Li Z, Huang C, Bao C, Chen L, Lin L, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264. doi:10.1038/nsmb.2959
Gao Y, Wang J, Zhao F (2015) CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 16:4. doi:10.1186/s13059-014-0571-3
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157. doi:10.1261/rna.035667.112
Chen L, Huang C, Wang X, Shan G (2015) Circular RNAs in eukaryotic cells. Curr Genomics 16:312–318. doi:10.2174/1389202916666150707161554
Ye CY, Chen L, Liu C, Zhu QH, Fan L (2015) Widespread noncoding circular RNAs in plants. New Phytol 208:88–95. doi:10.1111/nph.13585
Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, Luo J, Wang Y, Tian Q, Feng Q, Huang T, Han B (2015) Transcriptome-wide investigation of circular RNAs in rice. RNA 21:2076–2087. doi:10.1261/rna.052282.115
Lasda E, Parker R (2014) Circular RNAs: diversity of form and function. RNA 20:1829–1842. doi:10.1261/rna.047126.114
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. doi:10.1038/nature11993
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. doi:10.1038/nature11928
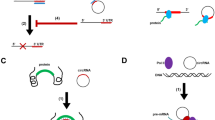
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. doi:10.1016/j.molcel.2014.08.019
Flores R, Grubb D, Elleuch A, Nohales MÁ, Delgado S, Gago S (2011) Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis δ virus: variations on a theme. RNA Biol 8:200–206
Abe N, Hiroshima M, Maruyama H, Nakashima Y, Nakano Y, Matsuda A, Sako Y, Ito Y, Abe H (2013) Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem Int Ed Engl 52:7004–7008. doi:10.1002/anie.201302044
Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H (2015) Rolling circle translation of circular RNA in living human. Cells Sci Rep 5:16435. doi:10.1038/srep16435
Danan M, Schwartz S, Edelheit S, Sorek R (2012) Transcriptome-wide discovery of circular RNAs in archaea. Nucleic Acids Res 40:3131–3142. doi:10.1093/nar/gkr1009
Plagens A, Daume M, Wiegel J, Randau L (2015) Circularization restores signal recognition particle RNA functionality in Thermoproteus. Elife 4. doi:10.7554/eLife.11623
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J (2014) Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9:e90859. doi:10.1371/journal.pone.0090859
Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Ohman M, Refojo D, Kadener S, Rajewsky N (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885. doi:10.1016/j.molcel.2015.03.027
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC (2014) Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9:1966–1980. doi:10.1016/j.celrep.2014.10.062
Shen T, Han M, Wei G, Ni T (2015) An intriguing RNA species—perspectives of circularized RNA. Protein Cell 6:871–880. doi:10.1007/s13238-015-0202-0
Lukiw WJ (2013) Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet 4:307. doi:10.3389/fgene.2013.00307
Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J (2015) Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 16:245. doi:10.1186/s13059-015-0801-3
You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, Wang X, Hou J, Liu H, Sun W, Sambandan S, Chen T, Schuman EM, Chen W (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18:603–610. doi:10.1038/nn.3975
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160:1125–1134. doi:10.1016/j.cell.2015.02.014
Abdelmohsen K, Panda AC, De S, Grammatikakis I, Kim J, Ding J, Noh JH, Kim KM, Mattison JA, de Cabo R, Gorospe M (2015) Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany NY) 7:903–910
Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16:126. doi:10.1186/s13059-015-0690-5
Wang Y, Wang Z (2015) Efficient back splicing produces translatable circular mRNAs. RNA 21:172–179. doi:10.1261/rna.048272.114
Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A (2015) Exon circularization requires canonical splice signals. Cell Rep 10:103–111. doi:10.1016/j.celrep.2014.12.002
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159:134–147. doi:10.1016/j.cell.2014.09.001
Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10:170–177. doi:10.1016/j.celrep.2014.12.019
Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28:2233–2247. doi:10.1101/gad.251926.114
Huang C, Shan G (2015) What happens at or after transcription: insights into circRNA biogenesis and function. Transcription 6:61–64. doi:10.1080/21541264.2015.1071301
Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE (2015) Combinatorial control of drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 29:2168–2182. doi:10.1101/gad.270421.115
Petkovic S, Muller S (2015) RNA circularization strategies in vivo and in vitro. Nucleic Acids Res 43:2454–2465. doi:10.1093/nar/gkv045
Ebbesen KK, Kjems J, Hansen TB (2016) Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta 1859:163–168. doi:10.1016/j.bbagrm.2015.07.007
Kelly S, Greenman C, Cook PR, Papantonis A (2015) Exon skipping is correlated with exon circularization. J Mol Biol 427:2414–2417. doi:10.1016/j.jmb.2015.02.018
Barrett SP, Wang PL, Salzman J (2015) Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 4:e07540. doi:10.7554/eLife.07540
Wilusz JE (2015) Repetitive elements regulate circular RNA biogenesis. Mob Genet Elements 5:1–7. doi:10.1080/2159256X.2015.1045682
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233. doi:10.1016/j.cell.2009.01.002
Taulli R, Loretelli C, Pandolfi PP (2013) From pseudo-ceRNAs to circ-ceRNAs: a tale of cross-talk and competition. Nat Struct Mol Biol 20:541–543. doi:10.1038/nsmb.2580
Thomas LF, Saetrom P (2014) Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics 30:2243–2246. doi:10.1093/bioinformatics/btu257
Hansen TB, Kjems J, Damgaard CK (2013) Circular RNA and miR-7 in cancer. Cancer Res 73:5609–5612. doi:10.1158/0008-5472.CAN-13-1568
Xu H, Guo S, Li W, Yu P (2015) The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 5:12453. doi:10.1038/srep12453
Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y (2015) Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 6:6001–6013
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, Liang L, Gu J, He X, Huang S (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215. doi:10.1038/ncomms11215
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44:2846–2858. doi:10.1093/nar/gkw027
Chen CY, Sarnow P (1995) Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268:415–417
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R (1993) Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73:1019–1030
Chao CW, Chan DC, Kuo A, Leder P (1998) The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med 4:614–628
Ghosal S, Das S, Sen R, Basak P, Chakrabarti J (2013) Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 4:283. doi:10.3389/fgene.2013.00283
Chen Y, Li C, Tan C, Liu X (2016) Circular RNAs: a new frontier in the study of human diseases. J Med Genet. doi:10.1136/jmedgenet-2016-103758
Pollock A, Bian S, Zhang C, Chen Z, Sun T (2014) Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Rep 7:1184–1196. doi:10.1016/j.celrep.2014.04.003
Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z, Fang X, Shorter J, Krogan NJ, Finkbeiner S, Farese RV Jr, Gitler AD (2012) Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models. Nat Genet 44:1302–1309. doi:10.1038/ng.2434
Cui X, Niu W, Kong L, He M, Jiang K, Chen S, Zhong A, Li W, Lu J, Zhang L (2016) hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in major depressive disorder. Biomark Med 12
Acknowledgments
This work was supported by a grant from the National Science Foundation of China (81471152), the Hirschl/Weill-Caulier Trust (T.S.), and an R01-MH083680-07 grant from the NIH/NIMH (T.S.).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Floris, G., Zhang, L., Follesa, P. et al. Regulatory Role of Circular RNAs and Neurological Disorders. Mol Neurobiol 54, 5156–5165 (2017). https://doi.org/10.1007/s12035-016-0055-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-0055-4