Abstract
Background
Forty years ago the actin cytoskeleton was determined to be disrupted in fibroblasts from persons with DNA repair-defective, hereditary colon cancer, with no clear connection between the cytoskeleton and DNA repair defects at that time. Recently, the large number of sequenced genomes has indicated that mammalian mutagenesis has a large stochastic component. As a result, large coding regions are large mutagen targets. Cytoskeletal protein-related coding regions (CPCRs), including extra-cellular matrix proteins, are among the largest coding regions in the genome and are indeed very commonly mutated in cancer.
Methods
To determine whether mutagen sensitivity of the actin cytoskeleton could be assessed experimentally, we treated tissue culture cells with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and quantified overall cytoskeleton integrity with rhodamine-phalloidin stains for F-actin.
Results
The above approach indicated cytoskeletal degradation with increasing mutagen exposure, consistent with increased mutagenesis of CPCRs in TCGA, smoker samples, where overall mutation rates correlate with CPCR mutation rates (R2 = 0.8694; p < 0.00001). In addition, mutagen exposure correlated with a decreasing cell perimeter to area ratio, raising questions about potential decreasing, intracellular diffusion and concentrations of chemotherapy drugs, with increasing mutagenesis and decreasing cytoskeleton integrity.
Conclusion
Determination of cytoskeletal integrity may provide the opportunity to assess mutation burdens in nonclonal cell populations, such as in intact tissues, where DNA sequencing for heterogeneous mutation burdens can be challenging.
Similar content being viewed by others
Background
Cytoskeleton protein-related coding regions (CPCRs) are among the most commonly mutated coding regions in the cancer genome atlas, consistent with the large stochastic component to mutagenesis and their large coding regions sizes [1]. In addition, various avenues of research have indicated that mutant CPCRs are likely to impact cancer development [2,3,4,5]. Regulatory anomalies related to cancer development have also implicated the cytoskeleton, although over the long history of this topic, there have been contradictory implications of the impact of the disorganized cytoskeleton on cancer progression [6,7,8,9,10]. And, it is clear numerous other conditions and medical issues are related to CPCR mutations [11, 12], potentially including heart and lung damage from mutagenic, cancer chemotherapy [13]. Yet, there has never been a direct test of the sensitivity of the cytoskeleton to mutagen exposure, in an experimental setting. In this report, we have examined the impact of in vitro mutagen treatments on the cytoskeleton, particularly in normal skin fibroblasts, with results indicating that cytoskeleton may provide an opportunity to assess mutation burdens in a highly cost-efficient manner, albeit with little specificity. Such an opportunity could be useful in settings where frequent (but cost-effective) monitoring of mutation burdens could reduce negative health consequences, such as the monitoring of children receiving chemotherapy.
Materials and methods
Melanoma cells, exome sequencing, phalloidin staining, and microscopy
WM9 melanoma cells [14] were maintained in a CO2-incubator, in tissue culture, with RPMI, penicillin, streptomycin, glutamine, pyruvate and 10% fetal bovine serum; treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK; Fluka), as follows. Ten milligrams (mg) of NNK were dissolved into DMSO to generate a 2 mg/ml (1000×) stock. A 30% confluent flask of WM9 cells were treated with 2 μg/ml of NNK, following use of NNK for tissue culture as described in Ref. [15]. After 4 days, cells were trypsinized and re-plated at low dilution for isolating individual colonies. The colonies were amplified and whole-exome sequencing was performed by the Functional Genomics Core Facility at the Moffitt Cancer Center and Research Institute (Tampa, FL). Briefly, one microgram of genomic DNA recovered from the NNK treated, WM9 subclones was used to generate a sequencing library using the Kapa Library Preparation Kit (Kapa Biosystems, Inc., Wilmington, MA). The size and quality of the library was evaluated using the Agilent BioAnalzyer, and equimolar amounts were used for a whole-exome enrichment using the Roche NimbleGen SeqCap EZ Exome Library v2.0 kit, which targets 64 Mb of genomic DNA sequence (Roche NimbleGen, Inc., Madison WI). Quantitative PCR was then used to quantitate the library with the Kapa Library Quantification Kit. Each enriched DNA library was sequenced on the Illumina NextSeq500 sequencer to generate 75–80 million, 75-base paired-end reads for a final average target coverage depth of about 100×. The raw sequence data were processed for alignment and variant calling using the BWA-Enrichment v2.10 software on the Illumina BaseSpace web application, which utilizes the BWA aligner and the GATK framework [16,17,18]. The verification of the WM9 cell line was obtained with the sequencing results representing the BRAF V600E mutation (rs113488022) as well as via previous study [14, 19]. For rhodamine-phalloidin staining, microscopy and quantification of rhodamine-phalloidin binding, the WM9 subclones were thawed in 10% FBS in glass bottom, 20 mm diameter micro-wells with #1 cover glass (0.13–0.0.16 mm) (In Vitro Scientific). The media was replaced the next day with 0.1% FBS after washing the cells twice with serum free media. After 48 h, cells were fixed with a 1 ml solution containing 50 µg/ml lysopalmitoylphosphatidylcholine in 3.7% formaldehyde. Ten units/ml of rhodamine-phalloidin (Biotium) were added immediately after fixation and cells were placed for 20 min at 4 °C, and washed twice with PBS for microscopy.
NNK treatment of foreskin fibroblasts
Pre-crises diploid fibroblasts were grown as described above for WM9 cells, plated at 10% confluence in 35 mm plates, and treated the following day with NNK in twofold serial dilutions, beginning with 4 μg/ml, twice the NNK concentration used for isolation of the WM9 subclones above, for a total of eleven NNK concentrations and one untreated sample. The process was duplicated to have one set of treatments for measuring cell viability and one set of treatments for cell expansion, storage and analysis. The cell expansion process was strictly limited prior to freezing to minimize overgrowth of any one variant until the rhodamine-phalloidin analysis was completed. Seven days post-NNK treatment, the cells were transferred to 75 cm2 flasks, and 7 days after that first transfer, four vials of the NNK treated fibroblasts were frozen. For phalloidin staining, microscopy and quantification of phalloidin binding, the fibroblasts were thawed in 10% FBS. The media was replaced the next day with 0.1% FBS after washing the cells twice with serum free media. The cells were cultured for an additional 48 h, fixed, and stained with rhodamine-phalloidin, as indicated above for the WM9 melanoma subclones. Microscopy is described below.
Raji B cell phalloidin assay
A saturated culture of Raji B-cells (2 million cells/ml) was diluted 1/6 and treated with NNK concentrations as indicated in Fig. 5. Cells were re-diluted 1/6 3 days after NNK treatment, diluted again 1/4 and treated with 2 μM Entinostat, which leads to apoptosis and perforation of the membranes for subsequent rhodamine-phalloidin and DAPI treatment prior to flow cytometry. After 48 h, cells are pelleted, re-suspended in PBS with 2% human serum, to which was added 1 μg/ml DAPI and 10 units/ml rhodamine-phalloidin. The cells were left at 4 °C for 20 min and then assayed by flow cytometry for rhodamine and DAPI absorbance.
Microscopic analyses
Cells in glass bottom wells were observed with a Zeiss Z1 Observer inverted microscope through a 20×/0.5NA Plan Apo objective (Carl Zeiss GmbH, Germany). An EXFO X-CITE 120 light source (Lumen Dynamics Group Inc, Mississauga, Canada) and filter cube (Chroma Technology Corporation, Vermont, USA) were used for viewing. Fluorescence and phase contrast images were captured with an AxioCam MRm3 CCD camera and Axiovision 4.8.2 software (Carl Zeiss GmbH, Germany).
ImageJ quantification
For the WM9 melanoma subclones, the ImageJ, Cell-Counter Analyzing tool was used to count each cell in the overlapping phase and rhodamine images; the cell counts were tabulated under “Results”. The Measure tool was used to obtain the mean value for brightness, per the user manual (http://imagej.nih.gov/ij/docs/guide/user-guide.pdf). The mean value for brightness was then divided by the number of cells in each image. For the fibroblasts, five images were obtained for each of the 12 wells with different concentrations of NNK treated fibroblasts. From each of these five images, five individual cells were exported as individual images. Each cell was selected on the basis of having a clear boundary, having no other overlying cells, and being completely included in the image. The individual cells were then exported as individual images, with a black background. Thus a total of three hundred images of individual cells were obtained. Each image was opened in ImageJ and the brightness and contrast controls were open. Auto button for Brightness and Contrast controls was selected to automatically optimize brightness and contrast based on an analysis of the image’s histogram. When selected, the entire image was optimized based on the ImageJ analysis of the selection. The ImageJ optimization was done by allowing a small percentage of pixels in the image to become saturated. To quantify the red color of the rhodamine-phalloidin, we selected Analyze, then Histogram, then RGB to obtain an optimized histogram. This histogram was termed “Red Histogram of [filename]” by the ImageJ software. The histogram values were then imported into Excel and the average was calculated. The average was then divided by the area of each cell. The ImageJ version 1.48v was used.
Analysis of the cancer genome atlas (TCGA) lung adenocarcinoma (LUAD) mutations from smokers
The TCGA (http://cancergenome.nih.gov/) smoker LUAD data was accessed as described [20]. Briefly, barcodes representing current smokers and total mutations were downloaded, and the Excel COUNTIF macro was used to determine the number of mutations per barcode for each of the current smoker barcodes. The mutation data for the HUGO symbols for the CPCR set were copied to a second Excel sheet and the COUNTIF macro was used to obtain CPCR mutations per barcode. The data was access with approval via National Institutes of Health Data Access Committee Request #27073-3 for Project #6300.
Exome sequences of Moffitt Cancer Center, bladder cancer patients
The exome sequences for the bladder cancer specimens were obtained exactly as described [21], using the Moffitt Cancer Center functional genomics core facility, USF IRB (Ethics) Approval Number Pro00022538 (Additional files 1, 2, 3 and 4).
Quantification of anti-F-actin staining of the bladder cancer samples
Formalin fixed, paraffin embedded (FFPE) bladder cancer samples (USF IRB Approval Number Pro00022536, July 16, 2015) were stained with a polyclonal anti-F-actin (Bioss, Catalog Number bs1571R) antibody at the Moffitt Cancer Center histology core facility. Image J (version 1.48v) was used to quantify F-actin binding. Images were opened into the ImageJ software. Using the brightness and contrast menu (image, adjust, brightness and contrast) the auto feature was selected. ImageJ was used to create a histogram of each cell (analyze, histogram—shortcut CRTL+H), per the Image J’s user guide: “With RGB images, the default histogram is calculated by converting each pixel to grayscale using the formula gray = (red + green + blue) ⁄ 3 or gray = 0.299 × red + 0.587 × green + 0.114 × blue” (https://imagej.nih.gov/ij/docs/guide/146-30.html#toc-Subsection-30.10). The grey-scale histograms were then copied into Excel for further analysis (see Additional file 5). Using images that contain large amounts of F-actin staining (brown), an area was selected that specifically represented F-actin binding. From this selection a plot profile was created representing the measurement of F-acting binding in each cell (see Additional files 5, 6).
Second harmonic generation (SHG) microscopy and image quantification of melanoma cell lines
Second harmonic generation [22, 23] images were captured through a 25×/0.95NA water objective lens with a Leica SP5 Multiphoton Microscope (Leica Microsystem GmbH, Wetzlar, Germany) equipped with a MaiTai DeepSee Ti–sapphire laser (Spectra-Physics Inc., Mountain View, CA) and HyD detectors. The MP laser was tuned to 880 nm and emissions were collected through a 440 nm band pass filter in order to achieve SHG imaging. In addition to SHG, bright field images of the identical fields were captured an Argon laser tune to 488 nm and transmitted light PMT. All images and overlays were prepared in Leica LASAF software (Leica Microsystems GmbH, Wetzlar, Germany). SHG signal from each image was evaluated using Definiens Developer version 2.4 (Definiens AG, Munich, Germany). Within this software an auto-threshold algorithm was used to segment the SHG signal from the background and determine total fluorescence signal from SHG in each image. Finally SHG signal per cell was calculated by using the bright field image to enumerate the total number of cells in each SHG image. Further details are in Additional file 7.
Results
We treated WM9 melanoma cells with NNK [15] and isolated five clones for phalloidin binding, to assess the integrity of the F-actin-based cytoskeleton [7] (Fig. 1) and for exome sequencing. The SNVs unique to each clone were identified (Additional file 8) and analyzed with the web based PROVEAN tool (PROVEAN v1.1.3, using a cutoff value of − 2.5, with “more negative” representing more deleterious; http://provean.jcvi.org/). Results indicated that the three subclones with the lowest level of rhodamine-phalloidin staining also had a large number of deleterious mutations among a CPCR set, previously defined by the frequency of mutated CPCRs in five cancer datasets [20]. The two subclones with the higher level of rhodamine-phalloidin staining had no deleterious mutations (Fig. 1; Table 1; p < 0.0004, Students t test). These results suggested that rhodamine-phalloidin binding could represent a measure of cytoskeleton integrity following mutagen exposure and prompted consideration of a different experimental design, which allowed for a more comprehensive opportunity for determination of statistical significance; and allowed for an opportunity to assess mutagen impact in a nonclonal cell population.
Quantification of rhodamine-phalloidin binding and summation of CPCR deleterious amino acids in NNK treated, WM9 subclones. Quantification of phalloidin binding is described in “Materials and methods”. See also Table 1 and Additional file 8. The p value for comparison of deleterious amino acids, for the high and low phalloidin-binding subclones is < 0.0004. However, there is no significant difference between the two types of subclones in terms of total unique and shared CPCR mutations, i.e., mutations shared with another subclone in the same high or low phalloidin-binding group. Thus, the high phalloidin-binding subclones had a total of 357 unique and shared mutations and the low phalloidin-binding subclones had a total of 569 mutations, in the CPCR set. The HUGO symbols for the CPCR set, for both Figs. 1 and 6, were obtained from Ref. [25] are: ANK2, APC, COL11A1, DNAH10, DNAH11, DNAH3, DNAH5, DNAH7, DNAH8, DSCAM, DST, FAT3, FAT4, FBN2, FGFR1, FLG, MUC16, MUC17, MUC4, NEB, NEFH, NF1, PCDH15, PCDHAC2, PCDHGC5, PCLO, PKHD1, PLEC, RELN, SPTA1, SPTAN1, SSPO, SYNE1, SYNE2, TTN, and XIRP2
The rhodamine-phalloidin detected cytoskeleton in pre-crises, foreskin fibroblasts (Fig. 2a) had more extensive structure than the phalloidin-detected cytoskeleton in the melanoma cells (Fig. 2b), not surprising, considering the association of disrupted cytoskeleton with tumorigenesis [this more extensive structure is in evidence when comparing the foreskin fibroblasts to either the low or high mutation WM9 subclones (Fig. 2a–c)]. The additional structure offered the opportunity of mutagen-sensitivity over a wider range. To meet the above goals, we treated the fibroblasts with eleven, twofold serial dilutions of NNK, allowed for several rounds of division, stained the fibroblasts with rhodamine-phalloidin, and quantified the rhodamine-phalloidin binding using the ImageJ software. Results indicated a statistically significant correlation in the reduction of rhodamine-phalloidin staining with increasing levels of mutagen exposure, whether calculated as rhodamine-phalloidin binding per cell (Fig. 3a; R2 = 0.7177; p < 0.004) or per unit area (of the cells) (Fig. 3b; R2 = 0.5983; p < 0.004).
Contrasting rhodamine-phalloidin stains of a WM9 melanoma subclone and foreskin fibroblasts. a Rhodamine-phalloidin stain of human, pre-crises foreskin fibroblasts used in Fig. 3. b Rhodamine-phalloidin stain of the subclone WM9-1F (a low mutation WM9 subclone). c WM9-3R, a high mutation, WM-9 subclone (Fig. 1; Table 1) shown here for comparison with the WM9-1F image (b). Scale bar, bottom right in all images, is 10 μm
The impact of the exposure to NNK on foreskin fibroblasts, rhodamine-phalloidin binding. a Correlation of mutagen exposure and overall red pixel count (rhodamine-phalloidin binding), per cell. R2 = 0.7177; p < 0.004. b Correlation of mutagen exposure and red pixel count (rhodamine-phalloidin binding), per cell-area. R2 = 0.5983; p < 0.004. For additional detail see “Materials and methods” and Additional file 9
We considered the possibility that the mutagen impact on the cytoskeleton could also impact cell shape. Thus, we obtained the perimeters and areas for 25 cells for each mutagen treatment level, and for the untreated cells, with results indicating a downward progression in the perimeter to area ratios (Fig. 4; R2 = 0.5043; p < 0.01).
Correlation of mutagen exposure and the decrease in the cell perimeter to area ratio. R2 = 0.5043; p < 0.01. For additional detail see “Materials and methods” and Additional file 10
To determine whether the impact of mutagen on rhodamine-phalloidin binding could be detected for non-adherent cells, we treated Raji B-cells with NNK, or mock-treated the cells, and assessed rhodamine-phallodin binding by flow cytometry, with results indicating reduced phalloidin binding after the NNK exposure (Fig. 5).
Raji B-cell, rhodamine-phalloidin binding following exposure to NNK. See Additional file 11
To obtain a better understanding of the CPCR set as a mutagen sentinel set in vivo, we assessed the mutation occurrences in this set, as well as total number of mutations, in smokers among the LUAD (lung adenocarcinoma) dataset in TCGA. Results indicated that the CPCR mutations correlate with overall mutation counts, with a high degree of statistical significance (Fig. 6; R2 = 0.8694; p < 0.00001).
Correlation of TCGA LUAD smoker mutations and CPCR mutations. The mutation counts were obtained as indicated in “Materials and methods”, with further detail in Additional file 12. R2 = 0.8694; p < 0.00001. The CPCRs included in the above analysis is identical to Ref. [20] and is included in the legend to Fig. 1
To determine whether the above indicated correlation of a high rate of mutagenesis and the degradation or loss of F-actin-based, cytoskeletal structures could be observed in a patient setting, we obtained the exome sequence of 16 Moffitt Cancer Center bladder cancer patients. After removal of SNPs, the number of remaining single nucleotide variants (SNVs) ranged from 1962 to 5054, with the majority of the samples very close to the lower number of SNVs. These two samples represented 53 (BLCA1) and 89 (BLCA-2) mutations in the above indicated CPCR set [20] (Fig. 1), respectively (see Additional files 2, 3, 4). We next stained FFPE tissues representing the two extreme bladder cancer samples, with respect to SNV occurrence, with anti-F-actin (Fig. 7a). Although only two patient samples were evaluated, quantification of the results clearly demonstrated continued consistency of the presence of F-actin structures and lower mutation counts (Fig. 7b).
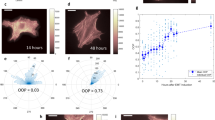
Anti-F-actin stains of MCC-BLCA1 and MCCBLCA-2. a FFPE, anti-F-actin stains of MCC-BLCA1 (top 2 panels; low mutation rates) and MCC-BLCA2 (bottom 2 panels; high mutation rates). b Higher magnification of arrowhead region of upper right panel of a, to show more precise image of anti-F-actin stain. c Higher magnification of the lower right panel of a, center of image of a lower right panel, emphasizing the reduced level of anti-F-actin staining. d Quantification of anti-F-actin binding to the bladder cancer cells (BLCA1, left side; BLCA2, right side). In a, the anti-F-actin binding to the cells is represented by the brown lines at the edges of the cells, indicated by arrowheads. See Additional files 5 and 6, particularly for explanation of c quantification units on y-axis
Finally, to identify a second approach, besides the F-actin dependent approach, for assessing the impact of mutations on ordered structure arising from large polypeptide subunits forming polymers, we obtained SHG images of the NNK exposed WM9 subclones with fewer or more mutations. We obtained images for two of the low mutation subclones, WM9-1F and -3F; and two high mutation subclones, WM9-5F and 3R (Fig. 8a). Quantification of the results (Fig. 8b) indicated a 25% decrease in pixel intensity representing ordered polymers in the higher mutation WM9 subclones, with p < 0.04 (Additional file 7).
Second harmonic generation (SHG) imaging of the NNK treated, WM9 subclones. a Example SHG image of the low mutation subclone, WM9-3F. b Quantification of the SHG signal (low mutation subclones, left bar; high mutation subclones, right bar) (see Additional file 7)
Discussion
Taken together, the above results are consistent with the conclusion that rhodamine-phalloidin binding reflects mutant cytoskeletal proteins that reduce the integrity of the F-actin based cytoskeleton. This type of “integrity reduction” is likely traceable to the fact that the cellular cytoskeleton is a polymeric structure composed of many subunits representing at least several, if not many distinct proteins. Thus, a large number of coding regions may be susceptible to mutation such that any one of the resultant mutant, cytoskeletal related proteins could corrupt the polymer, akin to what is seen in osteogenesis imperfecta, whereby mutant collagen subunits interfere with polymer and cartilage formation in a dominant negative fashion [11]. As an alternative possibility, it is conceivable that more mutagen would make it more likely that a point mutation could affect a regulatory pathway, in turn impacting the cytoskeleton [9, 10]. However, it is highly improbable that such rare mutagenic events would lead to a significant and strong correlation coefficient representing the mutagen increases and the impact on the phalloidin binding (Figs. 3, 5) and on the perimeter to area ratios (Fig. 4).
In 1977, Pollack and colleagues observed that the actin-cytoskeleton was disrupted in fibroblasts from persons with DNA repair-defective, hereditary colon cancer [24]. Given the above results and the points in the preceding paragraph, the most likely explanation for that observation is that the DNA repair defects led to mutations in CPCRs, which in turn lead to a disorganized, actin-cytoskeleton (Additional file 12).
Our results also indicate that more mutagen exposure leads to a reduced perimeter to area ratio, which raises questions about the basis for drug resistance in cancer. Many articles have indicated that the result of selecting for resistance to a variety of anticancer drugs is a spheroid cell, often invoking epithelial to mesenchymal transition as a process fundamental to both cell shape change and drug resistance. However, it is clear that the CPCR set is a large mutagen target and loss of cytoskeleton integrity may lead to reduced cell spreading and thus a reduced surface area to volume ratio, in our results represented as a reduced perimeter to area ratio. With such a shape alteration, reduced intra-cellular drug concentrations, and reduced intra-cellular drug diffusion should be considered as possible explanations for why drug resistance selects for spherical cells. In fact, given the vulnerability of the CPCR set to mutation, mutagenic drug treatments, common cancer therapies, may both create and select for drug resistance.
Finally, it is important to note the likely opportunity to take advantage of SHG microscopy for obtaining crude but potentially medically useful indications of mutation burdens in persons. For example, in some cases smoking may lead to an accumulation of mutation burdens easily assessable via a buccal swab and SHG microscopy. This type of approach may conveniently and inexpensively distinguish between persons who have suffered numerous mutations and those with low mutation burdens. A similar monitoring possibility could be useful for children treated with anti-cancer chemotherapy. An SHG based monitoring strategy would be far less expensive and less logistically complicated than DNA sequencing and thereby allow for very frequent, long-term monitoring. In particular, assessing mutation burdens via the impact of mutations on cytoskeletal or extra-cellular matrix structures would eliminate the need for a clonal population of cells for DNA sequencing or the need to do single cell sequencing. That is, assaying the mutation burdens via loss of subcellular structures need not require that mutations be in the same gene or at the same nucleotide position. This type of assay approach would allow for using a heterogeneous set of mutant cells as the basis of a mutation burden assessment.
Conclusion
The above work supports the idea that an indication of mutation burdens can be had by assessing the actin-cytoskeleton integrity, consistent with the idea that large coding regions are, based on probability, frequent mutagen targets and can thereby often express dominant negative mutant polypeptides that corrupt polymer formations. These results may lead to cost-effective approaches for monitoring mutation burdens where a high level monitoring frequency, or other circumstances, prevent relatively costly DNA sequencing approaches, for example for monitoring the impact of mutagenic chemotherapy.
Abbreviations
- CPCR:
-
cytoskeletal protein-related coding region
- ECM:
-
extra-cellular matrix
- NNK:
-
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- TCGA:
-
the cancer genome atlas
- SNV:
-
single nucleotide variant
- LUAD:
-
lung adenocarcinoma
- HUGO:
-
human genome organization
References
Parry ML, Ramsamooj M, Blanck G. Big genes are big mutagen targets: a connection to cancerous, spherical cells? Cancer Lett. 2015;356(2 Pt B):479–82.
Fawcett TJ, Parry ML, Blanck G. A novel approach to evaluating cancer driver gene mutation densities: cytoskeleton-related gene candidates. Cancer Genomics Proteomics. 2015;12(6):283–90.
Wang Y, Marino-Enriquez A, Bennett RR, Zhu M, Shen Y, Eilers G, Lee JC, Henze J, Fletcher BS, Gu Z, et al. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat Genet. 2014;46(6):601–6.
Fernandez K, Serinagaoglu Y, Hammond S, Martin LT, Martin PT. Mice lacking dystrophin or alpha sarcoglycan spontaneously develop embryonal rhabdomyosarcoma with cancer-associated p53 mutations and alternatively spliced or mutant Mdm2 transcripts. Am J Pathol. 2010;176(1):416–34.
Blighe K, Kenny L, Patel N, Guttery DS, Page K, Gronau JH, Golshani C, Stebbing J, Coombes RC, Shaw JA. Whole genome sequence analysis suggests intratumoral heterogeneity in dissemination of breast cancer to lymph nodes. PLoS ONE. 2014;9(12):e115346.
Pollack R, Lo A, Steinberg B, Smith K, Shure H, Blanck G, Verderame M. SV40 and cellular gene expression in the maintenance of the tumorigenic syndrome. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):681–8.
Verderame M, Alcorta D, Egnor M, Smith K, Pollack R. Cytoskeletal F-actin patterns quantitated with fluorescein isothiocyanate-phalloidin in normal and transformed cells. Proc Natl Acad Sci USA. 1980;77(11):6624–8.
Xu W, Mezencev R, Kim B, Wang L, McDonald J, Sulchek T. Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PLoS ONE. 2012;7(10):e46609.
Kim TY, Vigil D, Der CJ, Juliano RL. Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in regulation of the cytoskeleton and cell motility. Cancer Metastasis Rev. 2009;28(1–2):77–83.
Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28(1–2):65–76.
Ben Amor M, Rauch F, Monti E, Antoniazzi F. Osteogenesis imperfecta. Pediatric Endocrinol Rev. 2013;10(Suppl 2):397–405.
Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349(6251):982–6.
Blanck G. Letter to the editor: giant proteins and cancer chemotherapy cardiotoxicity. Am J Physiol Heart Circ Physiol. 2015;309(4):H718.
Goodwin BL, Xi H, Tejiram R, Eason DD, Ghosh N, Wright KL, Nagarajan U, Boss JM, Blanck G. Varying functions of specific major histocompatibility class II transactivator promoter III and IV elements in melanoma cell lines. Cell Growth Differ. 2001;12(6):327–35.
Dey N, Chattopadhyay DJ, Chatterjee IB. Molecular mechanisms of cigarette smoke-induced proliferation of lung cells and prevention by vitamin C. J Oncol. 2011;2011:561862.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8.
Cronin K, Escobar H, Szekeres K, Reyes-Vargas E, Rockwood AL, Lloyd MC, Delgado JC, Blanck G. Regulation of HLA-DR peptide occupancy by histone deacetylase inhibitors. Hum Vacc Immunother. 2013;9(4):784–9.
Parry ML, Blanck G. Flat cells come full sphere: are mutant cytoskeletal-related proteins oncoprotein-monsters or useful immunogens? Hum Vacc Immunother. 2016;12(1):120–3.
Gill TR, Samy MD, Butler SN, Mauro JA, Sexton WJ, Blanck G. Detection of productively rearranged TcR-alpha V-J Sequences in TCGA Exome Files: implications for tumor immunoscoring and recovery of antitumor T-cells. Cancer Inform. 2016;15:23–8.
Samim M, Prent N, Dicenzo D, Stewart B, Barzda V. Second harmonic generation polarization properties of myofilaments. J Biomed Opt. 2014;19(5):056005.
Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci USA. 2003;100(12):7075–80.
Kopelovich L, Conlon S, Pollack R. Defective organization of actin in cultured skin fibroblasts from patients with inherited adenocarcinoma. Proc Natl Acad Sci USA. 1977;74(7):3019–22.
Parry ML, Blanck G. Flat cells come full sphere: are mutant cytoskeletal-related proteins oncoprotein-monsters or useful immunogens? Hum Vacc Immunother. 2015;12:120–3.
Authors’ contributions
NMC, CAGG, VKP, MLP, EPP, EMJ, CMW conducted tissue culture and mutagen-treatment related laboratory procedures; employed ImageJ for analysis of microscopic images; and assisted in analyses of the data. Michele Parry was particularly responsible for the isolation of the WM9 subclones. Joseph Johnson conducted microscopy procedures and assisted in analysis of resulting data, particularly analyses of the SHG data. RJS, JMY contributed bioinformatics and computational analyses for mutation assessments, particularly for Table 1 and Fig. 4. WJS performed the clinical evaluations and surgery related to Fig. 7. DC performed the microscopy analysis related to Fig. 7. GB supervised the project at all stages and assisted in data analyses and manuscript preparation. All authors read and approved the final manuscript.
Acknowledgements
Authors wish to thank the Functional Genomics Core Facility at the H. Lee Moffitt Cancer Center and Research Institute, particularly Sean Yoder and Chaomei Zhang; the Moffitt Cancer Center Analytical Microscopy facility; the Moffitt Cancer Center Tissue Core facility, especially Michelle Fournier; USF research computing, especially Dr. Tony Green; the Morsani College of Medicine Flow cytometry Core facility, especially Dr. Karoly Szekeres;
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The additional files for this report includes all relevant, original data. Also, any additional data is freely available by email request to corresponding author.
Consent for publication
Not applicable (see Additional file 1).
Ethics approval and consent to participate
See Additional file 1.
Funding
Authors would like to acknowledge the financial contributions of the Anna Valentine Program and the taxpayers of the State of Florida.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Clark, N.M., Garcia Galindo, C.A., Patel, V.K. et al. The human, F-actin-based cytoskeleton as a mutagen sensor. Cancer Cell Int 17, 121 (2017). https://doi.org/10.1186/s12935-017-0488-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12935-017-0488-5