Abstract
Objective
Gene-diet interaction plays a key role in the inter-individual differences in lipid abnormalities as a major risk factor for cardiovascular diseases (CVDs). Thus, we explored the interaction between CETP TaqB1 polymorphism with dietary acid load (DAL) on lipid profile among type 2 diabetes mellitus (T2DM).
Method
This cross-sectional study conducted on 220 Iranian patients with T2DM. Dietary acid load (PRAL and NEAP) was calculated via a validated food-frequency questionnaire (FFQ). The polymerase chain reaction (PCR) used for genotyping Taq1B polymorphism. Biochemical markers were measured by standard protocol. The interaction between CETP Taq1B polymorphism and DAL (PRAL and NEAP) on lipid profile was performed by a generalized linear regression model (GLM).
Results
The overall prevalence of rs708272 genotypes was 8.6%, 72.7% and 18.6% for B1B1, B1B2 and B2B2 genotype respectively. This study showed that people with the B1B1 genotype had greater LDL, TC, LDL/HDL, and TG when they consumed diets that scored higher on the NEAP and PRAL indexes than those with the B1B2 and B2B2 genotypes. Besides, carriers of the B1B1 allele who were in the highest tertile of NEAP, had lower HDL (P Interaction < 0.05).
Conclusions
In summary, the lipid profile might be improved in B1B1 homozygotes by less adherence to DAL indexes, however, the findings should be validated in high-quality interventional studies.
Similar content being viewed by others
Introduction
Type 2 diabetes mellitus (T2DM) and cardiovascular diseases (CVDs) are known as common metabolic and multifactorial disorders which caused by a complex interaction between environmental factors, lifestyle and genetics [1, 2]. According to health forecasts, the global prevalence of diabetes will rise to 642 million by 2040 [3]. T2DM and its complications are recognized as major causes of mortality [4]. Over 1 million deaths per year directly caused by diabetes make it as the ninth leading cause of mortality [5]. Dyslipidemia, significantly contributes to the development of diabetes major complications including atherosclerosis and CVDs [6]. Thus, control of lipid abnormalities is of paramount importance in T2DM. Lipid profile can be modified by multiple factors includes genetic factors and environmental and their interactions as well [7]. Based on cost estimates, the expenditure of medical care for diabetes is at least 3.2 times larger than the average per capita healthcare cost which rises to 9.4 times when its complications occur [8].
Environmental factors contribute to the regulation of plasma level of lipoproteins as well as genetics [9]. Cholesteryl ester transfer protein (CETP) is one of those major factors which contributes to the metabolism of lipids via transferring cholesterol esters and triglycerides between very-low-density lipoprotein (VLDL), low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C) and increases atherogenicity [10]. This action leads to the reverse transportation of cholesterol from peripheral tissues to the liver and facilitating the CE transfer from HDL3 to HDL2 which may alter the susceptibility to atherosclerosis and CVDs [11]. Studies indicated that different polymorphisms of human CETP gene are associated with its concentration and activity and also elevated level of serum lipids [12, 13]. One notable polymorphism is Taq1B (rs708272, 279th nucleotide in the first intron) which is thought to play a role in the regulation of serum lipids [14]. The less common B2 allele (the restriction site is absent) has a reverse relationship with CETP concentration and its activity and an association with higher HDL-C compared to B1 allele [15]. Most of previous studies showed that individuals with B2B2 genotype had reduced CETP and elevated HDL levels [16, 17]. In contrast, there are a few researches which found a relationship between B1B1 genotype and lower HDL-C, enhanced C-reactive protein (CRP), and the development of cardiovascular disease [18, 19]. It is taught that environmental factors including physical activity, Body mass index (BMI), and diet may explain these controversial results [20]. Both animal and human studies have shown that high fat intake especially high saturated fat increases CETP activity and HDL-C level [21,22,23,24]. findings highlight the fact that even 1% increase in HDL-C level causes 2–3% reduction in CVDs [25]. Tricia Y Li et al. found a relationship between lower carbohydrate consumption and higher HDL-C concentration among B2 allele carriers [26]. However, Aitken et al. did not found noticable association [27].
Dietary acid load (DAL), determined by the potential renal acid load (PRAL) and net endogenous acid production (NEAP), reflects the dietary pattern acidity [28]. High consumption of animal protein, processed meat, and sweetened beverages and low consumption of fresh fruit and vegetables, known as western dietary pattern, lead to metabolic acidosis status and cardiometabolic abnormalities among diabetic patients [29]. Previous studies suggested a close link between DAL and lipid profile. Some of them considered hypertriglyceridemia as a major result of high DAL score [28, 30] while others showed a decrease in HDL-C level and increase in BMI level among individuals with higher tertiles of PRAL/NEAP [29, 31, 32].
Although different studies were conducted to find the impact of dietary intake on CETP Taq1B polymorphism and lipid profile, additional studies are needed to explore these findings and other gene-diet interactions [26]. We aimed to investigate the effect of CETP Taq1B polymorphism and dietary acid load determined by the Potential renal acid load (PRAL) and net endogenous acid production (NEAP) interaction in relation to lipid abnormalities among patients with T2DM.
Method and material
Participants
The current study is part of a larger cross-sectional research on 220 patients with T2DM Were recruited from the Tehran diabetes centers from individuals with fasting blood sugar (FBS) levels ≥ 126 mg/dl or consuming glucose-lowering medicines [33]. For this study we calculated minimum sample size in our first project [14] based on minor allele frequency of the CETP Taq1Bpolymorphism in the world and Iranian population.
Patients who were under 35 or over 65 years old, insulin-using, pregnant or lactating women were excluded. All participants gave their written informed consent and the ethical principles established in the Declaration of Helsinki and the protocol of study approved by the ethics committee of Tehran University of Medical Sciences (TUMS) (no.15060).
General information and physical activity assessment
All information about age, sex, job, smoking, medical information was collected using interview. Anthropometric information including physical activity (METs), waist circumference (WC), and body mass index (BMI) were measured according to standard protocols. Their weight was taken without shoes but with light clothes on. BMI was estimated as weight (kg) divided by height2 (m2). The narrowest part of the abdomen and the midway between the lowest rib and the iliac crest were used to quantify WC. Daily physical activity was assessed by the classified metabolic equivalent task (MET) questionnaire, which was validated in Iran by Kelishadi et al. [34].
Dietary assessment
Data on dietary consumption were gathered using a validated 147-item semi-quantitative food frequency questionnaire (FFQ) [35]. The PRAL and NEAP were used to compute DAL. The PRAL was calculated using the method recommended by Remer et al. as follows:. [36]
PRAL (mEq/d) is equal to 0.4888 g/day of protein plus 0.0366 mg/day of phosphorus, 0.205 mg/day of potassium, 0.0125 mg/day of calcium, and 0.0263 mg/day of magnesium.
Moreover, Frassetto et al.'s NEAP formula was as follows: [37]
NEAP (mEq/d) is equal to [54.5 potassium intake (mEq/d) protein intake (g/d)].—10.2
The categorized metabolic equivalent task (MET) questionnaire, which was validated in Iran by Kelishadi et al., was used to measure daily physical activity [38].
Biochemical evaluation and genotyping
To measure serum lipids, whole blood samples were collected following a 10- to 12-h fast. Enzymatic approach was used to evaluate the levels of triglycerides (TG) and total cholesterol (TC) in Pars Azmoon, Iran. Turbidimetry was used using a Roche Hitachi analyzer (Roche, Germany) to compute LDL-C and HDL-C. The levels of 8-isoprostane F2 (PGF2) and other inflammatory indicators, such as CRP, Interleukin-18 (IL-18), and Pentraxin 3 (PTX3), were also evaluated using the ELISA technique (Bioassay Technology Co, Germany and Shanghai Crystal Day Biotech Co., Ltd). Superoxide dismutase (SOD) serum enzymatic activity was assessed using colorimetric techniques, and total antioxidant capacity (TAC) was calculated using spectrophotometric techniques (Cayman Chemical Company, USA).
Whole blood was used to obtain genomic DNA using the salting-out extraction technique. Using a modified salting-out technique, genomic DNA was extracted from peripheral blood cells of ethylenediaminetetraacetic acid-anticogulated samples. Briefly stated, 500 ml of each blood sample were mixed with 1 mL of RBC-lysis buffer (0.32 M sucrose, 10 mM Tris–HCl, pH 8.2, 5 mM MgCl2, and 1% v/v Triton X-100), which was then incubated on ice for 15 min before centrifuging at 10,000 rpm for 2 min. After discarding the supernatant, the same procedure was repeated twice before addition of 300 ml of WBC-lysis buffer (0.45 M NaCl, 10 mM Tris–HCl, pH 8.2, and 2 mM ethylenediaminetetraacetic acid), 20 ml of 10% sodium dodecyl sulfate, and 10 ml of proteinase K (20 mg/mL), followed by incubation at 558C for 2 h. By adding 100 ml of 5 M NaCl and isopropanol (1v/v), gDNA was precipitated by mixing 100 ml of 5 M NaCl with 1v/v isopropanol. Following its removal using a thin glass rod, it was cleaned with 70% EtOH before being dissolved in 1TE buffer [39]. The CETP Taq1B polymorphism was genotyped by polymerase chain reaction (PCR) using primers (Forward: 50- CAC TAG CCC AGA GAG AGG AGTG-30 and Reverse: 50-TGA GCC CAG CCG CAC ACT AAC-30) and 8% polyacrylamide gel electrophoresis.
Statistical analysis
The normality of data was analyzed by the Kolmogorov–Smirnov test. The sample size was calculated given a type I error of α = 0.05 and type II error of β = 80%. By considering the median amount of PRAL and NEAP, participants were divided into 3 tertiles for evaluating their adherence to these indexes. One-way ANOVA was used for the comparison of the mean difference between three genotypes (B1B1, B1B2, and B2B2) groups. Moreover, a generalized linear regression model (GLM) was used for finding the interaction between CETP Taq1B polymorphism and DAL (PRAL and NEAP) on important risk factors (BMI, WC, HDL, LDL, LDL/HDL, TC, TG, CRP, IL-18, TAC, SOD, and PGF2α) in crude and adjusted models (age, physical activity, sex, smoking, alcohol and energy intake).
Result
In this cross-sectional investigation, associations between cardio-metabolic markers and the CETP Taq1B polymorphism were examined in 220 patients with T2DM. The overall prevalence of rs708272 genotypes was 8.6%, 72.7% and 18.6% for B1B1, B1B2 and B2B2 genotypes respectively. The genotype distributions were within HWE (P-value > 0.05).
Relationship between Cardio-metabolic Markers and NEAP and PRAL
Table 1 shows the basic data on diabetic individuals in the NEAP and PRAL groups. According to their NEAP and PRAL ranking, all patients were separated into three groups. The overall energy intake of patients in the third tertile of PRAL was greater (p = 0.01). Moreover, estimated energy intake (EER) has shown higher between men with higher NEAP (p = 0.02) and PRAL (p = 0.005). Patients in the third tertile of PRAL had a greater tendency of consuming more carbohydrates (P = 0.005). Diabetic patients who were in the last tertile of NEAP (p < 0.001) and PRAL (p = 0.001) consumed higher protein. Also, higher NEAP intake was associated with higher cholesterol consumption (p = 0.01). Other fundamental features and biological indicators were not significantly correlated between the NEAP and PRAL groups.
NEAP and PRAL interactions with the CETP Taq1B polymorphism and their effects on lipid profiles
Tables 1, 2 and 3 show the relationship between the CETP Taq1B polymorphism and the tertiles of the NEAP and PRAL scores on lipid profiles.
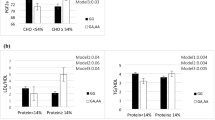
In terms of lipid profiles including (HDL, LDL, TC, LDL/HDL, and TG) in both crude and adjusted models, there were significant interactions between NEAP and PRAL scores and CETP Taq1B. This study found that individuals with the B1B1 genotype had higher LDL (P1-interaction = 0.01, P2-interaction = 0.01), TC (P1-interaction = 0.04, P2-interaction = 0.03), LDL/HDL (P1-interaction 0.001, P2-interaction 0.001), and TG (P1-interaction = 0.004, P2-interaction = 0.004) when they consumed diets that scored higher on the NEAP index. Furthermore, the adjustment model revealed a significant rs708272-NEAP interaction on HDL concentration (P Interaction = 0.03), and B1B1 allele carriers with the highest tertile of NEAP had lower HDL levels (Fig. 1).
The highest tertiles of the PRAL also showed higher levels of TG (P1-interaction = 0.001, P2-interaction = 0.001), LDL (P1-interaction = 0.009, P2-interaction = 0.009), TC (P1-interaction = 0.02, P2-interaction = 0.04), LDL/HDL (P1-interaction 0.001, P2-interaction 0.001), and TG (P1-interaction = 0.001, P2-interaction. Lower HDL concentration was seen in those with B1B1 genotypes in the upper tertile of PRAL (P1-interaction = 0.02, P2-interaction = 0.01) (Fig. 2).
Discussion
So far as we know, the effect of the interaction between CETP Taq1B and DAL on lipid profile was evaluated in this study for the first time. Here, the protein, carbohydrate, and cholesterol intake increased significantly by the adherence to DAL indexes (PRAL and NEAP). In the present study, not only did lipid profile have insignificant differences through tertiles of indexes, but also in genotypes. In agreement with the findings, various studies have noted a non-significant association between DAL indexes, genotypes, and lipid profile [10, 32, 40,41,42,43,44,45,46,47] whereas several reported either a direct or an inverse link between them [32, 42, 48,49,50,51]. Changes in lipid profile in relation to DAL might be interpreted by an increase in the production of cortisol which is affected by metabolic acidosis provoked by DAL. In this case, the lipase activity is aroused which leads to an intensifying production of very-low-density lipoproteins (rich in TG) [52, 53]. Employing the polymorphism, a greater level of HDL-C in B2B2 homozygotes has been indicated in the findings of the present study as well as some others despite whether the association between lipid profile and genotypes is significant. It’s important to underline that the other polymorphism in the region of the Taq1b might also affect the HDL-C level as well as the above-mentioned polymorphism [44, 54,55,56]. Still, the differences in health status, dietary assessment tool, sample size, ethnicity, age, and type of population participating in various studies should be taken into account for discussing the different findings with respect to the actual relationship between DAL, genotypes, and lipid profile.
The key finding of the present study was an increased level of LDL-C, TC, TG, and LDL-C/HDL-C ratio while HDL-C concentration dropped through the interaction between CETP Taq1B, NEAP, and PRAL, notably in B1B1 homozygotes (wild type) despite neither lipid profile nor indexes scores were statistically different in genotypes. Much less effort has been made in studying the polymorphism-dietary patterns interaction while many studies have investigated either the linkage of the CETP polymorphism and lipid profile or the interaction between genotypes and dietary components on lipid profile. Concerning this, Kalantar et al. [14, 47] reported that total fat intake modifies the association of CETP Taq1B polymorphism and HDL-C level in normolipidemic T2DM participants and B1B1 subjects are prone to have a decreased LDL/HDL ratio by the adherence to healthy eating index, dietary quality index, and dietary phytochemical index. Through the study of Abaj et al. [57], a greater following of dietary insulin index and dietary insulin load in T2DM patients was related to a lower level of TG, LDL/HDL ratio, and a higher level of HDL-C in B1B1 homozygotes. By the report of Campos-Perez et al. [58], subjects with the B1B2/B2B2 genotypes had elevated levels of TC and LDL-C by more sucrose intake. As stated by Gammon et al. [59], an improvement in TG/HDL-C ratio was observed by consuming two green kiwifruits a day along with a healthy dietary pattern in B1B1 homozygotes. An elevated level of HDL-C after consuming a high carbohydrate and low fat (HC/LF) diet was linked with the B2 allele by the findings of Du et al. [60] while men with B1B1 genotypes were more vulnerable to getting affected by HC/LF diet on their HDL-C concentration. Though the absolute underlying mechanism of the aforementioned relationship is still vague, some conceivable clarification might be available. The B1B1 subjects of the present study consuming fewer amounts of fruits and vegetables as opposed to B2B2 homozygotes, resulting in a lower intake of potassium, however, the scores of the indexes did not differ between genotypes. It is proposed that consuming high amounts of fruits and vegetables could drop DAL by various justifications [61]. The acidifying effect of vegetable proteins is not comparing the same as animal proteins and their phosphorus content is less bioavailable by the form of phytate [62]. Moreover, the metabolism of the potassium salts i.e. malate and citrate lead to the consumption of hydrogen ions which might give vegetables and fruits a neutral effect on the acid load and accordingly, they’re considered as sources of alkalizing foods which have negative effects on DAL indexes [62,63,64]. Besides, the accessibility of dietary fat could be limited via dietary fiber through fat absorption reduction which could lessen hepatic synthesis of cholesterol while binding to bile acid which facilitates lipid circulation modification [65,66,67,68]. Dietary fiber could additionally control the metabolic activity and metabolites of the intestinal flora [69] which could be another explanation for the elevated level of lipid profile in B1B1 genotypes with low consumption of vegetables and fruits, and high adherence to DAL. At the same time, the CETP Taq1B polymorphism is linked with the serum lipid profile that codes the CETP protein which is responsible for the reverse transfer of cholesterol ester. In this regard, it has been proposed that the B1 allele has been linked with the risk of dyslipidemia due to the linkage with further CETP activity and increased TG. On the other hand, the protective one (B2 allele) is related to a lower level of serum CETP which decreases the reverse transfer of cholesterol ester and carries out an increase in HDL-C level [18, 70, 71], yet, more investigations are required to provide insight into the interaction effect of the CETP Taq1b and DAL on lipid profile.
Provided the novelty of the study, still it has some limitations. Above all, it’s a cross-sectional study without measuring CETP concentration across genotypes and no causality was provided about detected interactions. Recall bias and over-or under-reporting of the dietary intake were other limitations of the present study which might happen by using the FFQ.
Conclusion
Conclusively, the amount of LDL-C, TC, TG, and LDL-C/HDL-C ratio elevated while HDL-C concentration dropped in B1B1 homozygotes by greater adherence to NEAP and PRAL which might be the result of consuming more fruits and vegetables. The findings could be utilized together with the genetic relation of the patients to yield appropriate nutritional advice to prevent or amend the cardiovascular risk factor of type 2 diabetes, nevertheless, it should be confirmed in high-quality interventional studies.
Comparisons with other studies and what does the current work add to the existing knowledge
The identification of the aforementioned interactions could be used in personalized nutritional recommendations for the refinement and prevention of type 2 diabetes complications and for ameliorating the lipid profile of T2DM patients.
Strengths
This is the first study attempt to explore the interaction effect of the DAL and CETP Taq1b polymorphism on lipid profile in type 2 diabetic patients.
Limitations
It is a cross-sectional study without measuring CETP concentration across genotypes and providing any causality about detected interactions. Recall bias and over-or under-reporting of the dietary intake was another limitation of the present study which might happen by using the FFQ.
Availability of data and materials
The corresponding author will provide the datasets used and/or analyzed during the current work upon reasonable request.
References
Aka TD, Saha U, Shati SA, et al. Risk of type 2 diabetes mellitus and cardiovascular complications in KCNJ11, HHEX and SLC30A8 genetic polymorphisms carriers: A case-control study. Heliyon. 2021;7:e08376.
Naeini Z, Toupchian O, Vatannejad A, et al. Effects of DHA-enriched fish oil on gene expression levels of p53 and NF-κB and PPAR-γ activity in PBMCs of patients with T2DM: A randomized, double-blind, clinical trial. Nutr Metab Cardiovasc Dis. 2020;30:441–7.
Mirzaei M, Rahmaninan M, Mirzaei M, et al. Epidemiology of diabetes mellitus, pre-diabetes, undiagnosed and uncontrolled diabetes in Central Iran: results from Yazd health study. BMC Public Health. 2020;20:1–9.
Feigin VL, Nichols E, Alam T, et al. Global, regional, and national burden of neurological disorders, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:459–80.
Khan MAB, Hashim MJ, King JK, et al. Epidemiology of type 2 diabetes–global burden of disease and forecasted trends. J Epidemiol Glob Health. 2020;10:107.
Hedayatnia M, Asadi Z, Zare-Feyzabadi R, et al. Dyslipidemia and cardiovascular disease risk among the MASHAD study population. Lipids Health Dis. 2020;19:1–11.
Qi Q, Durst R, Schwarzfuchs D, et al. CETP genotype and changes in lipid levels in response to weight-loss diet intervention in the POUNDS LOST and DIRECT randomized trials1. J Lipid Res. 2015;56:713–21.
Oh S-H, Ku H, Park KS. Prevalence and socioeconomic burden of diabetes mellitus in South Korean adults: a population-based study using administrative data. BMC Public Health. 2021;21:1–13.
Aminian O, Moinfar Z, Eftekhari S, et al. Association of plasma levels of lipid and polychlorinated biphenyls in Iranian adult. Heliyon. 2020;6:e03775.
Maroufi NF, Farzaneh K, Alibabrdel M, et al. Taq1B Polymorphism of Cholesteryl Ester Transfer Protein (CETP) and Its Effects on the Serum Lipid Levels in Metabolic Syndrome Patients. Biochem Genet. 2016;54:894–902.
Colombo GI, Bianconi V, Bonomi A, et al. The association between HDL-C and subclinical atherosclerosis depends on CETP plasma concentration: insights from the IMPROVE study. Biomedicines. 2021;9:286.
Kuivenhoven JA, de Knijff P, Boer JM, et al. Heterogeneity at the CETP gene locus: influence on plasma CETP concentrations and HDL cholesterol levels. Arterioscler Thromb Vasc Biol. 1997;17:560–8.
Dachet C, Poirier O, Fo C, et al. New functional promoter polymorphism, CETP/− 629, in cholesteryl ester transfer protein (CETP) gene related to CETP mass and high density lipoprotein cholesterol levels: role of Sp1/Sp3 in transcriptional regulation. Arterioscler Thromb Vasc Biol. 2000;20:507–15.
Kalantar Z, Eshraghian MR, Sotoudeh G, et al. Differences in the interaction between CETP Taq1B polymorphism and dietary fat intake on lipid profile of normolipedemic and dyslipidemic patients with type 2 diabetes mellitus. Clin Nutr. 2018;37:270–5.
Galati F, Colonna P, Galati A, Ciardiello C, Bozzetti MP, Massari S. CETP TaqIB Polymorphism, Serum Lipid Levels And Risk Of Atrial Fibrillation: A Case-Control Study. J Atr Fibrillation. 2014;6(6):964. https://doi.org/10.4022/jafib.964.
Kondo I, Berg K, Drayna D, et al. DNA polymorphism at the locus for human cholesteryl ester transfer protein (CETP) is associated with high density lipoprotein cholesterol and apolipoprotein levels. Clin Genet. 1989;35:49–56.
Wu JH, Lee Y-T, Hsu H-C, et al. Influence of CETP gene variation on plasma lipid levels and coronary heart disease: a survey in Taiwan. Atherosclerosis. 2001;159:451–8.
Thompson A, Di Angelantonio E, Sarwar N, et al. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA. 2008;299:2777–88.
Asselbergs FW, Moore JH, van den Berg MP, et al. A role for CETP TaqIB polymorphism in determining susceptibility to atrial fibrillation: a nested case control study. BMC Med Genet. 2006;7:1–9.
Abaj F, Rafiee M, Koohdani F. Interaction between CETP polymorphism and dietary insulin index and load in relation to cardiovascular risk factors in diabetic adults. Sci Rep. 2021;11:1–10.
Cheema SK, Agarwal-Mawal A, Murray CM, et al. Lack of stimulation of cholesteryl ester transfer protein by cholesterol in the presence of a high-fat diet. J Lipid Res. 2005;46:2356–66.
Fusegawa Y, Kelley KL, Sawyer JK, et al. Influence of dietary fatty acid composition on the relationship between CETP activity and plasma lipoproteins in monkeys. J Lipid Res. 2001;42:1849–57.
Chang C-K, Snook JT. The cholesterolaemic effects of dietary fats in cholesteryl ester transfer protein transgenic mice. Br J Nutr. 2001;85:643–8.
Babiak J, Gong EL, Nichols AV, et al. Characterization of HDL and lipoproteins intermediate to LDL and HDL in the serum of pedigreed baboons fed an atherogenic diet. Atherosclerosis. 1984;52:27–45.
Manninen V, Elo MO, Frick MH, et al. Lipid alterations and decline in the incidence of coronary heart disease in the Helsinki Heart Study. JAMA. 1988;260:641–51.
Li TY, Zhang C, Asselbergs FW, et al. Interaction between dietary fat intake and the cholesterol ester transfer protein TaqIB polymorphism in relation to HDL-cholesterol concentrations among US diabetic men. Am J Clin Nutr. 2007;86:1524–9.
Aitken WA, Alexandra W-AC, Duncan AW, et al. Variation in the cholesteryl ester transfer protein (CETP) gene does not influence individual plasma cholesterol response to changes in the nature of dietary fat. Nutr Metab Cardiovasc Dis. 2006;16:353–63.
Naeini Z, Abaj F, Esmaeily Z, et al. A Nutrigenetic Approach to Investigate ApoB EcoR1 Polymorphism-Dietary Acid Load Interactions on Lipid and Anthropometric-Related Outcomes in Adults with Dyslipidemic Type 2 Diabetes. Lifestyle Genomics. 2023;16:61–72.
Abaj F, Esmaeily Z, Naeini Z, et al. Dietary acid load modifies the effects of ApoA2–265 T> C polymorphism on lipid profile and serum leptin and ghrelin levels among type 2 diabetic patients. BMC Endocr Disord. 2022;22:190.
Wu T, Seaver P, Lemus H, et al. Associations between dietary acid load and biomarkers of inflammation and hyperglycemia in breast cancer survivors. Nutrients. 2019;11:1913.
Moghadam SK, Bahadoran Z, Mirmiran P, et al. Association between dietary acid load and insulin resistance: Tehran Lipid and Glucose Study. Prev Nutr Food Sci. 2016;21:104.
Abbasalizad Farhangi M, Nikniaz L, Nikniaz Z. Higher dietary acid load potentially increases serum triglyceride and obesity prevalence in adults: An updated systematic review and meta-analysis. PLoS ONE. 2019;14:e0216547.
Noorshahi N, Sotoudeh G, Djalali M, et al. APOA II genotypes frequency and their interaction with saturated fatty acids consumption on lipid profile of patients with type 2 diabetes. Clin Nutr. 2016;35:907–11.
Puchau B, Zulet MA, de Echávarri AG, et al. Dietary total antioxidant capacity is negatively associated with some metabolic syndrome features in healthy young adults. Nutrition. 2010;26:534–41.
Mirmiran P, Esfahani F, Azizi F. Relative validity and reliability of the food frequency questionnaire used to assess nutrient intakes: Tehran Lipid and Glucose Study. Iran J Diabetes Lipid. 2009;9:185–97.
Remer T, Manz F. Estimation of the renal net acid excretion by adults consuming diets containing variable amounts of protein. Am J Clin Nutr. 1994;59:1356–61.
Frassetto LA, Todd KM, Morris RC Jr, et al. Estimation of net endogenous noncarbonic acid production in humans from diet potassium and protein contents. Am J Clin Nutr. 1998;68:576–83.
Azizi F, Rahmani M, Ghanbarian A, et al. Serum lipid levels in an Iranian adults population: Tehran Lipid and Glucose Study. Eur J Epidemiol. 2003;18:311–9.
Alvandi E, Akrami SM, Chiani M, et al. Molecular Analysis of the RET Proto-Oncogene Key Exons in Patients with Medullary Thyroid Carcinoma: A Comprehensive Study of the Iranian Population. Thyroid. 2011;21:373–82.
Daneshzad E, Haghighatdoost F, Azadbakht L. Dietary acid load and cardiometabolic risk factors: a systematic review and meta-analysis of observational studies. Public Health Nutr. 2019;22:2823–34.
Mozaffari H, Namazi N, Larijani B, et al. Association of dietary acid load with cardiovascular risk factors and the prevalence of metabolic syndrome in Iranian women: A cross-sectional study. Nutrition. 2019;67–68:110570.
Kucharska AM, Szostak-Węgierek DE, Waśkiewicz A, et al. Dietary acid load and cardiometabolic risk in the Polish adult population. Adv Clin Exp Med. 2018;27:1347–54.
Ko B-J, Chang Y, Ryu S, et al. Dietary acid load and chronic kidney disease in elderly adults: Protein and potassium intake. PLoS ONE. 2017;12:e0185069.
Ghorban M, Karimpour F, Ghaffari MA et al. (2015) Association of the CETP TaqIB Polymorphism with Coronary Artery Disease in Type 2 Diabetic Patients %J Medical Laboratory Journal. 9, 53–59.
Ilanbey B, Meral K, Ebru DS, Sacide P, Ferhan GS, Ferda O, Eser YS. "The role of cholesteryl ester transfer protein TaqIB polymorphism in young atherosclerotic heart disease." Int J Med Biochem. 2020;3(1):8–13.
Porchay-Baldérelli I, Péan F, Bellili N, et al. The CETP TaqIB polymorphism is associated with the risk of sudden death in type 2 diabetic patients. Diabetes Care. 2007;30:2863–7.
Kalantar Z, Sotoudeh G, Esmaeily Z, Rafiee M, Koohdani F. Interaction between CETP Taq1B polymorphism and HEI, DQI and DPI on metabolic biomarkers in patients with type 2 diabetes. J Hum Nutr Diet. 2022;35(4):651-62. https://doi.org/10.1111/jhn.12958.
Haghighatdoost F, Najafabadi MM, Bellissimo N, et al. Association of dietary acid load with cardiovascular disease risk factors in patients with diabetic nephropathy. Nutrition. 2015;31:697–702.
Han E, Kim G, Hong N, et al. Association between dietary acid load and the risk of cardiovascular disease: nationwide surveys (KNHANES 2008–2011). Cardiovasc Diabetol. 2016;15:122.
Bahadoran Z, Mirmiran P, Khosravi H, et al. Associations between Dietary Acid-Base Load and Cardiometabolic Risk Factors in Adults: The Tehran Lipid and Glucose Study. Endocrinol Metab (Seoul). 2015;30:201–7.
Iwase H, Tanaka M, Kobayashi Y, et al. Lower vegetable protein intake and higher dietary acid load associated with lower carbohydrate intake are risk factors for metabolic syndrome in patients with type 2 diabetes: Post-hoc analysis of a cross-sectional study. J Diabetes Investig. 2015;6:465–72.
Djurhuus CB, Gravholt CH, Nielsen S, et al. Effects of cortisol on lipolysis and regional interstitial glycerol levels in humans. Am J Physiol Endocrinol Metab. 2002;283:E172-177.
Xu C, He J, Jiang H, et al. Direct effect of glucocorticoids on lipolysis in adipocytes. Mol Endocrinol. 2009;23:1161–70.
Collet X, Tall AR, Serajuddin H, et al. Remodeling of HDL by CETP in vivo and by CETP and hepatic lipase in vitro results in enhanced uptake of HDL CE by cells expressing scavenger receptor B-I. J Lipid Res. 1999;40:1185–93.
Freeman DJ, Griffin BA, Holmes AP, et al. Regulation of plasma HDL cholesterol and subfraction distribution by genetic and environmental factors. Associations between the TaqI B RFLP in the CETP gene and smoking and obesity. Arterioscler Thromb. 1994;14:336–44.
Seip RL, Moulin P, Cocke T, et al. Exercise training decreases plasma cholesteryl ester transfer protein. Arterioscler Thromb. 1993;13:1359–67.
Abaj F, Rafiee M, Koohdani F. Interaction between CETP polymorphism and dietary insulin index and load in relation to cardiovascular risk factors in diabetic adults. Sci Rep. 2021;11:15906.
Campos-Perez W, Perez-Robles M, Torres-Castillo N, et al. Physical inactivity and excessive sucrose consumption are associated with higher serum lipids in subjects with Taq1B CETP polymorphism. J Hum Nutr Diet. 2020;33(3):299–307. https://doi.org/10.1111/jhn.12747.
Gammon CS, Minihane AM, Kruger R, et al. TaqIB polymorphism in the cholesteryl ester transfer protein (CETP) gene influences lipid responses to the consumption of kiwifruit in hypercholesterolaemic men. Br J Nutr. 2014;111:1077–84.
Du J, Fang DZ, Lin J, et al. TaqIB polymorphism in the CETP gene modulates the impact of HC/LF diet on the HDL profile in healthy Chinese young adults. J Nutr Biochem. 2010;21:1114–9.
Osuna-Padilla IA, Leal-Escobar G, Garza-García CA, et al. Dietary Acid Load: mechanisms and evidence of its health repercussions. Nefrologia. 2019;39:343–54.
Cupisti A, D'Alessandro C, Gesualdo L, et al. Non-Traditional Aspects of Renal Diets: Focus on Fiber, Alkali and Vitamin K1 Intake. Nutrients. 2017;9(5):444. https://doi.org/10.3390/nu9050444.
Passey C. Reducing the Dietary Acid Load: How a More Alkaline Diet Benefits Patients With Chronic Kidney Disease. J Ren Nutr. 2017;27:151–60.
Rodrigues Neto Angéloco L, Arces de Souza GC, Almeida Romão E, et al. Alkaline Diet and Metabolic Acidosis: Practical Approaches to the Nutritional Management of Chronic Kidney Disease. J Ren Nutr. 2018;28:215–20.
Lattimer JM, Haub MD. Effects of dietary fiber and its components on metabolic health. Nutrients. 2010;2:1266–89.
Bordoni L, Petracci I, Zhao F, et al. Nutrigenomics of Dietary Lipids. Antioxidants (Basel). 2021;10(7):994. https://doi.org/10.3390/antiox10070994.
Mallillin AC, Trinidad TP, Raterta R, et al. Dietary fibre and fermentability characteristics of root crops and legumes. Br J Nutr. 2008;100:485–8.
Trinidad TP, Mallillin AC, Loyola AS, et al. The potential health benefits of legumes as a good source of dietary fibre. Br J Nutr. 2010;103:569–74.
Tuohy KM, Fava F, Viola R. ’The way to a man’s heart is through his gut microbiota’–dietary pro- and prebiotics for the management of cardiovascular risk. Proc Nutr Soc. 2014;73:172–85.
Chapman MJ, Le Goff W, Guerin M, et al. Cholesteryl ester transfer protein: at the heart of the action of lipid-modulating therapy with statins, fibrates, niacin, and cholesteryl ester transfer protein inhibitors. Eur Heart J. 2010;31:149–64.
Sandhofer A, Tatarczyk T, Laimer M, et al. The Taq1B-variant in the cholesteryl ester-transfer protein gene and the risk of metabolic syndrome. Obesity (Silver Spring, Md). 2008;16:919–22.
Acknowledgements
The researchers appreciate the participation of the study subjects.
Funding
The Tehran University of Medical Sciences [grant number 15061] provided funding for this investigation. The funding body played no role in the design of the study and collection, analysis, interpretation of data, and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
FA contributed to conception, design, data analyses and data interpretation. FA, ZE, EA and ZN have drafted the work or substantively revised it. FK and MR contributed to interpretation of data, design of the work.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All procedures are carried out in conformity with the applicable standards and laws. Before participating in the study, each subject signed a written permission form after receiving a thorough explanation of the procedure. The Tehran University of Medical Sciences (TUMS) ethics committee authorized the study plan with the following identification: IR.TUMS.VCR.REC.15061.
Consent for publication
‘Not applicable’.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Abaj, F., Esmaeily, Z., Naeini, Z. et al. Dietary acid load and its interaction with CETP TaqB1 polymorphisms on lipid profile among patients with Type 2 diabetes mellitus. BMC Endocr Disord 23, 138 (2023). https://doi.org/10.1186/s12902-023-01391-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12902-023-01391-6