Abstract
Background
Kruppel-like factor 4 (KLF4) is a zinc finger-containing transcription factor predominantly expressed in terminally differentiated epithelial tissues. Many studies have shown that KLF4 has various mechanisms in different tumours; however, the prognostic role of KLF4 remains unclear.
Methods and results
We searched the relevant literature that evaluated the prognostic value of KLF4 in different cancers, and the original survival data were obtained from the text, tables or Kaplan–Meier curves for both comparative groups. Thirty studies were included in this meta-analysis, and a total of 10 malignant tumours were involved. The expression of KLF4 was not associated with the prognosis for overall survival (hazard ratio(HR)0.86, 95% confidence interval (CI): 0.65–1.13, P = 0.28), disease-free survival/recurrence-free survival/metastasis-free survival (HR 0.87, 95% CI: 0.52–1.44, P = 0.58) or disease-specific survival (HR 1.13, 95% CI: 0.44–2.87, P = 0.8).
Conclusion
This study showed that the expression of KLF4 was not related to the prognosis of the tumours that were included in the study.
Similar content being viewed by others
Background
Cancer is a major public health problem worldwide and a heavy burden on society worldwide. In 2021, 1,898,160 new cancer cases and 608,570 cancer deaths are projected to occur in the United States, and cancer is the second leading cause of death in the United States [1]. Despite the rapid progression in the aetiology, diagnosis and treatment of cancer, the prognosis of malignant tumours remains unsatisfactory. Currently, in an era of precision therapy, the widespread use of molecular-targeted therapy has not only improved the survival rate of malignancies but also offered promising prospects in the treatment of cancer. The key to targeted therapy is to choose the most appropriate patients; thus, researchers should try their best to identify the ideal molecular markers. Therefore, the markers should represent both therapeutic value and prognostic value, as well as help the risk stratification and optimal treatment options for patients.
Kruppel-like factor 4 (KLF4), a member of the KLF family, is a zinc finger-containing transcription factor predominantly expressed in terminally differentiated epithelial tissues, including skin, lung and gastrointestinal tract. There are three functional domains in KLF4: a carboxyl-terminal DNA-binding domain containing three C2H2 zinc fingers, an activated or suppressed domain and nuclear location sequences [2, 3]. KLF4 binds to specific DNA sequences, including CACCC boxes and GC boxes, and regulates cellular proliferation.
KLF4 plays important roles in development, cellular reprogramming and cancer. It was initially thought to be a negative regulator of cell growth, with the ability to regulate the expression of a number of genes involved in cell cycle progression [4, 5]. A variety of studies have indicated that KLF-4 is a context-dependent oncogene or tumour suppressor gene that is regulated in many molecular pathways and cellular processes. Some studies have shown that KLF4 expression is decreased in gastric cancers, hepatocellular carcinoma and lung cancer and is a favourable prognostic factor [6,7,8,9]. However, some studies have shown that KLF4 expression is associated with poor survival in breast cancer, prostate cancer, colorectal cancer, and skin squamous cell carcinoma, which indicates that KLF4 may be an oncogene [6, 10,11,12]. Additionally, some recent studies have shown that patients with higher KLF4 expression have better overall survival and disease-free survival rates in breast cancer and prostate cancer [13, 14]. Moreover, there was no association between the KLF4 expression and pathological diagnosis and tumour–node–metastasis (TNM) stage [15]. Although most of the studies on KLF4 have focused on epithelial tumours, several studies have investigated the role of KLF4 in B cells and B cell malignancies and indicated that it is a tumour suppressor in non-Hodgkin’s lymphoma and a potential biomarker for inferior overall survival [16, 17].
The prognostic role of KLF4 remains unclear. In this study, we conducted a systematic review and meta-analysis to summarize the global findings in using KLF4 for the prediction of the clinical results of cancer patients.
Methods
Search strategy
A thorough search was carried out for all relevant literature that evaluated the prognostic value of KLF4 in different cancers until December 1, 2019, among the following electronic databases: PubMed, ISI Web of Science and Embase. Search terms were as follows: (KLF4 OR Krüppel-like factor 4 OR Gutenriched KLF OR GKLF OR ZEF OR Epithelial Zinc Finger Protein) AND (cancer OR tumour OR neoplasm OR carcinoma) AND (Prognosis OR prognostic OR survival OR outcome). The Cochrane Library was also reviewed for related papers. In addition, the citation lists of identified articles were manually reviewed to complete the search. Two authors independently performed this procedure. Any disagreement was resolved by mutual discussion.
Selection criteria
In this meta-analysis, the eligibility of candidate studies was determined based on the following criteria: (i) studied patients with all kinds of cancers; (ii) measured KLF4 expression using either semiquantitative immunohistochemistry (IHC) or quantitative reverse transcription PCR (RT-PCR); and (iii) evaluated the correlation between KLF4 expression and prognosis. Articles were not taken into account when the following criteria were met: (i) duplicated or overlapping studies; (ii) reviews, case reports, comments, or conference abstracts; and (iii) absence of key information for further quantification calculation. Two individuals separately carried out all evaluations, and any discrepancy was resolved by consensus.
Data extraction and conversion
Data retrieved from the reports included the following elements: author, publication year, origin of population, tumour type, follow-up time, sample size, KLF4 measurement method, cut-off value of the hazard ratios (HRs), and 95% confidence intervals (CIs) of KLF4 for overall survival (OS), disease-free survival (DFS), recurrence-free survival (RFS), progression-free survival (PFS), and disease-specific survival (DSS). The original survival data were obtained from the text, tables or Kaplan-Meier curves for both comparative groups. Engauge Digitizer 4.1 (downloaded from http://markummitchell.github.io/engauge-digitizer) helped us digitize and extract survival information from the Kaplan-Meier curves using the method established by Tierney et al. [18]
Statistical analysis
The HRs in combination with the corresponding 95% CIs of identified studies were combined to estimate the overall effective value following Tierney’s method. Cochran’s Q test and Higgin’s I2 statistics were simultaneously adopted to test the heterogeneity of combined HRs. A random effects model was adopted to aggregate the pooled HR when significant heterogeneity existed (I2 > 50%); in contrast, a fixed effects model was employed (I2 < 50%). The impact of decreased KLF4 expression on prognosis was measured by the combined HRs and their corresponding 95% CIs extracted from each included article. Indirect HRs with related 95% CIs were obtained via the method established by Tierney. Generally, a pooled HR of > 1 was assumed to indicate a significant association with poor prognosis and was interpreted as statistically significant when its 95% CI did not cross 1.
Results
Characteristics of the included studies
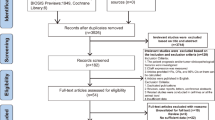
A flow diagram of the search process is given in Fig. 1. Ninety-two entries were identified from a primary literature search in PubMed, ISI Web of Science and Embase. Nineteen duplicates were removed. After manual screening of the titles and abstracts, we excluded 28 articles, such as basic studies, animal studies, noncancer subjects, non-KLF4 topics, or HRs or OS data that were unavailable. Thus, 30 studies [10, 19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45] were included in this meta-analysis. A total of 10 malignant tumours were involved.
Studies concerning hepatocellular carcinoma (HCC) are the largest group among all primary studies (n = 5), followed by colorectal cancer (CRC) (n = 4), breast cancer (BC) (n = 4), head and neck tumour (HNC) (n = 3), gastric cancer (GC) (n = 3), oesophageal squamous cell carcinoma (ESCC) (n = 3), pancreatic ductal adenocarcinoma (PDAC) (n = 3), urological cancer (n = 3), non-Hodgkin’s lymphoma (NHL) (n = 1), and cervical squamous cell carcinoma (CSCC) (n = 1). The majority of studies were carried out in China (n = 17), followed by Japan (n = 5) and other nations. The sample size of the identified articles ranged from 22 to 365, with a mean number of 128 patients. A total of 26 studies described the correlation of overall survival and KLF4 expression, one study reported only HR, and the other 25 studies reported both HR and 95% CI. The rest of the detailed features were recorded and summarized in Table 1.
Meta-analysis
The association between KLF4 expression and cancer prognosis is illustrated in Figs. 2, 3 and 4. Overall, the expression of KLF4 was not associated with the prognosis in those patients, with a pooled HR of 0.86 (95% CI: 0.65–1.13, P = 0.28) for OS via a random model because of the significant heterogeneity (I2 = 85%) (Fig. 2A). In the same way, the expression of KLF4 had nothing to do with DFS, RFS and metastasis-free survival (MFS), with a pooled HR of 0.87 (95% CI: 0.52–1.44, P = 0.58) calculated by a random model because of the presence of profound heterogeneity (I2 = 89%) (Fig. 2B). Additionally, KLF4 was not related to DSS, with a pooled HR of 1.13 (95% CI: 0.44–2.87, P = 0.8) through a random effects model for insignificant heterogeneity (I2 = 61%) (Fig. 2C).
Forest plot and pooled HR and 95% CI for OS and DFS of digestive system cancers (A: OS of digestive system cancers; B: DFS of digestive system cancers; C: OS of subgroups of gastric cancer (GC); D: OS of subgroups of colorectal cancer (CRC); E: OS of subgroups of pancreatic ductal adenocarcinoma (PDAC); F: OS of subgroups of hepatocellular cancer (HCC))
To explore the sources of heterogeneity, subgroup analyses for OS were conducted by ethnicity, measurement method, and cancer type. In the ethnicity subgroup analyses, the results showed that KLF4 expression had no impact on OS (HR = 0.77, 95% CI: 0.56–1.05, P = 0.1) in Asian cancer patients (Fig. 3B) or OS in non-Asian patients (HR = 1.44, 95% CI: 0.82–2.53, P = 0.2) (Fig. 3A). In the subgroup analyses by the measurement methods, high expression of KLF4 in the IHC group showed improved OS (HR = 0.72, 95% CI: 0.55–0.94, P = 0.02) (Fig. 3C). In the RT-PCR group, KLF4 was not associated with OS (HR =2.01, 95% CI: 0.76–5.33, P = 0.16) (Fig. 3D).
In the stratified analyses according to cancer type, high KLF4 expression indicated better prognosis in patients with GC, HCC, PDAC and CRC, with a pooled HR of 0.59 (95% CI: 0.53–0.65, P < 0.001) for OS (Fig. 4A). Disappointedly, no PFS/DFS/RFS benefits were observed (HR = 1.13, 95% CI: 0.48–2.66, P = 0.78) (Fig. 4B). For different types of digestive system cancers, our research showed that high expression of KLF4 was associated with a significantly prolonged OS in GC (HR = 0.64, 95% CI: 0.49–0.84, P = 0.001) (Fig. 4C) but not statistically significant in CRC (HR = 1.04, 95% CI: 0.61–1.76) (Fig. 4D), PDAC (HR = 1.18, 95% CI: 0.28–4.87) (Fig. 4E) or HCC for OS (HR = 2.37, 95% CI: 0.29–19.20, P = 0.42) (Fig. 4F).
In other cancers, high expression of KLF4 was associated with increased OS in BC (HR =0.49, 95% CI: 0.33–0.72) (Fig. 5A) and head and neck tumours (HR =0.41, 95% CI: 0.23–0.75) (Fig. 5B).
Discussion
The cell cycle regulates the proliferation and differentiation of tumour cells, and various transcription factors affect the progression of the cell cycle by activating or transcribing cell cycle-regulating genes. KLF4 is a transcription factor that is widely present in eukaryotic cells with zinc finger structures and can promote cancer or tumour suppressors in different types of tumours. A large number of researchers have studied the expression of KLF4 in different malignancies. In numerous clinical studies, overexpression of KLF4 inhibited cell growth, migration, invasion and metastasis in HCC, lung cancer and CRC and induced tumour cell apoptosis in oesophageal cancer and bladder cancer [6, 46]. These functional investigations suggest that decreased expression of KLF4 promoted tumour progression and poor prognosis.
A meta-analysis of 17 studies on malignant tumours of the digestive system showed that a decrease in KLF4 was associated with poorer survival outcomes [47]. The underlying mechanism of KLF4 downregulation in cancer remains to be elucidated. There are several mechanisms of KLF4 as an oncogene: (i) inhibitor of Wnt/beta – catenin pathway: Yu [48] has found that KLF4 could be combined with the β-catenin, and then inhibited beta-catenin getting into the nucleus and binding with T cell factor, leading to the suppression of the downstream target gene of Wnt/beta-catenin pathway; (ii) regulation of the Notch pathway to inhibit tumour development: Ghaleb [49] found that Klf4 was the downstream target gene of the Notch pathway, and its transcription activity was inhibited by Notch. In the tumour, the Notch gene was highly expressed, and KLF4 was downregulated by the C-side control element of Notch (ICN1); (iii) KLF4 plays a role in epithelial-to-mesenchymal transition (EMT): KLF4 can inhibit EMT through regulation of E-cadherin gene expression [50]; (iv) immune escape mechanism: tumour cell major histocompatibility complex (MHC) changes, affecting antigen presentation, thus avoiding immune surveillance. KLF4 inhibits the expression of MHC and thus inhibits tumour development [51]. (v) Inhibitor of cell cycle KLF4 cooperates with p53 to enhance p21 expression and inhibit the function of cyclin D1 and cyclin B1, which causes cell cycle arrest at the G1/S and/or G2/M checkpoints [52].
KLF4, in skin cancer and BC, appears to promote tumour progression, suggesting that KLF4 is an oncogene in these tumours [10, 12]. The mechanism of KLF4 as an oncogene is vague, but Hu’s study [53] found that the oestrogen receptor (ER) can affect the E3 ubiquitin ligase, causing the degradation of KLF4 and accumulation of KLF4 protein. On the other hand, KLF4, as an agonist of ER transcription factors, can promote the ER and its downstream target gene promoter region and promote cell growth and mitosis. In addition, when DNA is damaged, KLF4 plays an anti-apoptotic effect by inhibiting the expression of p53 [54]. The deletion of KLF4 in BC can lead to the recovery of p53 expression and the apoptosis of p53-dependent tumour cells [12, 55]. This also confirms that KLF4 in BC is used as a cancer gene to promote cancer by inhibiting the expression of p53.
Our results show that KLF4 expression is not related to the prognosis of various malignancies. There was no difference between Asian cancer patients and non-Asian patients (HR = 1.44, 95% CI: 0.82–2.53, P = 0.2). Our study indicated that KLF4 might be a new biomarker for GC with a HR of 0.64 (95% CI: 0.49–0.84, P = 0.001). Nevertheless, 2 Asian studies on GC showed that high expression of KLF4 was associated with better prognosis, which was opposite to the American study. The reason was considered to be the sample size or race. However, further studies are needed to support this point. In subgroup analyses, high expression of KLF4 in the IHC group showed a better prognosis (HR = 0.72, 95% CI: 0.55–0.94, P = 0.2), whereas the RT-PCR group did not. This is probably because the expression of KLF4 is regulated at both the transcriptional and posttranscriptional levels. High expression of KLF4 was associated with good prognosis in BC (HROS = 0.49, 95% CI: 0.33–0.72) and head and neck tumours (HROS = 0.41, 95% CI: 0.23–0.75), which is different from previous results.
There are limitations to this study. First, the HRs of most studies, extrapolated based on Tierney’s method, were less reliable than those directly provided in the original articles. Second, studies published in other languages were not included, which probably introduced bias. Third, different tumours and different treatments were included in our study, which could result in great heterogeneity. Fourth, the cut-off values and detection methods in the studies were not uniform, which was also a source of heterogeneity. Fifth, because of the limited number of included studies of each type of cancer, the results of some carcinomas were statistically insignificant and might be less powerful. Finally, KLF4 expression could be different depending on the TNM stage, whereas stratification analysis by TNM staging was not performed since the original data could not be obtained. Although we used a random-effects model and conducted subgroup analyses to explore the potential source of heterogeneity, there were still unexpected heterogeneities in those subgroups. Therefore, future research should focus on high-quality studies with comprehensive evaluation, thus resulting in more standardized research and more accurate conclusions.
Conclusion
Although our study showed that the expression of KLF4 is not related to the prognosis of the tumours included in the study, we found that high expression of KLF4 predicted a trend of better prognosis. Moreover, high expression of KLF4 is associated with prolonged OS in GC, BC and head and neck cancer and may be a predictive factor for these cancers. Numerous studies are needed to find out the role of KLF4, and the molecular basis of the transformation between tumour suppressor genes and oncogenes still needs to be solved.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- KLF4:
-
Kruppel-like factor 4
- HR:
-
Hazard ratio
- CI:
-
Confidence interval
- TNM:
-
Tumor–Node–Metastasis
- IHC:
-
Immunohistochemistry
- RT-PCR:
-
Quantitative reverse transcription PCR
- OS:
-
Overall survival
- DFS:
-
Disease-free survival
- RFS:
-
Recurrence-free survival
- PFS:
-
Progression free survival
- DSS:
-
Disease-special survival
- HCC:
-
Hepatocellular carcinoma
- CRC:
-
Colorectal cancer
- BC:
-
Breast cancer
- HNC:
-
Head and neck tumor
- GC:
-
Gastric cancer
- ESCC:
-
Esophageal squamous cell carcinomas
- PDAC:
-
Pancreatic ductal adenocarcinoma
- NHL:
-
Non-Hodgkin’s lymphoma
- CSCC:
-
Cervical squamous cell carcinoma
- MFS:
-
Metastasis-free survival
- ICN1:
-
C-side control element of Notch
- EMT:
-
Epithelial-to-mesenchymal transition
- MHC:
-
Major histocompatibility complex
- ER:
-
Estrogen receptor
References
Siegel R, Miller K, Fuchs H, et al. Cancer statistics. CA Cancer J Clin. 2020;71(1):7–33. https://doi.org/10.3322/caac.21654.
Flandez M, Guilmeau S, Blache P, Augenlicht LH. KLF4 regulation in intestinal epithelial cell maturation. Exp Cell Res. 2008;314(20):3712–23. https://doi.org/10.1016/j.yexcr.2008.10.004.
Camacho-Vanegas O, Till J, Mirandalorenzo I, et al. Shaking the family tree: identification of novel and biologically active alternatively spliced isoforms across the KLF family of transcription factors. FASEB J. 2013;27(2):432–6. https://doi.org/10.1096/fj.12-220319.
Shie JL, Chen ZY, Fu M, et al. Gut-enriched Krüppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 2000;28(15):2969–76. https://doi.org/10.1093/nar/28.15.2969.
Zhang W, Geiman DE, Shields JM, et al. The gut-enriched Krüppel-like factor (Krüppel-like factor 4) mediates the Transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275(24):18391. https://doi.org/10.1074/jbc.C000062200.
Wei D, Kanai M, Huang S, et al. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27(1):23–31. https://doi.org/10.1093/carcin/bgi243.
Wei D, Gong W, Kanai M, et al. Drastic Down-regulation of Krüppel-like factor 4 expression is critical in human gastric Cancer development and progression. Cancer Res. 2005;65(7):2746–54. https://doi.org/10.1158/0008-5472.
Xue M, Zhou C, Zheng Y, et al. The association between KLF4 as a tumor suppressor and the prognosis of hepatocellular carcinoma after curative resection. Aging (Albany NY). 2020;12(15):15566–80. https://doi.org/10.18632/aging.103592.
Wu Y, Lin L, Wang X, et al. Overexpression of Kruppel-like factor 4 suppresses migration and invasion of non-small cell lung Cancer through c-Jun-NH2-terminal kinase/epithelial-mesenchymal transition signaling pathway. Front Pharmacol. 2020;10:1512. https://doi.org/10.3389/fphar.2019.01512.
Pandya AY, Talley LI, Frost AR, et al. Nuclear localization of KLF4 is associated with an aggressive phenotype in early-stage breast cancer. Clin Cancer Res. 2004;10(8):2709–19. https://doi.org/10.1158/1078-0432.ccr-03-0484.
Feng F, Liu H, Chen A, et al. miR-148-3p and miR-152-3p synergistically regulate prostate cancer progression via repressing KLF4. J. Cell. Biochem. 2019, 120, 17228–17239. J Cell Biochem. 2019 Oct;120(10):17228–39. https://doi.org/10.1002/jcb.28984.
Foster KW, Liu Z, Nail CD, et al. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24(9):1491–500. https://doi.org/10.1038/sj.onc.1208307.
Nagata T, Shimada Y, Sekine S, et al. KLF4 and NANOG are prognostic biomarkers for triple-negative breast cancer. Breast Cancer. 2017;24(2):326–35. https://doi.org/10.1007/s12282-016-0708-1.
Jiang Z, Zhang Y, Chen X, et al. Long non-coding RNA LINC00673 silencing inhibits proliferation and drug resistance of prostate cancer cells via decreasing KLF4 promoter methylation. J Cell Mol Med. 2020;24(2):1878–92. https://doi.org/10.1111/jcmm.14883.
Nagata T, Shimada Y, Sekine S, et al. Prognostic significance of NANOG and KLF4 for breast cancer. Breast Cancer. 2014;21(1):96–101. https://doi.org/10.1007/s12282-012-0357-y.
Guan H, Xie L, Leithäuser F, et al. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. 2010;116(9):1469–78. https://doi.org/10.1182/blood-2009-12-256446.
Valencia-Hipόlito A, Hernández-Atenógenes M, Vega GG, et al. Expression of KLF4 is a predictive marker for survival in pediatric Burkitt lymphoma. Leuk Lymphoma. 2014;55(8):1806–14. https://doi.org/10.3109/10428194.2013.848437.
Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8(1):16. https://doi.org/10.1186/1745-6215-8-16.
Yin X, Yiwei LI, Jin J, et al. The clinical and prognostic implications of pluripotent stem cell gene expression in hepatocellular carcinoma. Oncol Lett. 2013;5(4):1155. https://doi.org/10.3892/ol.2013.1151.
Patel N, Ghaleb AM, Nandan MO, et al. Expression of the tumor suppressor Krüppel-like factor 4 as a prognostic predictor for Colon Cancer. Cancer Epidemiol Biomark Prev. 2010;19(10):2631–8. https://doi.org/10.1158/1055-9965.EPI-10-0677.
Wei D, Wang L, Kanai M, et al. KLF4α upregulation promotes cell cycle progression and reduces survival time of patients with pancreatic Cancer. Gastroenterology. 2010;139(6):2135. https://doi.org/10.1053/j.gastro.2010.08.022.
Chang Y. The significance of KLF4 expression and prognosis in the breast invasive ductal carcinoma tissue: Doctoral dissertation, HeiBei Medical University; 2016. Epub 2016 Sep 14.
Sun H, Tang H, Xie D, et al. Krüppel-like factor 4 blocks hepatocellular carcinoma dedifferentiation and progression through activation of hepatocyte nuclear Factor-6. Clin Cancer Res. 2016;22(2):502–12. https://doi.org/10.1158/1078-0432.CCR-15-0528.
Qi L, Yong G, Jia Z, et al. Dysregulated Krüppel-like factor 4 and vitamin D receptor signaling contribute to progression of hepatocellular carcinoma. Gastroenterology. 2012;143(3):799. https://doi.org/10.1053/j.gastro.2012.05.043.
Tseng WC, Chuang CW, Yang MH, et al. Krüppel-like factor 4 is a novel prognostic predictor for urothelial carcinoma of bladder and it regulates TWIST1-mediated epithelial-mesenchymal transition. Urol Oncol Semin Orig Investig. 2016;34(11):485.e15–24. https://doi.org/10.1016/j.urolonc.2016.07.002.
Yang Z, Li D, Liu Z, et al. BIRC7 and KLF4 expression in benign and malignant lesions of pancreas and their clinicopathological significance. Cancer Biomarkers. 2016;17(4):437–44. https://doi.org/10.3233/CBM-160660.
Liu Z, Yang H, Luo W, et al. Loss of cytoplasmic KLF4 expression is correlated with the progression and poor prognosis of nasopharyngeal carcinoma. Histopathology. 2013;63(3):362–70. https://doi.org/10.1111/his.12176.
Shimada Y, Okumura T, Sekine S, et al. Expression analysis of iPS cell - inductive genes in esophageal squamous cell carcinoma by tissue microarray. Anticancer Res. 2012;32:5507–14.
Tang W, Zhu Y, Gao J, et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br J Cancer. 2014;110(2):450–8. https://doi.org/10.1038/bjc.2013.724.
Sun H, Peng Z, Tang H, et al. Loss of KLF4 and consequential downregulation of Smad7 exacerbate oncogenic TGF-β signaling in and promote progression of hepatocellular carcinoma. Oncogene. 2017;36(21):2957–68. https://doi.org/10.1038/onc.2016.447.
Tai SK, Yang MH, Chang SY, et al. Persistent Krüppel-like factor 4 expression predicts progression and poor prognosis of head and neck squamous cell carcinoma. Cancer Sci. 2011;102(4):895–902. https://doi.org/10.1111/j.1349-7006.2011.01859.x.
Yoshihama R, Yamaguchi K, Imajyo I, et al. Expression levels of SOX2, KLF4 and brachyury transcription factors are associated with metastasis and poor prognosis in oral squamous cell carcinoma. Oncol Lett. 2016;11(2):1435–46. https://doi.org/10.3892/ol.2015.4047.
Funel N, Morelli M, Giovannetti E, et al. Loss of heterozygosity status of d9s105 marker is associated with downregulation of Krüppel-like factor 4 expression in pancreatic ductal adenocarcinoma and pancreatic intraepithelial lesions. Pancreatology. 2011;11(1):30–42. https://doi.org/10.1159/000322990.
Ma MQ, Zhang HD, Tang P, et al. Association of Kruppel-like factor 4 expression with the prognosis of esophageal squamous cell carcinoma patients. Int J Clin Exp Pathol. 2014;7(10):6679–85.
Hsu LS, Chan CP, Chen CJ, et al. Decreased Kruppel-like factor 4 (KLF4) expression may correlate with poor survival in gastric adenocarcinoma. Med Oncol. 2013;30(4):632. https://doi.org/10.1007/s12032-013-0632-6.
Lee HY, Ahn JB, Rha SY, et al. High KLF4 level in normal tissue predicts poor survival in colorectal cancer patients. World J Surg Oncol. 2014;12(1):232. https://doi.org/10.1186/1477-7819-12-232.
Hsu HT, Wu PR, Chen CJ, et al. High cytoplasmic expression of Krüppel-like factor 4 is an independent prognostic factor of better survival in hepatocellular carcinoma. Int J Mol Sci. 2014;15(6):9894–906. https://doi.org/10.3390/ijms15069894.
Xu J, Lü B, Xu F, et al. Dynamic down-regulation of Krüppel-like factor 4 in colorectal adenoma-carcinoma sequence. J Cancer Res Clin Oncol. 2008;134(8):891–8. https://doi.org/10.1007/s00432-008-0353-y.
Li H, Wang J, Xiao W, et al. Epigenetic inactivation of KLF4 is associated with urothelial cancer progression and early recurrence. J Urol. 2014;191(2):493–501. https://doi.org/10.1016/j.juro.2013.08.087.
Hashimoto I, Nagata T, Sekine S, et al. Prognostic significance of KLF4 expression in gastric cancer. Oncol Lett. 2017:819–26. https://doi.org/10.3892/ol.2016.5499.
Li H, Wang J, Xiao W, et al. Epigenetic alterations of krüppel-like factor 4 and its tumor suppressor function in renal cell carcinoma. Carcinogenesis. 2013;34(10):2262–70. https://doi.org/10.1093/carcin/bgt189.
Liu H, Li N, Wei L, et al. High expression of Kruppel-like factor 4 as a predictor of poor prognosis for cervical cancer patient response to radiotherapy. Tumour Biol. 2017 Jun;39(6):1010428317710225. https://doi.org/10.1177/1010428317710225.
Wei D, Gong W, Kanai M, et al. Drastic down-regulation of Kruppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65(7):2746–54. https://doi.org/10.1158/0008-5472.CAN-04-3619.
Chen CJ, Hsu LS, Lin SH, et al. Loss of nuclear expression of Krüppel-like factor 4 is associated with poor prognosis in patients with oral cancer. Hum Pathol. 2012;43(7):1119–25. https://doi.org/10.1016/j.humpath.2011.09.003.
Valencia-Hipólito A, Hernández-Atenógenes M, Vega GG, et al. Expression of KLF4 is a predictive marker for survival in pediatric Burkitt lymphoma. Leuk Lymphoma. 2014;55(8):1806–14. https://doi.org/10.3109/10428194.2013.848437.
Ohnishi S, Ohnami S, Laub F, et al. Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308(2):251–6. https://doi.org/10.1016/s0006-291x(03)01356-1.
Hu J, Li H, Wu C, et al. The prognostic value of decreased KLF4 in digestive system cancers: a Meta-analysis from 17 studies. Dis Markers. 2017;2017:1–11. https://doi.org/10.1155/2017/3064246.
Yu T, Chen X, Zhang W, et al. Regulation of the potential marker for intestinal cells, Bmi1, by β-catenin and the zinc finger protein KLF4: implications for colon cancer. J Biol Chem. 2012;287(6):3760–8. https://doi.org/10.1074/jbc.M111.316349.
Ghaleb AM, Aggarwal G, Bialkowska AB, et al. Notch inhibits expression of the Krüppel-like factor 4 tumor suppressor in the intestinal epithelium. Mol Cancer Res Mcr. 2008;6(12):1920–7. https://doi.org/10.1158/1541-7786.MCR-08-0224.
Yori JL, Johnson E, Zhou G, et al. Kruppel-like factor 4 inhibits epithelial-to-mesenchymal transition through regulation of E-cadherin gene expression. J Biol Chem. 2010;285(22):16854–63. https://doi.org/10.1074/jbc.M110.114546.
Ghorpade DS, Holla S, Sinha AY, et al. Nitric oxide and KLF4 protein epigenetically modify class II transactivator to repress major histocompatibility complex II expression during Mycobacterium bovis bacillus Calmette-Guerin infection. J Biol Chem. 2013;288(28):20592–606. https://doi.org/10.1074/jbc.M113.472183.
Ghaleb AM, Nandan MO, Chanchevalap S, et al. Krüppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005 Feb;15(2):92–6. https://doi.org/10.1038/sj.cr.7290271.
Hu D, Zhou Z, Davidson NE, et al. Novel insight into KLF4 proteolytic regulation in estrogen receptor signaling and breast carcinogenesis. J Biol Chem. 2012;287(17):13584–97. https://doi.org/10.1074/jbc.M112.343566.
Zhou Q, Hong Y, Zhan Q, et al. Role for Kruppel-like factor 4 in determining the outcome of p53 response to DNA damage. Cancer Res. 2009;69(21):8284–92. https://doi.org/10.1158/0008-5472.CAN-09-1345.
Raman V, Martensen SA, Reisman D, et al. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405(6789):974–8. https://doi.org/10.1038/35016125.
Funding
No
Author information
Authors and Affiliations
Contributions
YZ and XL contributed to the conception, design, data analysis and the manuscript writing of this study equally. YZ, XL, YM, MJ, and YW contributed to the conception and design the study. YZ and XL analyzed, interpreted the data and wrote the manuscript. XL approved the final submission of the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Luo, X., Zhang, Y., Meng, Y. et al. Prognostic significance of KLF4 in solid tumours: an updated meta-analysis. BMC Cancer 22, 181 (2022). https://doi.org/10.1186/s12885-022-09198-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-022-09198-9