Abstract
Background
To investigate associations between lower-grade glioma (LGG) mRNA-based subtypes (R1-R4) and MR features.
Methods
mRNA-based subtyping was obtained from the LGG dataset in The Cancer Genome Atlas (TCGA). We identified matching patients (n = 145) in The Cancer Imaging Archive (TCIA) who underwent MR imaging. The associations between mRNA-based subtypes and MR features were assessed.
Results
In the TCGA-LGG dataset, patients with the R2 subtype had the shortest median OS months (P < 0.05). The time-dependent ROC for the R2 subtype was 0.78 for survival at 12 months, 0.76 for survival at 24 months, and 0.76 for survival at 36 months. In the TCIA-LGG dataset, 41 (23.7%) R1 subtype, 40 (23.1%) R2 subtype, 19 (11.0%) R3 subtype and 45 (26.0%) R4 subtype cases were identified. Multivariate analysis revealed that enhancing margin (ill-defined, OR: 9.985; P = 0.003) and T1 + C/T2 mismatch (yes, OR: 0.091; P = 0.023) were associated with the R1 subtype (AUC: 0.708). The average accuracy of the ten-fold cross validation was 71%. Proportion of contrast-enhanced (CE) tumour (> 5%, OR: 14.733; P < 0.001) and necrosis/cystic changes (yes, OR: 0.252; P = 0.009) were associated with the R2 subtype (AUC: 0.832). The average accuracy of the ten-fold cross validation was 82%. Haemorrhage (yes, OR: 8.55; P < 0.001) was positively associated with the R3 subtype (AUC: 0.689). The average accuracy of the ten-fold cross validation was 87%. Proportion of CE tumour (> 5%, OR: 0.14; P < 0.001) was negatively associated with the R4 subtype (AUC: 0.672). The average accuracy of the ten-fold cross validation was 71%. For the prediction of the R2 subtype, the nomogram showed good discrimination and calibration. Decision curve analysis demonstrated that prediction with the R2 model was clinically useful.
Conclusions
Patients with the R2 subtype had the worst prognosis. We demonstrated that MRI features can identify distinct LGG mRNA-based molecular subtypes.
Similar content being viewed by others
Background
Primary brain tumours are one of the top ten causes of cancer-related deaths in the United States [1]. They are characterized by biological heterogeneity and can be classified into a variety of histological subtypes [2,3,4]. LGGs are currently classified by morphological criteria. However, this classification suffers from high interobserver and intraobserver variability [5, 6]. Therefore, clinicians increasingly rely on genetic classification to guide clinical decision making. The treatment of LGG could benefit from the incorporation of precision medicine. The majority of patients with high-risk LGG are treated with single-agent temozolomide (TMZ) and radiotherapy. Next-generation sequencing has definitively revealed that different LGG mRNA-based subtypes fundamentally differ in their underlying molecular pathways, despite being histologically similar [7,8,9].
A new direction in cancer research has emerged that focuses on the relationship between genomic data and imaging features [10,11,12]. Radiogenomic studies have indicated key imaging differences between certain LGG genetic groups and may aid in the diagnosis of LGG as well as the longitudinal assessment of treatment response and evaluation of tumour recurrence in patients with LGG [13,14,15,16]. The TCGA Research Network identifies four mRNA-based (R1–R4) subtypes [17] (N Engl J Med 2015). Core members in the four well-defined subtypes were identified and found to be distinctly enriched for the previously defined astrocytoma subtype and neural ontology signatures and correlated with specific genomic events.
Survival analysis revealed that the R2 subtype was significantly correlated with shorter overall survival. Both the R1 subtype and R3 subtype highly expressed an early progenitor-like astrocytoma gene signature. The R4 subtype highly expressed a neuroblastic astrocytoma signature and a neuron-specific signature [17]. This study aims to explore associations between LGG mRNA-based subtypes (R1-R4) and MR features. Our preliminary radiogenomics analysis may serve as a reference in the development of precision medicine for LGG patients.
Methods
Patient population
The clinical files of LGG patients were obtained from TCGA. MR data were provided by TCIA [18,19,20]. TCGA and TCIA are publicly available databases. The TCGA Research Network [17] classifies LGG into four categories (R1, R2, R3 and R4) according to mRNA expression patterns. The inclusion criteria of the study were as follows: (I) mRNA-based subtyping (R1-R4) was obtained from the LGG dataset in TCGA; and (II) MR data were available from TCIA (T1WI, T2WI, contrast enhancement). Unevaluable examinations and postsurgical patients were excluded. Finally, 145 patients met the inclusion criteria.
Assessment of MR features
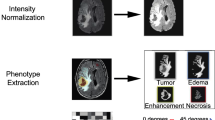
MR image analysis was performed as previously published by recent studies and our group [21,22,23]. The following lesion features were evaluated [21,22,23]: (A) volume (<median or > =median); (B) width (<median or > =median); (C) length (<median or > =median); (D) depth (<median or > =median); (E) proportion of CE tumour (no [<=5%] or yes [> 5%]); (F) enhancing margin (well-defined or poorly defined); (G) T1 + C/T2 mismatch (negative or positive); (H) extranodular growth (negative or positive); (I) shortest distance between the tumour centroid and the lateral edge [CS](<=30 mm or > 30 mm); (J) subventricular zone [SVZ] involvement (negative or positive); (K) location (frontal lobe or other); (XII) volume (> = 60 cm3 or < 60 cm3); (L) haemorrhage (negative or positive); (M) multifocal (negative or positive); (N) degree of enhancement (slight or obvious); and (O) necrosis/cystic change (negative or positive). Both neuroradiologists were blinded to LGG mRNA-based subtypes (R1-R4) as well as the clinical data.
Statistical analysis
We focused on the association of mRNA-based subtypes (R1-R4) and MR features. A colour heat map was drawn to show the correlation patterns between MR features and mRNA expression (R1-R4). Fisher’s exact test, the chi-square test and binary logistic regression analysis were used (version 23.0; SPSS Company) for each mRNA-based subtype. We use tenfold cross-validation test. Odds ratios (ORs) as well as their corresponding 95% confidence intervals (CIs) are reported. In the present study, binary logistic regression was repeated for the four LGG mRNA-based subtypes: R1, R2, R3 and R4. The area under the receiver operating characteristic curve (AUC) of each mRNA-based subtype (R1-R4) is reported. Survival analysis was conducted by using Kaplan-Meier analysis and the time-dependent ROC method (the worst prognostic subgroup; R package). A P-value of less than 0.05 (two-sided) was considered to indicate statistical significance.
Results
In the TCGA-LGG dataset, patients with the R2 subtype had the shortest median OS months (P < 0.05 Fig. 1a). The time-dependent ROC for the R2 subtype was 0.78 for survival at 12 months, 0.76 for survival at 24 months, and 0.76 for survival at 36 months (Fig. 1b).
In the TCIA-LGG dataset, a total of 145 LGG (grade II: 61 cases, grade III 68 cases) patients (IDHmut-non-codeletion: 70 cases; IDHmut-codeletion: 35 cases; IDHwt: 38 cases) were screened (female: 70 patients; male: 75 patients). The demographic and tumour characteristics of all 145 patients (astrocytoma: 49 cases; oligoastrocytoma: 31 cases; oligodendroglioma: 65 cases) are summarized in Table 1. The identified subtypes were as follows: 41 (23.7%) R1 subtype, 40 (23.1%) R2 subtype, 19 (11.0%) R3 subtype and 45 (26.0%) R4 subtype. The frequencies (heat map) of lesion features per mRNA-based subtype (R1-R4) are pictured in Fig. 2.
In univariate analysis (Tables 2 and 3), enhancing margin, extranodular growth and T1 + C/T2 mismatch were associated with the R1 subtype (all P < 0.05). Necrosis/cystic change, multifocality, degree of enhancement, proportion of CE tumour, volume, depth, extranodular growth and T1 + C/T2 mismatch were associated with the R2 subtype. Haemorrhage was associated with the R3 subtype (P < 0.05). The degree of enhancement, enhancing margin, proportion of CE tumour and extranodular growth were associated with the R4 subtype (all P < 0.05).
Multivariate analysis (Table 4) revealed that enhancing margin (ill-defined, OR: 9.985; P = 0.003) and T1 + C/T2 mismatch (yes, OR: 0.091; P = 0.023) were associated with the R1 subtype (AUC: 0.708). The average accuracy of the ten-fold cross validation was 71%. Proportion of contrast-enhanced (CE) tumour (> 5%, OR: 14.733; P < 0.001) and necrosis/cystic changes (yes, OR: 0.252; P = 0.009) were associated with the R2 subtype (Fig. 3a AUC: 0.832). The average accuracy of the ten-fold cross validation was 82%. Decision curve analysis (Fig. 3b) demonstrated that prediction with the R2 model was clinically useful. For the prediction of the R2 subtype, the nomogram showed good discrimination and calibration (Fig. 4). Haemorrhage (yes, OR: 8.55; P < 0.001) was positively associated with the R3 subtype (AUC: 0.689). The average accuracy of the ten-fold cross validation was 87%. Proportion of CE tumour (> 5%, OR: 0.14; P < 0.001) was negatively associated with the R4 subtype (AUC: 0.672). The average accuracy of the ten-fold cross validation was 71%.
We demonstrated (Fig. 5) that ill-defined margins and the absence of T1 + C/T2 mismatches were positively linked with the R1 subtype (AUC: 0.708). Proportion CE tumour (> 5%), volume < 60 cm3 and absence of necrosis/cystic change were positively associated with the R2 subtype (AUC: 0.832). Haemorrhage was positively associated with the R3 subtype (AUC: 0.689). Proportion of CE tumour > 5% was negatively associated with the R4 subtype (AUC: 0.672).
Discussion
Glioma is one of the most common primary central nervous system malignant tumours [24, 25]. Intratumoural genetic heterogeneity plays a pivotal role in driving disease progression and therapeutic resistance in LGG. Intratumoural heterogeneity has been linked to metastatic potential and is likely to be an important prognostic feature of human cancer [26, 27]. The TCGA Research Network [17] classifies LGG into four subgroups (R1–R4) based on mRNA expression. R1, R2, R3 and R4 tumours were found to be biologically and clinically distinct. Our previous published work has revealed that clinical and MR features may therefore be used to facilitate the preoperative prediction of LGG IDH/1p19q subtype. In this research, we revealed that MRI features can identify distinct LGG mRNA-based molecular subtypes.
Radiogenomic studies have revealed key imaging differences between certain LGG genetic groups and may aid in the diagnosis of patients with LGG as well as predict survival and guide treatment in patients with LGG [10, 28,29,30,31]. In this study, R2 tumours showed significantly worse overall survival than the other RNA subtypes (R1 subtype, R3 subtype and R4 subtype), which did not significantly differ from one another. The time-dependent ROC for the R2 subtype was 0.78 for survival at 12 months, 0.76 for survival at 24 months, and 0.76 for survival at 36 months.
The R2 subtype is mostly composed of GIII tumours (77%), tumours mostly of astrocytoma histology (68%), tumours enriched for the methylation subtype M2 (62%) and IDH wild type (67%) tumours. This subtype is correlated with GBM-related events such as PTEN mutation, chromosome 10 loss, and EGFR mutation and amplification. Our findings showed that the proportion of CE tumours (> 5%) and the absence of necrosis/cystic changes were positively associated with the R2 subtype (AUC: 0.832). This is the first article to show the connection between the R2 subtype and MR features. For the prediction of the R2 subtype, the nomogram showed good discrimination and calibration. Decision curve analysis demonstrated that prediction with the R2 model was clinically useful.
The other RNA subtypes (R1, R3, and R4) were populated with IDH-mutant gliomas. R1 lacked 1p/19q codeletion and was comprised of two methylation subtypes, M5 (70%) and M3 (30%), and the vast majority of R1 cases had TP53 and ATRX mutations17. We demonstrated that well-defined margins and the absence of T1 + C/T2 mismatches were positively associated with the R1 subtype (AUC: 0.708). The R3 subtype was entirely composed of IDHmut-codeletion gliomas and was equally distributed across the methylation subtypes M2 and M3. It was also enriched for oligodendrogliomas (85%), mutations in NOTCH1, FUBP1, and CIC, and oligodendrocyte progenitor-specific expression. Our findings showed that haemorrhage was positively associated with the R3 subtype (AUC: 0.689). The R4 subtype highly expressed a neuron-specific signature and a neuroblastic astrocytoma signature. The proportion of CE tumours (<=5%) was positively associated with the R4 subtype (AUC: 0.672). This is the first article to show that MRI features can identify distinct LGG mRNA-based molecular subtypes (R1, R3–4). Radiogenomics analysis allows researchers to explore the TCGA and TCIA databases for correlations between mRNA-based molecular subtypes and radiological phenotypes.
Our study has several limitations. The major limitation of this article was that the sample size of the R3 subtype was only 19 patients (11.0%). The disadvantages of a small sample size might have limited the statistical power to explore additional correlations of the R3 subtype. Our findings should be further investigated and externally validated in larger cohorts of LGG patients. In addition, the MR data are heterogeneous, and in most cases, the images were acquired as part of routine care and not as part of a controlled research study or clinical trial. Our results should be validated using standardized MR imaging.
Conclusions
Our results revealed connections between LGG mRNA-based subtypes (R1-R4) and MR lesion features. Our findings revealed that ill-defined margins and the absence of T1 + C/T2 mismatches were positively associated with the R1 subtype. The proportion of CE tumour > 5%, volume < 60 cm3 and absence of necrosis/cystic changes were positively associated with the R2 subtype. Haemorrhage was positively associated with the R3 subtype. Proportion of CE tumour > 5% was negatively associated with the R4 subtype (AUC: 0.672).
Availability of data and materials
No administrative permissions were required to access the raw data from The Cancer Genome Atlas Low Grade Glioma (TCGA-LGG) project. Public access to the databases is open. All data generated or analysed in this study are included in this published article. TCIA: https://wiki.cancerimagingarchive.net/; TCGA: https://portal.gdc.cancer.gov/.
Abbreviations
- LGG:
-
Lower-grade glioma
- CI:
-
Confidence interval
- OR:
-
Odds ratio
- TCIA:
-
The Cancer Imaging Archive
- CE:
-
Contrast-enhanced
- OS:
-
Overall survival
- AUC:
-
Area under the receiver operating characteristic curve
- TCGA:
-
The Cancer Genome Atlas
- SVZ:
-
Subventricular zone
- CS:
-
The shortest distance between the lateral edge and the tumour centroid
References
Mawson AR. Retinoids in the treatment of glioma: a new perspective. Cancer Manag Res. 2012;4:233–41.
Wang Y, Liu S, Fan X, Li S, Wang R, Wang L, Ma J, Jiang T, Ma W. Age-associated brain regions in gliomas: a volumetric analysis. J Neuro-Oncol. 2015;123:299–306.
Inano R, Oishi N, Kunieda T, Arakawa Y, Yamao Y, Shibata S, Kikuchi T, Fukuyama H, Miyamoto S. Voxel-based clustered imaging by multiparameter diffusion tensor images for glioma grading. NeuroImage Clin. 2014;5:396–407.
Raja R, Sinha N, Saini J, Mahadevan A, Rao KN, Swaminathan A. Assessment of tissue heterogeneity using diffusion tensor and diffusion kurtosis imaging for grading gliomas. Neuroradiology. 2016;58:1217–31.
Bai Y, Lin Y, Tian J, Shi D, Cheng J, Haacke EM, Hong X, Ma B, Zhou J, Wang M. Grading of Gliomas by using Monoexponential, Biexponential, and stretched exponential diffusion-weighted MR imaging and diffusion kurtosis MR imaging. Radiology. 2016;278:496–504.
Ma L, Song ZJ. Differentiation between low-grade and high-grade glioma using combined diffusion tensor imaging metrics. Clin Neurol Neurosurg. 2013;115:2489–95.
Appin CL, Brat DJ. Biomarker-driven diagnosis of diffuse gliomas. Mol Asp Med. 2015;45:87–96.
Siegal T. Clinical relevance of prognostic and predictive molecular markers in Gliomas. Adv Tech Stand Neurosurg. 2016;(43):91–108.
Weller M, Stupp R, Hegi ME, van den Bent M, Tonn JC, Sanson M, Wick W, Reifenberger G. Personalized care in neuro-oncology coming of age: why we need MGMT and 1p/19q testing for malignant glioma patients in clinical practice. Neuro Oncol. 2012;14(Suppl 4):iv100–8.
Jansen RW, van Amstel P, Martens RM, Kooi IE, Wesseling P, de Langen AJ, Menke-Van der Houven van Oordt CW, BHE J, Moll AC, Dorsman JC, Castelijns JA, de Graaf P, de Jong MC. Non-invasive tumor genotyping using radiogenomic biomarkers, a systematic review and oncology-wide pathway analysis. Oncotarget. 2018;9:20134–55.
Kai C, Uchiyama Y, Shiraishi J, Fujita H, Doi K. Computer-aided diagnosis with radiogenomics: analysis of the relationship between genotype and morphological changes of the brain magnetic resonance images. Radiol Phys Technol. 2018;11(3):265–73.
Hong EK, Choi SH, Shin DJ, Jo SW, Yoo RE, Kang KM, Yun TJ, Kim JH, Sohn CH, Park SH, Won JK, Kim TM, Park CK, Kim IH, Lee ST. Radiogenomics correlation between MR imaging features and major genetic profiles in glioblastoma. Eur Radiol. 2018;28(10):4350–61.
Alessandrino F, Shinagare AB, Bosse D, Choueiri TK, Krajewski KM. Radiogenomics in renal cell carcinoma. Abdom Radiol. 2019;44(6):1990–98.
Bai HX, Lee AM, Yang L, Zhang P, Davatzikos C, Maris JM, Diskin SJ. Imaging genomics in cancer research: limitations and promises. Br J Radiol. 2016;89:20151030.
Zhang YA, Zhou Y, Luo X, Song K, Ma X, Sathe A, Girard L, Xiao G, Gazdar AF. SHOX2 is a potent independent biomarker to predict survival of WHO grade II-III diffuse Gliomas. EBioMedicine. 2016;13:80–9.
Huang SP, Chiou J, Jan YH, Lai TC, Yu YL, Hsiao M, Lin YF. Over-expression of lysyl oxidase is associated with poor prognosis and response to therapy of patients with lower grade gliomas. Biochem Biophys Res Commun. 2018;501(3):619–27.
Cancer Genome Atlas Research N, Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA, Rheinbay E, Miller CR, Vitucci M, Morozova O, Robertson AG, Noushmehr H, Laird PW, Cherniack AD, Akbani R, Huse JT, Ciriello G, Poisson LM, Barnholtz-Sloan JS, Berger MS, Brennan C, Colen RR, Colman H, Flanders AE, Giannini C, Grifford M, Iavarone A, Jain R, Joseph I, Kim J, Kasaian K, Mikkelsen T, Murray BA, O'Neill BP, Pachter L, Parsons DW, Sougnez C, Sulman EP, Vandenberg SR, Van Meir EG, von Deimling A, Zhang H, Crain D, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton T, Sherman M, Yena P, Black A, Bowen J, Dicostanzo K, Gastier-Foster J, Leraas KM, Lichtenberg TM, Pierson CR, Ramirez NC, Taylor C, Weaver S, Wise L, Zmuda E, Davidsen T, Demchok JA, Eley G, Ferguson ML, Hutter CM, Mills Shaw KR, Ozenberger BA, Sheth M, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Ayala B, Baboud J, Chudamani S, Jensen MA, Liu J, Pihl T, Raman R, Wan Y, Wu Y, Ally A, Auman JT, Balasundaram M, Balu S, Baylin SB, Beroukhim R, Bootwalla MS, Bowlby R, Bristow CA, Brooks D, Butterfield Y, Carlsen R, Carter S, Chin L, Chu A, Chuah E, Cibulskis K, Clarke A, Coetzee SG, Dhalla N, Fennell T, Fisher S, Gabriel S, Getz G, Gibbs R, Guin R, Hadjipanayis A, Hayes DN, Hinoue T, Hoadley K, Holt RA, Hoyle AP, Jefferys SR, Jones S, Jones CD, Kucherlapati R, Lai PH, Lander E, Lee S, Lichtenstein L, Ma Y, Maglinte DT, Mahadeshwar HS, Marra MA, Mayo M, Meng S, Meyerson ML, Mieczkowski PA, Moore RA, Mose LE, Mungall AJ, Pantazi A, Parfenov M, Park PJ, Parker JS, Perou CM, Protopopov A, Ren X, Roach J, Sabedot TS, Schein J, Schumacher SE, Seidman JG, Seth S, Shen H, Simons JV, Sipahimalani P, Soloway MG, Song X, Sun H, Tabak B, Tam A, Tan D, Tang J, Thiessen N, Triche T Jr, Van Den Berg DJ, Veluvolu U, Waring S, Weisenberger DJ, Wilkerson MD, Wong T, Wu J, Xi L, Xu AW, Yang L, Zack TI, Zhang J, Aksoy BA, Arachchi H, Benz C, Bernard B, Carlin D, Cho J, DiCara D, Frazer S, Fuller GN, Gao J, Gehlenborg N, Haussler D, Heiman DI, Iype L, Jacobsen A, Ju Z, Katzman S, Kim H, Knijnenburg T, Kreisberg RB, Lawrence MS, Lee W, Leinonen K, Lin P, Ling S, Liu W, Liu Y, Liu Y, Lu Y, Mills G, Ng S, Noble MS, Paull E, Rao A, Reynolds S, Saksena G, Sanborn Z, Sander C, Schultz N, Senbabaoglu Y, Shen R, Shmulevich I, Sinha R, Stuart J, Sumer SO, Sun Y, Tasman N, Taylor BS, Voet D, Weinhold N, Weinstein JN, Yang D, Yoshihara K, Zheng S, Zhang W, Zou L, Abel T, Sadeghi S, Cohen ML, Eschbacher J, Hattab EM, Raghunathan A, Schniederjan MJ, Aziz D, Barnett G, Barrett W, Bigner DD, Boice L, Brewer C, Calatozzolo C, Campos B, Carlotti CG Jr, Chan TA, Cuppini L, Curley E, Cuzzubbo S, Devine K, DiMeco F, Duell R, Elder JB, Fehrenbach A, Finocchiaro G, Friedman W, Fulop J, Gardner J, Hermes B, Herold-Mende C, Jungk C, Kendler A, Lehman NL, Lipp E, Liu O, Mandt R, McGraw M, McLendon R, McPherson C, Neder L, Nguyen P, Noss A, Nunziata R, Ostrom QT, Palmer C, Perin A, Pollo B, Potapov A, Potapova O, Rathmell WK, Rotin D, Scarpace L, Schilero C, Senecal K, Shimmel K, Shurkhay V, Sifri S, Singh R, Sloan AE, Smolenski K, Staugaitis SM, Steele R, Thorne L, Tirapelli DP, Unterberg A, Vallurupalli M, Wang Y, Warnick R, Williams F, Wolinsky Y, Bell S, Rosenberg M, Stewart C, Huang F, Grimsby JL, Radenbaugh AJ, Zhang J. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015;372:2481–98.
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C. Advancing the Cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017;4:170117.
Colen RR, Vangel M, Wang J, Gutman DA, Hwang SN, Wintermark M, Jain R, Jilwan-Nicolas M, Chen JY, Raghavan P, Holder CA, Rubin D, Huang E, Kirby J, Freymann J, Jaffe CC, Flanders A, Group TGPR, Zinn PO. Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA glioma phenotype research group project. BMC Med Genet. 2014;7:30.
Gutman DA, Cooper LA, Hwang SN, Holder CA, Gao J, Aurora TD, Dunn WD Jr, Scarpace L, Mikkelsen T, Jain R, Wintermark M, Jilwan M, Raghavan P, Huang E, Clifford RJ, Mongkolwat P, Kleper V, Freymann J, Kirby J, Zinn PO, Moreno CS, Jaffe C, Colen R, Rubin DL, Saltz J, Flanders A, Brat DJ. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267:560–9.
Gutman DA, Dunn WD Jr, Grossmann P, Cooper LA, Holder CA, Ligon KL, Alexander BM, Aerts HJ. Somatic mutations associated with MRI-derived volumetric features in glioblastoma. Neuroradiology. 2015;57:1227–37.
Liu S, Wang Y, Fan X, Ma J, Ma W, Wang R, Jiang T. Anatomical involvement of the subventricular zone predicts poor survival outcome in low-grade Astrocytomas. PLoS One. 2016;11:e0154539.
Pope WB, Chen JH, Dong J, Carlson MR, Perlina A, Cloughesy TF, Liau LM, Mischel PS, Nghiemphu P, Lai A, Nelson SF. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology. 2008;249:268–77.
Inoue T, Ogasawara K, Beppu T, Ogawa A, Kabasawa H. Diffusion tensor imaging for preoperative evaluation of tumor grade in gliomas. Clin Neurol Neurosurg. 2005;107:174–80.
Shan W, Wang XL. Clinical application value of 3.0T MR diffusion tensor imaging in grade diagnosis of gliomas. Oncol Lett. 2017;14:2009–14.
Brooks SA, Brannon AR, Parker JS, Fisher JC, Sen O, Kattan MW, Hakimi AA, Hsieh JJ, Choueiri TK, Tamboli P, Maranchie JK, Hinds P, Miller CR, Nielsen ME, Rathmell WK. ClearCode34: A prognostic risk predictor for localized clear cell renal cell carcinoma. Eur Urol. 2014;66:77–84.
Tang W, Duan J, Zhang JG, Wang YP. Subtyping glioblastoma by combining miRNA and mRNA expression data using compressed sensing-based approach. EURASIP J Bioinform Syst Biol. 2013;2013:2.
Lam A, Bui K, Hernandez Rangel E, Nguyentat M, Fernando D, Nelson K, Abi-Jaoudeh N. Radiogenomics and IR. J Vasc Interv Radiol. 2018;29:706–13.
Kickingereder P, Andronesi OC. Radiomics, metabolic, and molecular MRI for brain tumors. Semin Neurol. 2018;38:32–40.
Tamez-Pena JG, Rodriguez-Rojas JA, Gomez-Rueda H, Celaya-Padilla JM, Rivera-Prieto RA, Palacios-Corona R, Garza-Montemayor M, Cardona-Huerta S, Trevino V. Radiogenomics analysis identifies correlations of digital mammography with clinical molecular signatures in breast cancer. PLoS One. 2018;13:e0193871.
Diaz Candamio MJ, Jha S, Martel Villagran J. Overdiagnosis in imaging. Radiologia. 2018;60(5):362–7.
Acknowledgements
None.
Funding
Supported by a grant from the Medical and Health Science and Technology Project of Guangzhou (No. 20161A011022) and partially supported by the fund from Guangzhou Institute of Pediatrics/Guangzhou Women and Children’s Medical Center/(NO:IP-2016-002). The funding body did not play any role in design, in the collection, analysis, and interpretation of data; in the writing of the manuscript; and in the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
ZYL performed the statistical analysis and drafted the manuscript, JZ and ZYL participated in the design of the study and helped to draft the manuscript, JZ and ZYL collected the data, JZ and ZYL helped to perform the statistical analysis, JZ conceived of the study and participated in the design of the study. All authors read the approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All patient data was acquired from the published The Cancer Genome Atlas Low Grade Glioma (TCGA-LGG) project and within this publication it is stated “Specimens were obtained from patients, with appropriate consent from institutional review boards”. (http://cancergenome.nih.gov/).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, Z., Zhang, J. Radiogenomics correlation between MR imaging features and mRNA-based subtypes in lower-grade glioma. BMC Neurol 20, 259 (2020). https://doi.org/10.1186/s12883-020-01838-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12883-020-01838-6