Abstract
Background
Antibiotics have long been the first line of defense to prevent Escherichia coli infections, but they have lost their potency since bacteria have grown increasingly resistant to treatment. The present research aimed to study the drug resistance and the prevalence of tetracycline resistance genes in E. coli isolated from broilers with colibacillosis.
Results
The results showed that the most prevalent type of drug resistance was to tetracycline at 95.0%, and the least was to gentamicin at 21.7%. The prevalences of antimicrobial resistance among the tested antibiotics were significantly different (p < 0.001). A statistically significant difference was observed between the prevalence of the tet genes (p < 0.001). The tetD positive isolates and antibiotic sensitivity to tetracycline showed statistical significant differences (p = 0.017).
Conclusions
Considering the results, tetA is the most common tetracycline resistance gene, and the presence of tetD and antibiotic sensitivity to tetracycline had a significant relationship in E. coli isolated from colibacillosis infections.
Similar content being viewed by others
Background
Antibiotic are mostly used in animals, including chickens, for the treatment or prevention of diseases [1,2,3]. Some of these agents, such as avilamycin, avoparcin, flavomycin, monensin, and salinomycin, have also been used in a few countries to increase chick growth rates [1]. The high consumption of antibiotics by livestock is a global problem that can increase the antibiotic resistance of human and animal bacteria such as Escherichia coli [4,5,6]. Antibiotic resistance in poultry bacteria and transfer of this acquired ability to human bacteria can disrupt the treatment of human infections [4].
Tetracyclines are broad-spectrum antibiotics that are widely used against gram-negative and gram-positive bacteria. These drugs prevent the binding of aminoacyl-tRNA to the 30s ribosomal subunit and disrupt protein synthesis, which prohibits the growth of sensitive bacteria [7]. Because of the numerous advantages of tetracyclines, such as widespread availability, low cost, and few side effects, the use of these kinds of antibiotics for the treatment of animal and human infections has been increasing in recent years [8]. This has led to the emergence of tetracycline-resistant bacteria, which is now limiting the use of these antibiotics [9].
Tetracycline resistance genes are generally coded in plasmids and transposons and are transmitted through conjugation. However, the relevant genes are also found in the chromosome in some isolates [10, 11]. Mechanisms of resistance to tetracycline through the acquisition of tet genes mainly include efflux pumps, ribosomal protection, and enzymatic deactivation. Mutations also contribute to the antibiotic resistance [12]. The tet genes found at the highest frequency in gram-negative bacteria are related to efflux pumps, which are coded by the tetA, tetB, tetC, tetD, and tetG genes [13, 14].
Resistance to tetracycline can be used for evaluating antibiotics resistance genes. Also, the epidemiological-molecular evaluation of resistant strains to these compounds can broaden the knowledge of appropriate treatments and prevent loss of capital [15]. Strategies to prevent the spread of antibiotic resistance require surveillance of its encoding genes. This study provides new insight for explaining the combination tetracycline-resistant encoding genes that may synergistically enhance the antimicrobial resistance against tetracycline in E. coli isolates.
Hence, the present research aimed to study the resistance to antimicrobial agents and the prevalence of tetracycline resistance genes (tetA, tetB, tetC, and tetD) in E. coli isolated from broiler chickens affected with colibacillosis on farms in Sistan, Sistan and Baluchestan Province, Iran.
Results
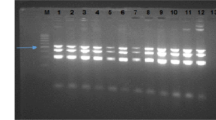
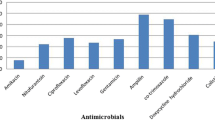
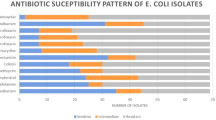
The antimicrobial drug susceptibility patterns of the E. coli isolates are presented in Table 1. The frequency of resistance to tetracycline, ciprofloxacin, co-trimoxazole, lincospectin, cefuroxime, and gentamicin were 95.0, 88.3, 86.7, 53.3, 46.7, and 21.7%, respectively (Supplementary materials). A total of 52/60 (86.6%) isolates were identified as multidrug resistance strains. The prevalence of antimicrobial resistance among the tested antibiotics was significantly different (p < 0.001). The gel photograph of the amplified products is presented in Fig. 1; Additional file 1: Fig. 1. Antibiotic sensitivity to tetracycline dependent on the presence or absence of the tetA, tetB, tetC, and tetD genes is shown in Table 2. The tetD positive isolates and antibiotic sensitivity to tetracycline showed statistical significant differences (p = 0.017). There was no observed statistical association between tet genes and other antibiotics sensitivity, including to gentamicin, lincospectin, cefuroxime, ciprofloxacin and co-trimoxazole (statistical results not shown). The prevalence of the tetA, tetB, tetC and tetD genes was 96.7, 38.3, 31.7, and 8.3%, respectively. A statistically significant difference was observed between the prevalence of the tet genes (p < 0.001). The results showed that all isolates had at least one of the tetracycline resistance genes. The distribution of E. coli isolates harboring one, two, three, and four tet genes were 43.3, 43.3, 13.3, and 0.0%, respectively.
a: Electrophoresis results for detecting the tetA and tetD genes. Lanes: 1, marker; 2, positive control; 3, negative control; 4, isolate contains tetA gene; 5 & 6, isolates contain tetA and tetD genes. b: Electrophoresis results for detecting the tetB and tetC genes. Lanes: 1, marker; 2, positive control; 3, negative control; 4 & 8, isolates contain tetB gene; 6 & 9, isolates contain tetC gene; 5 & 7, isolates contain tetB and tetC genes
Discussion
Tetracycline is the most frequently used antibiotic in the treatment and control of various poultry diseases. Tetracycline can be administered orally and has only a very low risk of major side effects. Furthermore, tetracycline is one of the cheapest antimicrobial agents available [16, 17]. In this study, resistance to more than one antimicrobial agent was observed for many of the E. coli isolates (52/60; 88.6%), and 95.0% of the isolates showed tetracycline resistance.
In another study, the frequency of tetracycline resistance of E. coli isolated from colibacillosis in Iran was reported at 96.0% [5]. Besides, the antimicrobial resistance prevalence of E. coli isolated from turkey in Iran, for gentamicin, tetracycline, co-trimoxazole, norfloxacin and cefuroxime, were 5, 51.7, 23.3, 5 and 48.3%, respectively [18]. Adesiyun et al. (2007) reported similar levels of resistance to antimicrobial agents among isolates of E. coli to tetracycline (58.5%), enrofloxacin (25.4%), gentamicin (9.3%), and sulphamethoxazole/trimethoprim (15.3%) [19]. The excessive use of tetracycline compounds for the treatment of poultry diseases could be the reason for their high rate of resistance to this type of antibiotic [5, 20,21,22,23].
Fluroquinolone compounds like ciprofloxacine are critically important antimicrobials for treating severe infections in human and reduced susceptibility to quinolone can lead to treatment failures and is considered a public health risk [24]. According to the results of the current study, the frequency of tetracycline resistance (95.0%) was only slightly higher than the frequency of quinolone resistance (88.3%).
In this study, we observed that 43.30 and 13.3% of the E. coli isolates harbored two and three tetracycline resistance genes, respectively. The presence of multiple tetracycline resistance genes in E. coli has been previously reported by Sandalli et al. (2010). They detected that 1.9% of E. coli isolates harbored both the tetA and tetB genes. They observed that the tetA and tetB genes could be well expressed in E. coli and subsequently impart resistance to tetracycline [25]. Furthermore, Koo and Woo (2011) also observed that 1.6% of E. coli isolates had both tetA and tetB simultaneously [12].
According to the results of our research, tetD showed a significant relationship with antibiotic sensitivity to tetracycline. This correlation may be due to the influence of the environment, different loci, mRNA copy numbers, different processes at the transcriptional/translational levels, and the expression patterns of the genes. However, Chopra and Roberts (2001) have reported that tetB provides additional resistance against doxycycline, while tetA induces resistance against tetracycline, oxytetracycline, and chlortetracycline [16].
In the current study, all isolates of E. coli had at least one tet gene, and tetA (96.7%) and tetB (38.3%) were observed at the highest frequencies. Similar to our results, in a study of meat and meat products, Koo and Woo (2011) observed that most of the isolates (98.3%) had at least one tet gene, with tetA (52.4%) and tetB (41.3%) showing the highest frequencies while the lowest frequencies were found for tetC (1.7%) and tetD (0.8%) [12]. Guerra et al. (2003) reported the prevalences of tetA and tetB were 66 and 42%, respectively, in E. coli isolated from cattle, swine, and poultry [26]. Maynard et al. (2004), in a study on E. coli isolated from human and animal samples, showed that the frequency of tetA was greater than tetB in isolates from both sources [27]. Seifi and Khoshbakht (2016) reported that 73% of E. coli strains isolated from Iranian broiler flocks were tetracycline resistant. Moreover, 46 and 41% of the isolates contained tetA and tetB genes, respectively [28].
Conclusions
The evaluation of the tetracycline resistance pattern can be helpful in choosing proper antibiotic agents for the treatment of poultry diseases. According to our results, tetA is the most common tetracycline resistance gene, and the presence of tetD and antibiotic sensitivity to tetracycline has significant relationships in E. coli isolated from colibacillosis. By identifying the types of genes responsible for resistance, more effective approaches may be developed to treat infectious diseases. The results of this study emphasize the need for cautious use of tetracycline in poultry production to decrease the prevalence of tetracycline-resistant E. coli. However, it is also recommended that antibiotics from different classes will need to be used to reduce antimicrobial resistance, and more efficiently treat infectious diseases of poultry.
Methods
Sample collection and bacterial isolation
The study population consisted of commercial broilers affected by colibacillosis. Sampling was conducted on eight broiler farms during 2017 to 2018 in Sistan, Iran. Verbal informed consent was obtained from the all farm owners for sample collection. This study was approved by the Ethics Committee of the University of Zabol with the approval number IR.UOZ.REC.1395.11.
After necropsy and identification of the respective lesions, the samples were taken from the infected chicks. Lesions were sampled by using a sterile swab and then the swab was placed in 5 ml of tryptic soy broth (TSB) medium and transferred to the microbiology laboratory of the Faculty of Veterinary Medicine of Zabol University. The TSB media were incubated at 37 °C for 24 h and then sub-cultured on MacConkey agar and eosin methylene blue (EMB) agar media (Merck, Germany).
A total of 60 isolates of E. coli were procured from the colibacillosis-related lesions and identified using precise microbiological and biochemical methods. In summary, bacterial isolates with typical colony morphology on MacConkey and EMB agar media that were lactose, indole, and methyl red positive, while Voges–Proskauer, citrate, urease, and H2S negative, were considered as E. coli [29]. The isolated E. coli were not serotyped in this study.
Antimicrobial susceptibility testing
To study the resistance pattern of the isolates, the disk diffusion method on Mueller-Hinton agar with six antibacterial paper disks was used (HamoonTeb, Iran; Table 1). In summary, a number of pure bacterial colonies were suspended in tubes containing sterilized 0.9% saline in order to make its opacity equal to the 0.5 McFarland standard tube. They were then cultured on Mueller-Hinton agar. The antibacterial paper disks were placed on the agar and after 24 h of incubation at 37 °C, the diameter of the zone of inhibition was measured. The results were interpreted based on the Clinical and Laboratory Standards Institute guidelines [30].
DNA extraction and detection of tet genes
The genomes of the isolated bacteria were extracted through boiling [31], using a thermomixer (Eppendorf, Germany), at 95 °C. In this study we investigated just the DNA genome, and the extracted DNA was stored at − 20 °C in 200 μl tubes until further analysis. Specific primers were used in order to identify the tetA, tetB, tetC, and tetD genes (Pishgam, Iran; Table 3). The Multiplex-polymerase chain reaction (PCR) mixture, with a final volume of 25 μl, contained 12 μl of 2X Master Mix (2X PCR Master Mix Red; Pishgam, Iran), 3 μl of DNA, 1 μl of forward primers (0.5 μl of tetA + 0.5 μl of tetD or 0.5 μl of tetB + 0.5 μl of tetC), 1 μl of reverse primers (0.5 μl of tetA + 0.5 μl of tetD or 0.5 μl of tetB + 0.5 μl of tetC), and 8 μl of sterilized distilled water. E. coli ATCC 25922 was used as a control strain [32]. The PCR reaction was performed using a thermocycler (Eppendorf, Germany) under the following program: one cycle of initial denaturation at 94 °C for 5 min and 35 cycles for the other steps including a denaturation step at 94 °C for 30 s, annealing of primers at 55 °C for 30 s, and an extension step at 72 °C for 30 s, followed by one cycle of final extension at 72 °C for 5 min [12]. The PCR products were electrophoresed on a 1% agarose gel and then studied in a Gel Doc Machine (Cambridge, Germany) after 20 min of exposure to ethidium bromide (CinnaGen, Iran).
Statistical analysis
The data were analyzed statistically using SPSS® (version 20) software by Chi-square and Fisher’s exact tests. Differences were considered to be statistically significant at p < 0.05.
Availability of data and materials
The data used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- E. coli :
-
Escherichia coli
- TSB:
-
Tryptic soy broth
- EMB:
-
Eosin methylene blue
- CLSI:
-
Clinical and laboratory standards institute
- ml:
-
Milliliter
- PCR:
-
Polymerase chain reaction
References
Aarestrup FM. Occurrence, selection and spread of resistance to antimicrobial agents used for growth promotion for food animals in Denmark. APMIS Suppl. 2000;101:1–48.
Miranda JM, Guarddon M, Mondragon A, Vazquez BI, Fente CA, Cepeda A, Franco CM. Antimicrobial resistance in Enterococcus spp. strains isolated from organic chicken, conventional chicken, and Turkey meat: a comparative survey. J Food Prot. 2007;70(4):1021–4.
Miranda JM, Guarddon M, Vázquez BI, Fente CA, Barros-Velázquez J, Cepeda A, Franco CM. Antimicrobial resistance in Enterobacteriaceae strains isolated from organic chicken, conventional chicken and conventional Turkey meat: a comparative survey. Food Control. 2008;19(4):412–6.
Hammerum AM, Heuer OE. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin Infect Dis. 2009;48:916–21.
Jahantigh M, Dizaji RE. Antimicrobial drug resistance pattern of Escherichia coli isolated from chickens farms with colibacillosis infection. Open J Med Microbiol. 2015;5:159–62.
Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep. 2012;127(1):4–22.
Schnappinger D, Hillen W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch Microbiol. 1996;165(6):359–69.
Garcia PG, Silva VL, Diniz CG. Occurrence and antimicrobial drug susceptibility patterns of commensal and diarrheagenic Escherichia coli in fecal microbiota from children with and without acute diarrhea. J Microbiol. 2011;49(1):46–52.
Speer BS, Shoemaker NB, Salyers AA. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev. 1992;5(4):387–99.
Guillaume G, Verbrugge D, Chasseur-Libotte M, Moens W, Collard J. PCR typing of tetracycline resistance determinants (Tet A-E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol Ecol. 2000;32(1):77–85.
Oppegaard H, Steinum TM, Wasteson Y. Horizontal transfer of a multi-drug resistance plasmid between coliform bacteria of human and bovine origin in a farm environment. Appl Environ Microbiol. 2001;67(8):3732–4.
Koo HJ, Woo GJ. Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int J Food Microbiol. 2011;145(2–3):407–13.
Schwaiger K, Holzel C, Bauer J. Resistance gene patterns of tetracycline resistant Escherichia coli of human and porcine origin. Vet Microbiol. 2010;142(3–4):329–36.
Skockova A, Cupakova S, Karpiskova R, Janstova B. Detection of tetracycline resistance genes in Escherichia coli from raw cow’s milk. J Microbiol Biotech Food Sci. 2012;1:777–84.
Gow SP, Waldner CL, Harel J, Boerlin P. Associations between antimicrobial resistance genes in fecal generic Escherichia coli isolates from cow-calf herds in western Canada. Appl Environ Microbiol. 2008;74(12):3658–66.
Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–60.
Miranda JM, Rodriguez JA, Galan-Vidal CA. Simultaneous determination of tetracyclines in poultry muscle by capillary zone electrophoresis. J Chromatogr A. 2009;1216(15):3366–71.
Shahbazi P, Jahantigh M, Salari S. Antibiotial resistance pattern and prevalence of some extende-spectrum beta-lactamase genes in Escherichia coli isolated from Turkey. Vet Res Biol Prod. 2018;15:647–79.
Adesiyun A, Offiah N, Seepersadsingh N, Rodrigo S, Lashley V, Musai L. Antimicrobial resistance of Salmonella spp. and Escherichia coli isolated from table eggs. Food Control. 2007;18:306–11.
Bryan A, Shapir N, Sadowsky MJ. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl Environ Microbiol. 2004;70(4):2503–7.
Karami N, Nowrouzian F, Adlerberth I, Wold AE. Tetracycline resistance in Escherichia coli and persistence in the infantile colonic microbiota. Antimicrob Agents Chemother. 2006;50(1):156–61.
Saenz Y, Brinas L, Dominguez E, Ruiz J, Zarazaga M, Vila J, Torres C. Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob Agents Chemother. 2004;48(10):3996–4001.
Tuckman M, Petersen PJ, Howe AY, Orlowski M, Mullen S, Chan K, Bradford PA, Jones CH. Occurrence of tetracycline resistance genes among Escherichia coli isolates from the phase 3 clinical trials for tigecycline. Antimicrob Agents Chemother. 2007;51(9):3205–11.
Hang BPT, Wredle E, Borjesson S, Sjaunja KS, Dicksved J, Duse A. High level of multidrug-resistant Escherichia coli in young dairy calves in southern Vietnam. Trop Anim Health Prod. 2019;51(6):1405–11.
Sandalli C, Özgümüş OB, Sevim A. Characterization of tetracycline resistance genes in tetracycline-resistant Enterobacteriaceae obtained from a coliform collection. World J Microb Biot. 2010;26(11):2099–103.
Guerra B, Junker E, Schroeter A, Malorny B, Lehmann S, Helmuth R. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J Antimicrob Chemother. 2003;52:489–92.
Maynard C, Bekal S, Sanschagrin F, Levesque RC, Brousseau R, Masson L, Lariviere S, Harel J. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J Clin Microbiol. 2004;42(12):5444–52.
Seifi S, Khoshbakht R. Prevalence of tetracycline resistance determinants in broiler isolated Escherichia coli in Iran. Br Poult Sci. 2016;57(6):729–33.
Quinn PJ, Markey BK, Carter ME, Donnelly WJC, Leonar FC. Veterinary microbiology and microbial disease. USA: Wiley-Blackwell; 2002.
CLSI. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. CLSI document M100-S21. Wayne, PA: Clinical and Laboratory Standards Institute; 2011.
Sambrook J, Russell DW. Molecular cloning. In: Cold Springs Harb. 3rd ed: Lab Press; 2001.
Tavakoli M, Pourtaghi H. Molecular detection of virulence genes and multi-drug resistance patterns in Escherichia coli (STEC) in clinical bovine mastitis: Alborz province, Iran. Iran J Vet Res. 2017;18:208–11.
Acknowledgements
Not applicable.
Funding
This study was only financially supported by the Vice Chancellor of Research and Technology of University of Zabol (Grant No. UOZ-GR-9618-56). They had no role in the design of the study and data collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
MJ has been contributed in necropsy, bacterial isolation, antibiogram tests, PCR, literature search, manuscript preparation and manuscript editing; KS has been contributed in antibiogram, PCR, literature search, and data collection; RED has been contributed in necropsy, bacterial isolation, antibiogram, data collection, literature search and manuscript preparation; SS has been contributed in antibiogram, PCR, statistical analysis and manuscript preparation. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Verbal informed consent was obtained from the all farm owners for sample collection and approved by the Ethics Committee of the University of Zabol. The address of the farm owners was recorded. The samples were transferred to the Faculty of Veterinary Medicine, University of Zabol, by farm owners for experimental purposes. Because the samples were transported by the owners themselves, the Ethics Committee approved this study and they felt the need for written consent was not necessary. The study was approved by the Ethics Committee with the approval number IR.UOZ.REC.1395.11.
Consent for publication
Not applicable.
Competing interests
Authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Fig. 1 The original full-length gel image of the amplified products is presented.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jahantigh, M., Samadi, K., Dizaji, R.E. et al. Antimicrobial resistance and prevalence of tetracycline resistance genes in Escherichia coli isolated from lesions of colibacillosis in broiler chickens in Sistan, Iran. BMC Vet Res 16, 267 (2020). https://doi.org/10.1186/s12917-020-02488-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-020-02488-z