Abstract
Background
Odontogenic mesenchymal stem cells (MSCs) isolated from tooth tissues are a reliable resource that can be utilized for dental tissue regeneration. Exploration of the mechanisms underlying the regulation of their differentiation may be helpful for investigating potential clinical applications. The stem cell niche plays an important role in maintaining cell functioning. Previous studies found that Wnt inhibitory factor 1 (WIF1) is more highly expressed in apical papilla tissues than in stem cells from apical papilla (SCAPs) using microarray analysis. However, the function of WIF1 in SCAPs remains unclear. In the present study, we investigated the function of WIF1 during dentinogenic differentiation in SCAPs.
Methods
A retrovirus containing HA-WIF1 was used to overexpress WIF1 in SCAPs. Using Western blot analysis, we verified the expression of HA-WIF1. Alkaline phosphatase (ALP) activity assays, Alizarin Red staining and quantitative calcium analysis were performed to investigate the in vitro potential for dentinogenic differentiation in SCAPs. The expression of dentinogenesis-associated genes DSPP, DMP1, Runx2 and OSX were assayed using real-time RT-PCR. Transplantation experiments were used to measure dentinogenesis potential in vivo.
Results
The real time RT-PCR results showed that WIF1 was more highly expressed in apical papilla tissues than in SCAPs, and its expression was increased during the process of dentinogenic differentiation. Overexpression of WIF1 enhanced ALP activity and mineralization in vitro, as well as the expression of DSPP, DMP1 and OSX in SCAPs. Moreover, in vivo transplantation experiments revealed that dentinogenesis in SCAPs was enhanced by WIF1 overexpression.
Conclusion
These results suggest that WIF1 may enhance dentinogenic differentiation potential in dental MSCs via its regulation of OSX and identified potential target genes that could be useful for improving dental tissue regeneration.
Similar content being viewed by others
Background
Due to technological improvements in tissue engineering, stem cell therapy holds great promise for the advancement of regenerative medicine. Mesenchymal stem cells (MSCs) are considered a good cell source for use in tissue regeneration [1]. From tooth tissues, it is possible to isolate several types of MSCs, including periodontal ligament (PDLSCs), dental pulp (DPSCs), and dental follicular stem cells (DFSCs), as well as stem cells from apical papilla (SCAPs) [1,2,3]. These MSCs are more intimately associated with dental tissues and can regenerate bone/dentin-like tissues and repair damaged dental tissues [1,2,3,4,5]. However, their potential for use in clinical applications is restricted since the mechanisms underlying dentinogenic differentiation remain unclear.
The stem cell niche supports and maintains the functioning of MSCs. This niche cannot be maintained when MSCs are isolated and cultured in vitro using current methods. An impaired niche will not be beneficial for dental MSC-mediated tooth tissue regeneration [6,7,8]. Therefore, the identification of the key regulator of the niche that supports dental MSCs will aid in the enhancement of dental tissue regeneration. A previous study examined gene expression profiles in apical papilla tissues, SCAPs and SCAP cell sheets and revealed several candidate key genes within the SCAP niche using microarray and bioinformatic analysis [9]. In their analysis, one canonical Wnt/β-catenin inhibitor, Wnt inhibitory factor 1 (WIF1), was found to be more highly expressed in apical papilla tissues than in SCAPs. Wnt proteins are important regulators of differentiation in stem cells; the Wnt/β-catenin pathway enhances osteogenesis of MSCs and osteoprogenitor cells by upregulating osteoblast-related genes [10]. However, another study found that the Wnt/β-catenin pathway inhibited osteogenesis of PDLSCs [11]. This suggests that the Wnt/β-catenin pathway may play different roles in MSC regulation. WIF1 belongs to a family of secreted modulators of Wnt proteins. Other members of the Wnt modulator family, secreted frizzled-related proteins (sFRPs), play different roles in Wnt signaling depending on the cell subtype and model used [12,13,14,15,16]. In fact, WIF1 also has the effect of modulating Wnt signaling. For example, a previous study found that WIF1 inhibits osteoblastic differentiation in mouse mesenchymal C3H10T1/2 cells [17]. Another study found that WIF1 is highly expressed at cartilage-mesenchyme interfaces in the skeleton during early development and that it blocks the classical Wnt signaling pathway and impairs Wnt3a-mediated inhibition of chondrogenesis in embryonic chick limb-bud cells [18]. However, the function of WIF1 during dentinogenic differentiation in dental MSCs remains unclear.
In the present study, we utilized SCAPs to investigate the functioning of WIF1 during dentinogenic differentiation. We found that WIF1 could enhance the dentinogenic differentiation capacity in SCAPs. These findings provide novel insights into the mechanisms underlying directed differentiation of dental tissue-derived MSCs and have valuable clinical applications.
Methods
Cell culture of SCAPs and human embryonic kidney (293 T) cells
All our research involving human stem cells complies with the ISSCR “Guidelines for the Conduct of Human Embryonic Stem Cell Research.” Wisdom teeth were obtained from patients who provided written informed consent according to guidelines approved by the ethical committee (Ethical Committee Agreement, Beijing Stomatological Hospital, Ethics Review No. 2011–02). We disinfected the teeth in 75% ethanol and then washed them with phosphate buffered saline (PBS). The cell culture procedures used, as well as the phenotype, lineage markers and lineage differentiation potentials of SCAPs were identified and described in our previous study [19]. Briefly, apical papilla were gently separated from the apical root and then digested in a solution containing 3 mg/mL collagenase type I (Worthington Biochemical Corp., Lakewood, NJ, USA) and 4 mg/mL dispase (Roche Diagnostics Corp., Indianapolis, IN, USA) for 1 h at 37 °C. A single-cell suspension was produced by passing the solution through a 70 μm strainer (Falcon, BD Biosciences, San Jose, CA, USA). The SCAPs were grown in DMEM alpha modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) that was supplemented with 15% fetal bovine serum (FBS; Invitrogen), 2 mmol/L glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen) at 37 °C in a humidified incubator containing 5% CO2. The culture medium was changed every 3 days.
Human embryonic kidney 293 T cells were maintained in complete DMEM medium containing 10% fetal bovine serum (FBS; Invitrogen), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen).
Plasmid construction and viral infection
Full-length human WNT1 cDNA containing a hemagglutinin (HA) tag was produced using a standard gene synthesis method and subcloned into the pQCXIN retroviral vector (BD Clontech, Mountain View, CA, USA) between the Age I and EcoR1 restriction sites and verified by genetic sequencing. The viral packaging was then performed in 293 T cells according to the manufacturer’s protocol (BD Clontech). Prior to viral infections, the SCAPs were subcultured overnight and then infected with retroviruses in the presence of polybrene (6 μg/ml; Sigma-Aldrich, St. Louis, MO, USA) for 12 h. After 48 h, infected cells were selected using 600 mg/ML G418 (Sigma-Aldrich).
Reverse transcriptase-polymerase chain reaction (RT-PCR) and real-time RT-PCR
Total RNA was isolated from SCAPs using Trizol reagent (Invitrogen). cDNA was synthesized from a 2 μg aliquot of RNA containing oligo(dT), and reverse transcriptase(Invitrogen) according to the manufacturer’s protocol. Real-time PCRs were performed using the QuantiTect SYBR Green PCR kit (Qiangen, Hilden, Germany) and the Bio-Rad Real-time PCR Detection System. The changes in gene expression were determined using the 2-△△CT method. The primers used to specific genes are shown in Table 1.
In vitro cell differentiation assay
Osteo/dentinogenesis differentiation assays were performed as previously reported [4]. The expression of the DSPP, DMP1, OSX and Runx2 genes was determined using real-time RT-PCR.
Alkaline phosphatase and alizarin red detection
Mineralization was induced in SCAPs using the STEMPRO Osteogenesis Differentiation Kit (Invitrogen). ALP activity was assayed at 5 days using an ALP activity kit according to the manufacturer’s protocol (Sigma-Aldrich). For the mineralization assay, the cells were induced for 2 weeks, fixed in 70% ethanol and then stained with 2% Alizarin red (Sigma-Aldrich). To quantitatively measure calcium, the Alizarin Red-stained cells were then destained with 10% cetylpyridinium chloride in 10 mM sodium phosphate for 30 min at room temperature and measured at 562 nm in a multiplate reader; the amount of calcium was determined via comparison to a standard calcium curve obtained using calcium dilutions prepared in the same solution. The final calcium level in each group was normalized to the total protein concentration measured in a duplicate plate.
Western blot analysis
The cells were lysed in RIPA buffer (10 mM Tris-HCL, 1 mM EDTA, 1% sodium dodecylsulfate (SDS), 1% NP-40, 1:100 proteinase inhibitor cocktail, 50 mM β-glycerophosphate, 50 mM sodium fluoride). The samples were separated on a 10% SDS polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane using a semidry transfer apparatus (Bio-Rad, Hercules, CA,USA). The membranes were blotted with 5% dehydrated milk for 2 h and then incubated with primary antibodies overnight. The membranes then were incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (Promega, Madison, WI, USA) and visualized using SuperSignal reagents (Pierce, Rockford, IL, USA). The primary antibodies used were anti-HA (Clone No. C29F4, Cat No. 3724, Cell Signaling Technology, Beverly, MA, USA) and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Clone No. GAPDH-71.1, Cat No. G8795, Sigma-Aldrich), which is a housekeeping gene.
Transplantation in nude mice
These procedures were performed with the approval of the Ethics Committee and Animal Care and Use Committee of Beijing Stomatological Hospital at Capital Medical University (AEEI-2015-085). The animals were purchased from the Institute of Animal Science at Vital River Co., Ltd. and were not treated with drugs or subject to any other procedures prior to transplantation. Approximately 2.0 × 106 cells were mixed with 20 mg of HA/tricalcium phosphate ceramic particles (Engineering Research Center for Biomaterials, Sichuan University, Chengdu, China) and then transplanted subcutaneously beneath the dorsal skin of 10-week-old immunocompromised beige mice (nu/nu nude mice). Eight weeks later, the tissues were harvested, fixed in 10% formalin, decalcified using 10% EDTA buffer (pH 8.0), and embedded in paraffin.
Immunohistochemical staining
Five μm sections were cut from tissue blocks, deparaffinized and hydrated. The sections were then soaked in 3% H2O2 for 10 min and washed 3 times in distilled water for 5 min each. Epitope retrieval was conducted for 10 min in a microwave oven using 1.0 mmol/L EDTA buffer (pH 8.0), followed by a 1-h cool down period. The sections were then washed 5 times with phosphate-buffered saline (PBS) for 5 min each time and incubated with 10% goat serum for 60 min to block nonspecific antibody binding. After washing with PBS, the sections were incubated with a primary monoclonal antibody against DSPP (sc-33,586, Santa Cruz) overnight at 4 °C. After washing 3 times in PBS for 5 min each, the sections were incubated with horseradish peroxidase–conjugated anti-rabbit secondary antibody (Promega, Madison, WI, USA) for 30 min at room temperature. The sections were washed 3 times in PBS for 5 min each and then subjected to coloration using a diaminobenzidine (DAB) kit (Cat No. 8059S, Cell Signaling Technology, Inc. Beverly, MA, USA). Finally, the sections were counterstained with hematoxylin, dehydrated in a graded series of alcohol washes and mounted for light microscopy with neutral gum.
Statistical analyses
All data are presented as the mean ± standard deviation (SD). All statistical calculations were performed using SPSS13.0 statistical software. Student’s t-test was performed to determine the statistical significance. A P-value ≤0.05 was considered to denote statistical significance.
Results
WIF1 expression was increased during dentinogenic differentiation
We first examined the expression of WIF1 in SCAPs and apical papilla tissues. The real-time RT-PCR results showed that WIF1 expression was much higher in apical papilla tissues than in SCAPs (Fig. 1a). We then measured the expression of WIF1 in differentiated SCAPs upon the induction of mineralization. We found that, when the osteo/dentinogenic differentiation process was induced in SCAPs, WIF1 expression was upregulated during dentinogenic differentiation in a time-dependent manner (Fig. 1b).
WIF1 expression was increased in apical papilla tissue and in SCAPs during the induction of mineralization. (a) Real-time RT-PCR results showing that the expression of WIF1 was much higher in apical papilla tissues than in SCAPs. (b) Real-time RT-PCR results showing that the expression of WIF1 was increased during osteo/dentinogenic differentiation in SCAPs. GAPDH was used as an internal control. Student’s t-test was performed to determine the statistical significance. All error bars represent the s.d. (n = 3). *P ≤ 0.05; **P ≤ 0.01
WIF1 enhanced the dentinogenic differentiation potential of SCAPs in vitro
To understand the function of WIF1 during the differentiation of SCAPs, we inserted the WIF1 sequence along with an HA tag into a retroviral vector. This construct resulted in the overexpression of ectopic HA-WIF1 when transduced into SCAPs via retroviral infection. After selection with 600 μg/mL G418 for 14 days, ectopic WIF1 expression was confirmed using real-time RT-PCR and Western blot analysis (Fig. 2a, b). Next, the transduced SCAPs were cultured in mineralization-inducing medium to investigate their dentinogenic differentiation potential. At 5 days after induction, the results indicated that the overexpression of wild-type WIF1 promoted the activity of ALP, which is an early marker of dentinogenic differentiation in SCAPs (Fig. 2c). This was confirmed by the observation that mineralization was significantly enhanced in SCAPs that overexpressed WIF1 after 2 weeks of induction compared to cells infected with the empty vector, as determined using Alizarin Red staining and quantitative calcium measurement (Fig. 2d, e).
Overexpression of WIF1 enhanced osteo/dentinogenic differentiation in SCAPs. (a, b) Real-time RT-PCR result (a) and Western blot result (b) showing that WIF1 was overexpressed in SCAPs due to retrovirus infection. GAPDH was used as an internal control. (c) ALP activity assays showing that ALP activity was enhanced in SCAPs that overexpressed WIF1 5 days after mineralization induction. (d, e) Alizarin Red staining (d) and calcium quantitative analysis (e) showing that overexpression of WIF1 enhanced mineralization in SCAPs 2 weeks after mineralization induction. Student’s t-test was performed to determine the statistical significance. All error bars represent the s.d. (n = 3). *P ≤ 0.05; **P ≤ 0.01
The dentinogenic marker genes, DSPP and DMP1, were subsequently measured. The real-time RT-PCR results showed that DSPP was induced to a greater extent in SCAP-WIF1 cells than in SCAP-Vector cells after 2 weeks of induction (Fig. 3a), and DMP1 was induced to a greater extent in SCAP-WIF1 cells than in SCAP-Vector cells at 1 and 2 weeks after induction (Fig. 3b). We also examined the expression of key transcription factors involved in dentinogenic differentiation in SCAPs, including runt-related transcription factor 2 (known as RUNX2) and OSX. Our results showed that the mRNA levels of OSX were significantly higher in SCAP-WIF1 cells than in SCAP-Vector cells (Fig. 3c), but that the expression of RUNX2 was not significantly altered (Fig. 3d).
Overexpression of WIF1 increased the expressions of DPM1, DSPP and OSX in SCAPs. Real-time RT-PCR results showing that the expression of DSPP (a) DMP1 (b), and OSX (c) were increased in WIF1 overexpressing SCAPs compared to control SCAPs. (d) Real-time RT-PCR results showing that the expression of RUNX2 was not significantly altered by WIF1 overexpression. GAPDH was used as an internal control. Student’s t-test was performed to determine the statistical significance. All error bars represent the s.d. (n = 3). *P ≤ 0.05; **P ≤ 0.01
WIF1 enhanced in vivo dentinogenesis in SCAPs
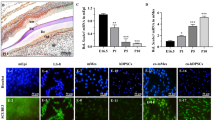
We investigated whether the overexpression of WIF1 would affect SCAP dentinogenesis in vivo. We transplanted control or WIF1 SCAPs subcutaneously into nude mice. The transplanted tissues were harvested eight weeks after transplantation. H&E staining revealed that a greater amount of bone/dentin-like tissues were present in the mice transplanted with WIF1 SCAPs than in mice transplanted with control SCAPs (Fig. 4a). Qualitative measurements also revealed a greater amount of bone/dentin-like tissues in mice transplanted with WIF1 SCAPs than in those transplanted with control SCAPs (Fig. 4b). These transplantation experiments demonstrated that WIF1 SCAPs generated a greater amount of bone/dentin-like tissues than control SCAPs in vivo.
Overexpression of WIF1 enhanced dentinogenesis in SCAPs. SCAPs were transplanted subcutaneously into immunocompromised mice and monitored for 8 weeks. (a) H&E-stained micrographs showing bone/dentin-like tissue formation. (b) Qualitative measurement of bone/dentin-like tissues in tissue samples. (c) Immunohistochemical results demonstrating DSPP expression. (d) Immunohistochemical results showing negative control staining in the absence of primary antibody. All error bars represent the s.d. (n = 5). Bar: 100 μm. **P ≤ 0.05. B/D, bone/dentin-like tissues; HA, hydroxyapatite tricalcium carrier
Immunohistochemical staining was then used to measure the expression of DSPP. The results revealed that more DSPP protein was present in tissues transplanted with SCAP-WIF1 cells than those transplanted with SCAP-Vector cells (Fig. 4c). Additionally, negative control staining was shown to occur in the absence of primary antibody (Fig. 4d). Overall together, these results showed that the overexpression of WIF1 significantly enhanced the formation of dentin-like tissues.
Discussion
MSCs derived from dental tissues have proved to be promising source of cells with therapeutic potential, especially for use in dental tissue treatment. However, little is known about the molecular mechanisms underlying the stability and differentiation potential of dental MSCs. SCAPs are important for tooth development and regeneration. A key factor that ensures successful dental tissue regeneration is the niche in which the dental cells and tissues are grown [20], which makes it important to identify the key genes that support the functioning of MSCs in the niche. The commitment and differentiation of MSCs is reliant upon Wnt/β-catenin signaling [21,22,23,24]. Therefore, we chose to investigate the Wnt/β-catenin modulator WIF1, which has been found to be more highly expressed in apical papilla tissues than in SCAPs using microarray analysis. First, we confirmed that WIF1 expression was decreased in cultured SCAPs after isolation from apical papilla and then verified that it was increased during the dentinogenic differentiation process; this indicated that increased levels of WIF1 may play a role in dentinogenic differentiation.
Based on these observations, we investigated the function of WIF1 during the dentinogenic differentiation process in SCAPs. The results showed that WIF1 promoted ALP activity and mineralization in vitro. ALP is as an indicator of early differentiation during the osteo/dentinogenic process [25]. The presence of the mineralization phenotype is an indicator of the end stage of the osteo/dentinogenic differentiation process. Moreover, transplantation experiments demonstrated that newly formed bone/dentin-like tissues were deposited by transplanted SCAPs-Vector and SCAPs-WIF1 cells and revealed that WIF1 promoted osteo/dentinogenesis in vivo. These results indicated that WIF1 enhanced osteo/dentinogenic differentiation in SCAPs. To clarify the role of WIF1 in dentinogenic differentiation, we also investigated dentinogenic differentiation indicators. DSPP and DMP1 are classic odontogenic markers; DSPP is a key gene involved in the process of dentin formation, while DMP1 has been shown to regulate DSPP [26,27,28]. We found that the expression of DSPP and DMP1 were enhanced by WIF1 in SCAPs in vitro. Additionally, a greater amount of DSPP protein was found in tissues, transplanted with SCAPs-WIF1 cells. These results indicated that WIF1 was able to promote dentinogenic differentiation in SCAPs.
In addition, we found that expression of the transcription factor OSX was also enhanced by WIF1. OSX is known to be an essential transcription factor that contains three C2H2-type zinc finger DNA binding domains. Osx is expressed during the entire process of tooth development [29,30,31]. The amount of cementum has been found to be reduced due to Osx deletion in mice [32]. An in vitro study found that Osx increases Dspp transcription in odontoblast-like cells [33]. This evidence suggests that Osx plays a critical role in dentinogenic differentiation and formation. We also found that the mRNA expression level of RUNX2, a transcription factor, was not significantly different in SCAP-WIF1 and SCAP-Vector cells. An in vitro study by Han found that Wnt/β-catenin could enhance dentinogenic differentiation in DPSC cells by activating RUNX2 [34]. There are no reports suggesting that RUNX2 upregulation is not required for dentinogenic differentiation. Overall, these findings suggested that WIF1 may enhance dentinogenic differentiation via enhancement of OSX expression in SCAPs.
Conclusion
Our results showed that WIF1 enhanced dentinogenic differentiation in SCAPs by activating the transcription factor OSX. Our work explored the mechanisms underlying the effects of WIF1 on directed differentiation in dental MSCs and provided potential target genes that could be useful in improving dental tissue regeneration using dental tissue-derived MSCs.
Abbreviations
- ALP:
-
Alkaline phosphatase
- DFSCs:
-
Stem cells from dental follicles
- DMP1:
-
Dentin matrix protein 1
- DPSCs:
-
Dental pulp stem cells
- DSPP:
-
Dentin sialophosphoprotein
- HA:
-
Hemagglutinin
- MSCs:
-
Mesenchymal stem cells
- OSX:
-
Osterix
- PBS:
-
Phosphate-buffered saline
- PDLSCs:
-
Periodontal ligament
- RT-PCR:
-
Reverse transcriptase-polymerase chain reaction
- SCAPs:
-
Stem cells from apical papilla
- sFRPs:
-
Secreted frizzled-related proteins
- SPSS:
-
Statistical Package for Social Science
- WIF1:
-
Wnt inhibitory factor 1
References
Park YJ, Cha S, Park YS. Regenerative Applications Using Tooth Derived Stem Cells in Other Than Tooth Regeneration: A Literature Review. Stem Cells Int. 2016;2016:9305986.
Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. 2009;88(9):792–806.
Saito MT, Silvério KG, Casati MZ, Sallum EA, Nociti FH Jr. Tooth-derived stem cells: Update and perspectives. World J Stem Cells. 2015;7(2):399–407.
Wei F, Song T, Ding G, Xu J, Liu Y, Liu D, Fan Z, Zhang C, Shi S, Wang S. Functional tooth restoration by allogeneic mesenchymal stem cell-based bio-root regeneration in swine. Stem Cells Dev. 2013;22(12):1752–62.
Liu O, Xu J, Ding G, Liu D, Fan Z, Zhang C, Chen W, Ding Y, Tang Z, Wang S. Periodontal ligament stem cells regulate B lymphocyte function via programmed cell death protein 1. Stem Cells. 2013;31(7):1371–82.
Zhao H, Li S, Han D, Kaartinen V, Chai Y. Alk5-mediated transforming growth factor β signaling acts upstream of fibroblast growth factor 10 to regulate the proliferation and maintenance of dental epithelial stem cells. Mol Cell Biol. 2011;31(10):2079–89.
Palosaari H, Wahlgren J, Larmas M, Rönkä H, Sorsa T, Salo T, Tjäderhane L. The expression of MMP-8 in human odontoblasts and dental pulp cells is down-regulated by TGF-beta1. J Dent Res. 2000;79(1):77–84.
Iohara K, Zheng L, Ito M, Tomokiyo A, Matsushita K, Nakashima M. Side population cells isolated from porcine dental pulp tissue with self-renewal and multipotency for dentinogenesis, chondrogenesis, adipogenesis, and neurogenesis. Stem Cells. 2006;24(11):2493–503.
Diao S, Lin X, Wang L, Dong R, Du J, Yang D, Fan Z. Analysis of gene expression profiles of apical papilla tissues, stem cells from apical papilla and cell sheet. Cell Prolif. 2017;50(3).
Guo AJ, Choi RC, Cheung AW, Chen VP, Xu SL, Dong TT, Chen JJ, Tsim KW. Baicalin, a flavone, induces the differentiation of cultured osteoblasts: an action via the Wnt/beta-catenin signaling pathway. J Biol Chem. 2011;286(32):27882–93.
Liu W, Liu Y, Guo T, Hu C, Luo H, Zhang L, Shi S, Cai T, Ding Y, Jin Y. TCF3, a novel positive regulator of osteogenesis, plays a crucial role in miR-17 modulating the diverse effect of canonical Wnt signaling in different microenvironments. Cell Death Dis. 2013;4:e539.
Ehrlund A, Mejhert N, Lorente-Cebrián S, Aström G, Dahlman I, Laurencikiene J, Rydén M. Characterization of the Wnt inhibitors secreted frizzled-related proteins (SFRPS) in human adipose tissue. J Clin Endocrinol Metab. 2013;98(3):E503–8.
Pomduk K, Kheolamai P, U-Pratya Y, Wattanapanitch M, Klincumhom N, Issaragrisil S. Enhanced human mesenchymal stem cell survival under oxidative stress by overexpression of secreted frizzled-related protein 2 gene. Ann Hematol. 2015;94(2):319–27.
Alfaro MP, Vincent A, Saraswati S, Thorne CA, Hong CC, Lee E, Young PP. sFRP2 suppression of bone morphogenic protein (BMP) and Wnt signaling mediates mesenchymal stem cell (MSC) self-renewal promoting engraftment and myocardial repair. J Biol Chem. 2010;285(46):35645–53.
Yu GX, Wang JS, Lin X, Diao S, Cao Y, Dong R, Wang LP, Yang DM, Wang SL, Fan ZP. Demethylation of SFRP2 by Histone Demethylase KDM2A Regulated the Osteo-/dentinogenic Differentiation of Stem Cells from Apical Papilla. Cell Prolif. 2016;49(3):330–40.
Lin X, Dong R, Diao S, Yu G, Wang L, Li J, Fan Z. SFRP2 enhanced the adipogenic and neoronal differentiation potentials of stem cells from apical papilla. Cell Biol Int. 2017;41(5):534–43.
Cho SW, Yang JY, Sun HJ, Jung JY, Her SJ, Cho HY, Choi HJ, Kim SW, Kim SY, Shin CS. Wnt inhibitory factor (WIF)-1 inhibits osteoblastic differentiation in mouse embryonic mesenchymal cells. Bone. 2009;44(6):1069–77.
Surmann-Schmitt C, Widmann N, Dietz U, Saeger B, Eitzinger N, Nakamura Y, Rattel M, Latham R, Hartmann C, von der Mark H, Schett G, von der Mark K, Stock M. Wif-1 is expressed at cartilagemesenchyme interfaces and impedes Wnt3a-mediated inhibition of chondrogenesis. J Cell Sci. 2009;122(Pt20):3627-3637.
Cao Y, Xia DS, Qi SR, Du J, Ma P, Wang SL, Fan ZP. Epiregulin can promote proliferation of stem cells from the dental apical papilla via MEK/Erk and JNK signalling pathways. Cell Prolif. 2013;46(4):447–56.
Kaigler D, Cirelli JA, Giannobile WV. Growth factor delivery for oral and periodontal tissue engineering. Expert Opin Drug Deliv. 2006;3(5):647–62.
D'Alimonte I, Lannutti A, Pipino C, Di Tomo P, Pierdomenico L, Cianci E, Antonucci I, Marchisio M, Romano M, Stuppia L, Caciagli F, Pandolfi A, Ciccarelli R. Wnt signaling behaves as a "master regulator" in the osteogenic and adipogenic commitment of human amniotic fluid mesenchymal stem cells. Stem Cell Rev. 2013;9(5):642–54.
Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280(39): 33132-33140.
Visweswaran M, Pohl S, Arfuso F, Newsholme P, Dilley R, Pervaiz S. Dharmarajan A5. Multi-lineage differentiation of mesenchymal stem cells - To Wnt, or not Wnt. Int J Biochem Cell Biol. 2015;68:139–47.
Yano F, Kugimiya F, Ohba S, Ikeda T, Chikuda H, Ogasawara T, Ogata N, Takato T, Nakamura K, Kawaguchi H, Chung UI. The canonical Wnt signaling pathway promotes chondrocyte differentiation in a Sox9-dependent manner. Biochem Biophys Res Commun. 2005;333(4):1300–8.
Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7(1-2):1–16.
Yao KL, Reynaldo Todescan JR, Sodek J. Temporal changes in matrix protein synthesis and mRNA expression during mineralized tissue formation by adult rat bone marrow cells in culture. J Bone Miner Res. 1994;9(2):231–40.
Zhu Q, Gibson MP, Liu Q, Liu Y, Lu Y, Wang X, Feng JQ, Qin C. Proteolytic processing of dentin sialophosphoprotein (DSPP) is essential to dentinogenesis. J Biol Chem. 2012;287(36):30426–35.
Gibson MP, Zhu Q, Wang S, Liu Q, Liu Y, Wang X, Yuan B, Ruest LB, Feng JQ, D'Souza RN, Qin C, Lu Y. The rescue of dentin matrix protein 1 (DMP1)-deficient tooth defects by the transgenic expression of dentin sialophosphoprotein (DSPP) indicates that DSPP is a downstream effector molecule of DMP1 in dentinogenesis. J Biol Chem. 2013;288(10):7203–14.
Zhang H, Jiang Y, Qin C, Liu Y, Ho SP, Feng JQ. Essential role of Osterix for tooth root but not crown dentin formation. J Bone Miner Res. 2015;30(4):742–6.
Kim TH, Bae CH, Lee JC, Kim JE, Yang X, de Crombrugghe B, Cho ES. Osterix regulates tooth root formation in a site-specific manner. J Dent Res. 2015;94(3):430–8.
Cao Z, Liu R, Zhang H, Liao H, Zhang Y, Hinton RJ, Feng JQ. Osterix Controls Cementoblast Differentiation through Downregulation of Wnt-signaling via Enhancing DKK1 Expression. Int J Biol Sci. 2015;11(3):335–44.
Cao Z, Zhang H, Zhou X, Han X, Ren Y, Gao T, Xiao Y, de Crombrugghe B, Somerman MJ, Feng JQ. Genetic evidence for the vital function of Osterix in cementogenesis. J Bone Miner Res. 2012;27(5):1080–92.
Chen S, Gluhak-Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L, Yuan GH, Dong J, Gay I, MacDougall M. Runx2, osx, and dspp in tooth development. J Dent Res. 2009;88(10):904–9.
Sharp T, Wang J, Li X, et al. A Pituitary Homeobox 2 (Pitx2): microRNA-200a-3p:β-catenin Pathway Converts Mesenchymal Cells to Amelogenin-expressing Dental Epithelial Cells. J Biol Chem. 2014;289(39):27327–41.
Acknowledgements
We would like to acknowledge Pro. Zhipeng Fan from the Capital Medical University School of Stomatology for his assistance in the study design.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81670948; provided to YC) and the China Rehabilitation Research Center Foundation (2017ZX-03; provided to HW).
Availability of data and materials
The datasets used for the current study are available from the corresponding author by request.
Author information
Authors and Affiliations
Contributions
HW collected and analyzed the data and wrote the first manuscript. YC contributed to the study design, supervised the data collection, participated in manuscript preparation and critically reviewed the intellectual content of the paper. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Beijing Stomatological Hospital at the Capital Medical University (Ethics Review No. 2011–02); all patients provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this paper. All authors read and approved the final manuscript prior to publication.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, H., Cao, Y. WIF1 enhanced dentinogenic differentiation in stem cells from apical papilla. BMC Oral Health 19, 25 (2019). https://doi.org/10.1186/s12903-018-0700-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12903-018-0700-6