Abstract
Background
Variants in the DEPDC5 have been proved to be main cause of not only various dominant familial focal epilepsies, but also sporadic focal epilepsies. In the present study, a novel variant in DEPDC5 was detected in the patient with focal epilepsy and his healthy father. We aimed to analyze the pathogenic DEPDC5 variant in the small family of three.
Case presentation
A 5-month-old male infant presented with focal epilepsy. Whole exome sequencing identified a novel heterozygous variant c.1696delC (p.Gln566fs) in DEPDC5, confirmed by Sanger sequencing. The variant was inherited from healthy father.
Conclusions
Our study expands the spectrum of DEPDC5 variants. Moreover, We discuss the relation between the low penetrance of DEPDC5 and the relatively high morbidity rate of DEPDC5-related sporadic focal epilepsy. Besides, due to interfamilial phenotypic and genetic heterogeneity, we speculate the prevalence of familial focal epilepsy with variable foci might be underestimated in such small families. We emphasize the importance of gene detection in patients with sporadic epilepsy of unknown etiology, as well as their family members. It can identify causative mutations, thus providing help to clinicians in making a definitive diagnosis.
Similar content being viewed by others
Background
The DEPDC5 (OMIM #614,191) is located on chromosome 22q12.2–12.3. It contains 43 exomes and encodes the disheveled, Egl-10 and pleckstrin domain-containing protein 5 (DEPDC5), a full-length protein composed of 1603 amino acids [1]. DEPDC5 is a part of the GAP Activity Toward Rags 1 complex, which also contains nitrogen permease regulator-like-2 and nitrogen permease regulator-like-3. They work together and regulate mammalian target of rapamycin complex 1 (mTORC1) negatively [2]. The mTORC1 participates in the regulation of neuronal growth, homeostasis and metabolism in neurons. Besides, mTORC1 plays an important role in the development of neurons, such as the differentiation of neurons, synapse formation and neurite outgrowth [3].
To date, variants in the DEPDC5 have been proved to be main cause of various dominant familial focal epilepsies, such as autosomal dominant sleep-related hypermotor epilepsy, familial focal epilepsy with variable foci (FFEVF), familial temporal lobe epilepsy and focal epilepsy caused by various cortical developmental malformations [4]. According to the reports, 13% of familial lateral temporal lobe epilepsies caused by DEPDC5 variants [5]. The DEPDC5 is also related with 13% of autosomal dominant sleep-related hypermotor epilepsies [6]. What’s more, more than 80% FFEVF patients were caused by DEPDC5 variants [4]. Besides, variants in DEPDC5 have been detected in many patients with non-familial focal epilepsies [7]. Here we describe a 5-month-old male infant with focal epilepsy, and discuss what role a novel DEPDC5 variant plays in the small family of three.
Case presentation
The male infant was born to healthy non-consanguineous parents. There was no discovered family history of epilepsy or other nervous system diseases. The patient was the only child of his family. He was born at term with no asphyxia after an uneventfully pregnancy. His weight and body length were within the normal range at birth. The neonatal period was unremarkable. And up to now, there has been no problem with the patient’s developmental milestones.
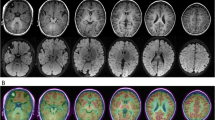
At the age of 5 months, the infant appeared the first afebrile epileptic seizure, which happened when he was sleeping. It lasted for 10 s approximately. And it was characterized by eyes blinking frequently, followed by binocular gaze and the tonic seizure of right upper limb. Asking for past medical history, we found it worth noting that the patient showed short-lasting symptom of blinking about half of a month ago. While, it failed to draw the attention of his parents. Physical examinations and the laboratory examinations showed no abnormality. The blood and urine metabolic screenings were normal as well. The brain magnetic resonance imaging (MRI) showed widened extracerebral space and widening of the beginning of left lateral fissure cistern. A video-electroencephalogram (EEG) revealed epileptiform discharges with spikes and sharp waves over the right frontal area (Fig. 1).
Before the infant received effective treatment, he experienced 9 times focal seizures with similar symptoms. The epileptic seizure usually happened before sleep or during sleeping period. In order to control the epileptic seizure, the patient received a treatment of phenobarbital (5 mg/kg) at first. However, the seizure wasn’t controlled until topiramate was applied. 5 months later, a follow-up review found that epileptic seizure had not occurred to the infant since he discharged from the hospital. And 6 months later, the re-examine of video-EEG showed nothing unusual. The psychomotor development didn’t show retardation or decline. So far, the patient has been seizure-free for 7 months.
We performed whole exome sequencing (WES) to the patient and detected a novel heterozygous variant in DEPDC5 (NM_001242896.1), c.1696delC. The variant was autosomal dominant and could result in a frameshift in translation (p.Gln566fs) and a premature termination codon (PVS1). Besides, it was absent in healthy controls in the 1000 Genomes Project Database [8], the Exome Aggregation Consortium Database [9] or National Heart, Lung, and Blood Institute Exome Sequencing Project [10] (PM2). And it was not recorded in Human Gene Mutation Database [11]. As a result, the variant c.1696delC in DEPDC5 was classified as “likely pathogenic” based on the above evidence according to the 2015 American College of Medical Genetics and Genomics variant classification guidance [12]. The variant was confirmed by Sanger sequencing (the primers used for amplifying were forward 5’—TCTTCAGGCAGTGTCCTTC—3’ and reverse 5’—AGCAACCAACTTACCCACA—3’) using the DNA of the family members (see Fig. 2). And the result revealed that the variant was inherited from healthy father. The mother didn’t carry this alteration. The pedigree chart in this family is shown in Fig. 3.
Pedigree chart in this family. The father (I1) was healthy (marked as a dark dot in a blank background) even though he carried the heterozygous variant c.1696delC in DEPDC5. The proband (II1, pointed out by the arrow) inherited the variant from his father and developed the corresponding disease (marked as dark)
Discussion and conclusions
DEPDC5 variants have been demonstrated to be the most common cause of familial focal epilepsies. While in clinical work, most patients have no family history of epilepsy. In 2017, a study reported that 2 in 220 (0.9%) patients with sporadic non-lesiona focal epilepsy, had disease-causing variants in the DEPDC5 [7]. It suggested DEPDC5 also have a significant effect on sporadic focal epilepsy. FFEVF, proved mainly caused by DEPDC5 variants, is a type of autosomal dominant epilepsy. It is characterized by a obvious intrafamilial variation that focal seizures happen in different cortical regions within the family members. Seizure type varies considerably between individuals, rather than showing phenotypic homogeneity [13]. Because of its interfamilial phenotypic and genetic heterogeneity, we speculate that the prevalence of FFEVF might be underestimated in small families. Interestingly, in the case we reported, the infant’s variant was inherited from his healthy father who carries the same variant as the infant. In addition to considering that variable expressivity could be caused by the combination of genetic, environmental, and lifestyle factors, this is accordant with the low penetrance (45–67%) which has been reported in previous studies [7]. Besides, viewing Table 1 which shows clinical expressions of various focal epilepsies caused by DEPDC5 variants as well as that of proband, it is impossible to exclude the probability of FFEVF. Though this family is so small that we can’t diagnose the epilepsy as FFEVF clinically yet. In view of this situation, the misdiagnosed families may contribute to the relatively high morbidity rate of sporadic focal epilepsy. To some degree, it can explain why DEPDC5 variants account for nearly 1% of patients with sporadic focal epilepsy. It suggests that gene detection is necessary for the patients with sporadic epilepsy of unknown etiology, as well as their family members. The identification of DEPDC5 variants in these small families enables the possibility of molecular diagnosis with FFEVF. In the present study, WES played a critical role in the differentiation and diagnosis of disease. Considering the clinical and genetic heterogeneity of FFEVF, WES represents a rapid, cost-effective and accurate method for the diagnosis of the disease.
Previous studies have generally believed that focal epilepsies caused by DEPDC5 variants are non-lesional [13]. In recent years, many teams have found focal cortical dysplasia (FCD) in epilepsy patients through brain MRI and neuropathology [7, 14]. Most of them agreed that FCD results from a second-hit somatic variant [15]. It is explained that DEPDC5 somatic variants trigger FCD through mammalian target of rapamycin hyperactivation in dysmorphic neurons [15]. Common MRI features of FCD include T2 hyperintensity of the white matter, abnormally deep sulcus, transmantle sign, cortical thickening, and blurring of the gray-white matter border [16]. In the case we reported, the abnormal brain MRI may be a common physiological manifestation in infants. Besides, the patient’s seizure is controlled well by topiramate monotherapy. While the patients with FCD usually presented frequent and refractory seizures. Therefore, it is unlikely that the patient will be complicated with FCD. In view of the abnormal MRI, a further observation is still needed. High resolution MRI can be considered to detect subtle cortical malformations. Moreover, Regular follow-up and the re-examination of EEG should be arranged.
In rat models, heterozygous Depdc5± rats had altered cortical neuron excitability and firing patterns but without developmental abnormalities or spontaneous seizures. However, homozygous Depdc5−/− embryos died in utero caused by global growth delay. The study revealed a potential quantitative correlation between genetic impairment and phenotype severity. And heterozygous variants would probably cause susceptibility alterations or mild phenotype [17]. Febrile seizures plus/febrile seizures and FCD was defined as phenotypes of DEPDC5 variants, through evaluating evidence from five clinical-genetic aspects [18]. Further analysis revealed that FCD was more frequently associated with null variants. In contrast, febrile seizures plus/febrile seizures had a high frequency of missense variants. It was coincident with the findings from gene knockout rat models. Besides, the study proposed that variants closer to the binding site of DEPDC5 to nitrogen permease regulator-like-2/nitrogen permease regulator-like-3 complex could lead to more severe phenotype like FCD.
In conclusion, we found a novel heterozygous variant in DEPDC5, c.1696delC (p.Gln566fs) in a family of three. The variant spectrum of DEPDC5 is expanded. Due to interfamilial phenotypic and genetic heterogeneity, the prevalence of FFEVF might be underestimated in such small families. Considering the existence of an unaffected carrier, FFEVF is possible to be diagnosed clinically. In clinical practice, gene detection can provide support to a definitive diagnosis and it is necessary for the patients with sporadic epilepsy of unknown etiology, as well as their family members.
Availability of data and materials
The datasets used and/or analysed during the current study are available in ClinVar, accession number is SCV002025780.
Abbreviations
- DEPDC5:
-
Disheveled, Egl-10 and pleckstrin domain-containing protein 5
- mTORC1:
-
Mammalian target of rapamycin complex 1
- FFEVF:
-
Familial focal epilepsy with variable foci
- MRI:
-
Magnetic resonance imaging
- EEG:
-
Electroencephalogram
- WES:
-
Whole exome sequencing
- FCD:
-
Focal cortical dysplasia
References
Myers KA, Scheffer IE. DEPDC5 as a potential therapeutic target for epilepsy. Expert Opin Ther Targets. 2017;21(6):591–600.
Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340(6136):1100–6.
Bockaert J, Marin P. mTOR in Brain Physiology and Pathologies. Physiol Rev. 2015;95(4):1157–87.
Dibbens LM, de Vries B, Donatello S, Heron SE, Hodgson BL, Chintawar S, Crompton DE, Hughes JN, Bellows ST, Klein KM, et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat Genet. 2013;45(5):546–51.
Pippucci T, Licchetta L, Baldassari S, Palombo F, Menghi V, D’Aurizio R, Leta C, Stipa C, Boero G, D’Orsi G, et al. Epilepsy with auditory features: A heterogeneous clinico-molecular disease. Neurol Genet. 2015;1(1):e5.
Picard F, Makrythanasis P, Navarro V, Ishida S, de Bellescize J, Ville D, Weckhuysen S, Fosselle E, Suls A, De Jonghe P, et al. DEPDC5 mutations in families presenting as autosomal dominant nocturnal frontal lobe epilepsy. Neurology. 2014;82(23):2101–6.
Tsai MH, Chan CK, Chang YC, Yu YT, Chuang ST, Fan WL, Li SC, Fu TY, Chang WN, Liou CW, et al. DEPDC5 mutations in familial and sporadic focal epilepsy. Clin Genet. 2017;92(4):397–404.
The 1000 Genomes Project Database. https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/. Accessed 24 Jan 2021.
The Exome Aggregation Consortium Database. http://exac.broadinstitute.org/. Accessed 24 Jan 2021.
National Heart, Lung, and Blood Institute Exome Sequencing Project. https://evs.gs.washington.edu/EVS/. Accessed 23 Jan 2021.
Human Gene Mutation Database. http://www.hgmd.cf.ac.uk/ac/index.php. Accessed 24 Jan 2021.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24.
Martin C, Meloche C, Rioux MF, Nguyen DK, Carmant L, Andermann E, Gravel M, Cossette P. A recurrent mutation in DEPDC5 predisposes to focal epilepsies in the French-Canadian population. Clin Genet. 2014;86(6):570–4.
Baldassari S, Picard F, Verbeek NE, van Kempen M, Brilstra EH, Lesca G, Conti V, Guerrini R, Bisulli F, Licchetta L, et al. The landscape of epilepsy-related GATOR1 variants. Genet Med. 2019;21(2):398–408.
De Fusco A, Cerullo MS, Marte A, Michetti C, Romei A, Castroflorio E, Baulac S, Benfenati F. Acute knockdown of Depdc5 leads to synaptic defects in mTOR-related epileptogenesis. Neurobiol Dis. 2020;139:104822.
Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD, Dobyns WB. A developmental and genetic classification for malformations of cortical development: update 2012. Brain. 2012;135(Pt 5):1348–69.
Marsan E, Ishida S, Schramm A, Weckhuysen S, Muraca G, Lecas S, Liang N, Treins C, Pende M, Roussel D, et al. Depdc5 knockout rat: A novel model of mTORopathy. Neurobiol Dis. 2016;89:180–9.
Liu L, Chen Z, Xu H, Liu D, Mao Y, Liu H, Liu X, Zhou P, Lin S, Li B, et al. DEPDC5 Variants Associated Malformations of Cortical Development and Focal Epilepsy With Febrile Seizure Plus/Febrile Seizures: The Role of Molecular Sub-Regional Effect. Front Neurosci-Switz. 2020;14:821.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant number 81771589), the Program of Tianjin Science and Technology Plan (grant number 18ZXDBSY00170) and the Health Science and Technology Plan of Tianjin (grant number ZC20120). The role of the funding body: Genetic analysis was supported by the fundings.
Author information
Authors and Affiliations
Contributions
CG, XL and LP participated in the conception of the study and writing of the manuscript. JM made substantial contributions to the the acquisition, analysis and interpretation of EEG data. CG, XL, LP, XZ and JS performed the experiments. JS, DL and CC analyzed the experimental results. JM and CC revised the manuscript and CC submitted the manuscript. All of the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from all family members and the study was approved by the ethics committee of Tianjin Children's Hospital (Tianjin, China).
Consent for publication
Written informed consent was obtained from the patient’s parents for publication.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gu, C., Lu, X., Ma, J. et al. What is the impact of a novel DEPDC5 variant on an infant with focal epilepsy: a case report. BMC Pediatr 22, 459 (2022). https://doi.org/10.1186/s12887-022-03515-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-022-03515-8