Abstract
Background
As antimicrobial resistance (AMR) has become a global health crisis, new strategies against AMR infection are urgently needed. Quorum sensing (QS), responsible for bacterial communication and pathogenicity, is among the targets for anti-virulence drugs that thrive as one of the promising treatments against AMR infection.
Methods
We identified a natural compound, Harmine, through virtual screening based on three QS receptors of Pseudomonas aeruginosa (P. aeruginosa) and explored the effect of Harmine on QS-controlled and pathogenicity-related phenotypes including pyocyanin production, exocellular protease excretion, biofilm formation, and twitching motility of P. aeruginosa PA14. The protective effect of Harmine on Caenorhabditis elegans (C. elegans) and mice infection models was determined and the synergistic effect of Harmine combined with common antibiotics was explored. The underlaying mechanism of Harmine’s QS inhibitory effect was illustrated by molecular docking analysis, transcriptomic analysis, and target verification assay.
Results
In vitro results suggested that Harmine possessed QS inhibitory effects on pyocyanin production, exocellular protease excretion, biofilm formation, and twitching motility of P. aeruginosa PA14, and in vivo results displayed Harmine’s protective effect on C. elegans and mice infection models. Intriguingly, Harmine increased susceptibility of both PA14 and clinical isolates of P. aeruginosa to polymyxin B and kanamycin when used in combination. Moreover, Harmine down-regulated a series of QS controlled genes associated with pathogenicity and the underlying mechanism may have involved competitively antagonizing autoinducers’ receptors LasR, RhlR, and PqsR.
Conclusions
This study shed light on the anti-virulence potential of Harmine against QS targets, suggesting the possible use of Harmine and its derivates as anti-virulence compounds.
Similar content being viewed by others
Background
There is a desperate need for new therapies to overcome the imminent post-antibiotic era as bacterial pathogens with antimicrobial resistance (AMR) become extremely complex to deal with [1]. A list of AMR organisms published by the World Health Organization suggests that AMR is rising to be one of the greatest challenges for public health [2, 3]. The infections of AMR bacteria cause around 700,000 deaths yearly and are estimated to reach over 10 million per year in 2050 [4]. Moreover, AMR towards widely used drugs is evolving rapidly while the pipeline of new antibiotics has gone into a decline. Therefore, new therapies or alternative treatments are urgently needed, including anti-virulence and anti-biofilm treatments, phage therapies, antimicrobial peptides, and nanoparticle formulations [5, 6].
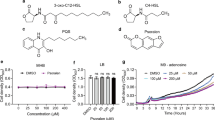
Among all the new therapies under development, the anti-virulence treatment draws the most attention. Instead of killing or halting bacterial growth like antibiotics do, the anti-virulence therapy disarms pathogens by targeting virulence that imposes none or less selection pressure on bacteria, hence, reducing the probability of AMR development [7, 8]. With a deeper understanding of pathogenetic mechanisms, Quorum-sensing (QS) was found to be responsible for a variety of physiological behaviors in bacteria by building a team that is proficient in both communication and cooperation. QS systems among pathogens such as ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa (P. aeruginosa), and Enterobacter sp.) regulate processes like virulence factors and toxin productions, motility, biofilm formation and even drug resistance [9, 10]. As a key regulatory hub for virulence, QS is one of the most intensively studied targets for anti-virulence therapy. P. aeruginosa, a ubiquitous Gram-negative bacterium, has become one of the model organisms in QS research due to the complexity of its QS networks [7, 11]. P. aeruginosa produces and releases three main small chemical signals (Fig. 1a-c) at a high population density, which are N-3-oxo-dodecanoyl-homoserine lactone (C12-HSL), N-butanoyl-homoserine lactone (C4-HSL) and 2-heptyl-3-hydroxy-4-quinolone (PQS). The accumulated signals then interact with cognate receptors LasR, RhlR, and PqsR, respectively, to induce the transcriptional expression of various target genes including those encoding production of virulence factors. The three main QS systems form a complicated hierarchy with las system sitting atop and accounting for a majority of the physiological processes and virulence phenotypes [12, 13]. The LasR-HSL complex regulates the expression of lasI, rhlR, pqsR and pqsH positively; the RhlR-HSL complex regulates rhlI positively and pqsR negatively; and the PqsR-PQS activates the transcription of the pqsABCDE and phnAB operons and rhlR. Other factors such as VfR, VqsR, QscR and GacA can positively or negatively regulate las or rhl system, which makes the QS system more intricate [14].
Recently, developments have been made in QS inhibitors (QSIs), the most extensively explored anti-virulence therapy. Chloroacetamide and maleimide analogues were examples of LasR antagonists; synthetic halogenated furanone derivatives exhibited antagonist activity against both LasR and RhlR [15]; natural compounds like cinnamaldehyde, curcumin, and andrographolide were reported to lower a variety of virulence factors by targeting QS [16]. Computer-aided drug discovery has emerged as a powerful technology for the faster identification, design, and development of cheaper but more efficient drug candidates [17]. Harmine, a naturally occurring alkaloid known for its medicinal properties, is of significant interest due to its diverse pharmacological potential, including psychoactive, anti-inflammatory, neuroprotective, antidiabetic, antitumor, and antibacterial properties [18]. Recent studies highlight Harmine's potential as a QSI, disrupting bacterial communication systems that regulate virulence and biofilm formation. For example, a Harmine-derived compound has demonstrated antimicrobial activity against carbapenem-resistant Acinetobacter baumannii clinical isolates [19]. In this study, we employed virtual screening of receptor-based (LasR, RhlR and PqsR) method to identify promising compounds from the DrugBank database. Among the selected compounds, including Prednisone acetate, Dioxybenzone, Glipizide, Haloperidol, Meticrane, Irsogladine and Harmine (Fig. 1d and Fig. S1), Harmine was found to be capable of inhibiting the QS system of the hypervirulent P. aeruginosa isolate PA14 and curbed the virulence-related phenotypes controlled by QS. Moreover, we demonstrated the protective effect of Harmine on infection models and suggested possible mechanism of Harmine’s QS inhibitory effect.
Methods
Bacterial strains, media, and chemicals
The bacterial strains we used in this study including P. aeruginosa PA14, PAO1, PAO1-ΔlasI (lasI mutant), PAO1-ΔrhlI (rhlI mutant), PAO1-ΔpqsA (pqsA mutant) of P. aeruginosa, and the uracil nutrition-deficient Escherichia coli OP50 were all kept in our laboratory [20, 21]. The collection of samples, isolation of bacterial strains, and preparation of culture media for the clinical isolates were conducted using the identical methods outlined by Zhao et al. [20], as these procedures are part of the same experimental framework. The used media included lysogeny broth (LB), Mueller–Hinton broth (MHB), M9 minimum growth medium with adenosine (0.1%, w/v) or skim milk powder (0.5%, w/v), LB with agar (0.3% and 0.5%, w/v), nematode growth medium (NGM), and peptone-glucose-sorbitol agar medium (PGS). Harmine (CSA: 442-51-3), C12-HSL (CAS: 168982-69-2), C4-HSL (CAS: 67605–85-0) and PQS (CAS: 108985-27-9) were purchased from MedChemExpress (LLC, Shanghai, China). All the molecules were dissolved in dimethyl sulfoxide (DMSO) to the concentration of 100 mM for storage. Overnight cultures from a single colony were grown in LB, and their optical density at 600 nm (OD600) was adjusted to 1 using saline for subsequent procedures.
Molecular docking
Molecular docking was performed by using Dockey, a tool built upon Autodock4 [22]. The crystal structures of LasR (PDB ID: 3IX3) and PqsR (PDB ID: 6B8A) were obtained from the Protein Data Bank (PDB). The structure of RhlR (P54292) was retrieved from AlphaFold Protein Structure Database. Each compound was docked for 10 iterations to obtain its energy-minimized best pose. The 3D visualization mapped by Pymol (version 2.5.5) of the molecular docking outcomes facilitated the analysis of interactions between ligands and receptors, along with assessment of docking energy. The interaction patterns of a ligand and receptor are determined with protein–ligand Interaction Profiler and displayed in Pymol [23].
Growth curve assay
Growth curve assay was employed based on previous description with modifications [24]. Briefly, PA14 (OD600 = 1) was cultivated (1:100) in 200 µL of LB broth or M9-adenosine medium supplemented with different concentrations of Harmine (50 µM, 100 µM and 200 µM) at 37℃ without shaking. The cell densities were monitored by measuring the absorbance at OD600 every hour throughout cultivation. All the experiments were independently repeated for 3 times.
Pyocyanin assay
Pyocyanin assay utilized absorbance measurement as previously described [25]. Briefly, PA14 (OD600 = 1) was cultivated (1:100) in 2 mL of LB medium supplemented with different concentrations of Harmine (50 µM, and 100 µM) at 37℃ with shaking (180 rpm). After 24 h, the cultures were centrifuged for 2 min (12000 rpm). The supernatants were collected and followed by a two-step sequential extraction involving 1.2 mL of chloroform and 300 µL of 0.2N HCl. The resulting HCl layer was gathered in a fresh 1.5 mL centrifuge tube and centrifuged (12000 rpm) for 5 min. Next, 200 µL of the upper HCl layer was transferred to a new 96-well plate, and the OD520 was measured. Bacterial cells were resuspended by 1 mL of saline and the cell density was measured at OD600.
Exocellular proteases assay
Exocellular proteases assay was employed by M9-skim milk (0.5%, w/v) agar plates as previous described [26]. Briefly, PA14 (OD600 = 1) was inoculated (2 µL) onto the surface of agar plates supplemented with 50 µM and 100 µM of Harmine, with 3 distinct spots on each plate. Incubation of plates was carried out at 37℃. Proteolytic zone diameters were measured after 24 h. All the experiments above were independently repeated for 3 times.
Biofilm assay
Biofilm assay was employed by a crystal violet method within 96-well polystyrene plates as previously described [27]. PA14 (OD600 = 1) was inoculated (1:100) into 200 µL of LB medium supplemented with Harmine (50 µM and 100 µM) of each well in 96-well plates. The plates were cultivated at 37℃ for 24 h without shaking. After measuring the cell densities at OD600, the culture liquids were removed, and the wells were washed with phosphate buffer solution (PBS) 2 or 3 times. The attached biofilms were stained with 0.01% (wt/v) of crystal violet (220 µL) for 20 min. Following crystal violet removal, wells underwent 3 PBS washes. Crystal violet within attached biofilms was dissolved using 95% (v/v) ethanol (200 µL). Subsequently, 100 µL of the solution was transferred to a new 96-well plate, and the OD595 was measured. All the experiments were independently repeated for 3 times.
Motility assay
Twitching motility was employed by using LB medium containing 1% of agar as previously described [28]. Plates were supplemented with Harmine (50 µM and 100 µM), and PA14 (OD600 = 1) was inoculated (2 µL) at the central bottom area of plates using a sterile pipette tip. Plates were incubated at 37℃ for 24 h, and experiments were independently repeated for 3 times.
Transcriptomic analysis
TRIzol reagents (Thermo) and the Total RNA Isolation Kit with gDNA removal (Foregene Biothchnology, Co. Ltd, China) were employed to extract total RNAs from both Harmine (50 µM)-treated and untreated PA14 samples. RNA-sequencing (RNA-seq) was conducted using the Illumina NovaSeq 6000 platform (Novogene Bioinformatics Technology, China). The obtained reads with high quality were aligned to the reference genome of PA14 using Bowtie2 (version 2.3.4.3). DESeq 2 and HTSeq (version 0.9.1) were used to quantify gene expression level and calculate the Fragments Per Kilobase of transcript sequence per Million base pairs sequenced (FPKM). P values were adjusted using the Benjamini & Hochberg method, and Padj < 0.05 and |log2(foldchange)|> 0 were set as the threshold for significantly differential expression. The ClusterProfiler R package (version 3.8.1) was used to conduct Gene Ontology (GO) enrichment analysis of differentially expressed genes and to assess the statistical enrichment of these genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Differentially expressed QS-regulated genes in P. aeruginosa were screened by mapping the genes to previously established list of QS-induced genes [29] by using Venn Diagrams. (https://bioinformatics.psb.ugent.be/webtools/Venn/).
Quantitative real-time PCR
Total RNA samples from both Harmine-treated and untreated PA14 were collected using the same method from transcriptomic analysis section. Quantitative real-time PCR was conducted using iTaqTM universal SYBR Green Supermix (Bio-Rad) and the CFX Connect Real-Time PCR Detection System. The expression levels of typical genes positively controlled by QS system of PA14, including lasB, lasR, rhlA, rhlR, pqsA, pqsD, pqsR, phzA1, hcnA, and pslA, were validated by using 16 s rRNA as the internal reference. Primers used in q-PCR assay were displayed in Table S1 and were sourced from Wen [19]. Gene expression levels were determined by utilizing the 2−∆∆ct method.
Target verification assay
P. aeruginosa mutants PAO1-ΔlasI, PAO1-rhlI and PAO1-ΔpqsA, which were autoinducers negative isolates, were utilized to verify the targets of Harmine [21]. Mutant strains were treated with (a) exogenous QS signal molecules (50 µM), (b) Harmine (50 µM), and (c) exogenous QS signal molecules (50 µM) and Harmine (50 µM). Each experiment contained a DMSO group as control, and all experiments were independently repeated for 3 times. The expression levels of downstream genes lasB, rhlA, and phzA1, which were regulated by the three QS regulators were detected by qPCR. Primers used in this assay were also displayed in Table S1 and were sourced from Wen [21].
Caenorhabditis elegans killing assay
The Fast-killing assay was conducted on PGS agar medium as previously described [30]. Thin lawn plates were made by spreading 20 µL of PA14 (OD600 = 1) with 50 µM Harmine. The plates were incubated first at 37℃ for 24 h and then at 25℃ for another 24 h. Each experiment consisted of a DMSO negative control and an uninfected control using E. coli OP50. All experiments were conducted in triplicate. In the fast-killing assay, 10 age-matched L4 stage nematodes were added to each prepared assay plate. The plates were subsequently incubated at 25℃.
Mouse infection model
The mice infection model was employed as previously described [31, 32]. C57BL/6 female mice were anesthetized by intraperitoneal injection of ketamine (50 µg/ml) in sterile saline. A total of 3.0 × 107 CFU bacterial cells in 30 µL of sterile saline supplemented with or without Harmine (50 µM) were intranasally instilled into the lungs of mice. The Harmine-treated group was then intranasally instilled with 50 µM (30 µL) of Harmine every 12 h, and the untreated group was intranasally instilled with an equal amount of DMSO.
Antibiotic synergy test
Synergy effects of Harmine combined with levofloxacin, polymyxin B, meropenem, and kanamycin on P. aeruginosa PA14 and clinical isolates were evaluated through the checkerboard assay [33, 34]. PA14 and clinical isolates were inoculated in MH broth to reach a final CFU of 105 mL−1. Harmine (0 µM, 6.25 µM, 12.5 µM, 25 µM, 50 µM and 100 µM) was then combined with antibiotics across a range of concentrations. Each combination was performed in triplicate. Incubation of the 96-well plate was carried out at 37℃ for 16 h, followed by measurement of OD600. The identical procedure was applied for synergy tests involving levofloxacin-intermediate clinical isolate 3-100-1, polymyxin B-susceptible clinical isolate 7-61-28, meropenem-resistant clinical isolate PA2 and kanamycin-resistant clinical isolate W8a. The degree of synergy (S) was calculated using the Bliss independence model [35], employing the formula S = fX,0 · f0,Y = fX,Y. A value of S = 0 signifies independent drug actions. Conversely, S < 0 implies an antagonistic interaction, while S > 0 suggests synergy.
Statistical analysis
The creation of graphs and the analysis of significance, including Mann–Whitney test, one-way analysis of variance (ANOVA), two-sample t-test, and Log-rank test, were performed using GraphPad Prism (version 9.5.1). Significance was denoted by a P-value below 0.05, signifying a difference between the treated and control groups.
Results
Molecular docking of Harmine and QS receptor proteins
Molecular docking was employed to predict the interactions of molecules with QS receptors LasR, RhlR and PqsR of P. aeruginosa. Results suggested that Harmine was capable of binding to the ligand binding domain of LasR, RhlR, and PqsR (Fig. 1e). Compared to native ligands on hydrophobic interactions with receptors, Harmine exhibited a similar pattern with C12-HSL and PQS while having fewer interactions with amino acid residues. Notably, Harmine engaged in hydrogen bond interactions with amino acid residues Trp60, Arg61 of LasR, Trp68 of RhlR, and Leu197, Ile236 of PqsR and in π-stacking interactions with Tyr56, Tyr64 of LasR, Tyr64 of RhlR and Phe221 of PqsR due to the conjugated system of Harmine’s structure. The results of best pose energy suggested that Harmine was more likely to bind with LasR and RhlR when competing with C12-HSL and C4-HSL, respectively, while it was less likely to bind with PqsR when competing with PQS.
Harmine inhibits the growth of PA14 in QS-required medium
In this study, growth curves of P. aeruginosa PA14 in LB and M9-adenosine with different concentrations of Harmine were determined. In LB broth, the nutritional sufficiency medium, different concentrations of Harmine did not significantly inhibit the growth of PA14 compared to the DMSO groups (Fig. 2a). However, there was a tendency of growth inhibition when in 200 µM. In M9-adenosine medium, the growth of PA14 depends on the degradation of adenosine by the QS-controlled enzyme Nuh [36], and Harmine’s disruption of QS and enzymatic degradation of adenosine in all concentrations is shown through a dose-dependent inhibitory effect on the growth of PA14 (Fig. 2b). Results suggested that Harmine exhibited an inhibitory effect on QS system of PA14 while not curbing its growth in LB broth. Hence, the concentration values of 50 µM and 100 µM were set to the working concentration in subsequent experiments.
Harmine inhibited the virulence-related phenotypes of P. aeruginosa PA14. The growth status of PA14 in LB (a) and M9-adenoisne (b), the cell density was measured in OD600 and was blank corrected. The pyocyanin assay (c), exocellular protease assay (d), biofilm assay (e), and motility assay (f) of PA14 treated with 50 µM and 100 µM of Harmine. Each experiment included a DMSO negative control. Data shown were means ± SD of three independent replicates. Two-tailed Mann–Whitney test and One-way ANOVA test were used to determine P values. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001
Harmine inhibits the QS-related phenotypes of PA14
Orchestrated by QS systems during infection, virulence factors, such as pyocyanin and proteases, and phenotypes, such as biofilm formation and cell motility, significantly contribute to P. aeruginosa’s pathogenicity. Compared to the control group, 50 µM and 100 µM of Harmine treatment decreased pyocyanin production to about 60% and 31%, respectively (Fig. 2c). For extracellular proteases assay, both Harmine-treated groups exhibited significantly lower extracellular proteases production (Fig. 2d) and significantly curbed biofilm formation (Fig. 2e) when compared to the control group. However, in the twitching motility assay, only the 100 µM Harmine treated group showed significant inhibition of the twitching motility of PA14 (Fig. 2f). Results suggested that Harmine, especially in high concentrations, could significantly inhibit the phenotypes associated with the pathogenicity of PA14. Among all the selected compounds, Haloperidol and Meticrane also exhibited inhibitory effects on all tested phenotypes (Fig. S2), but compared with Harmine, Haloperidol did not inhibit pyocyanin production and biofilm formation in a dose-dependent manner and was less inhibitory of exocellular protease production in 50 µM concentrations (Fig. S2) while Meticrane exhibited a lower inhibitory effect on biofilm formation and twitching motility (Fig. S2).
Harmine inhibits the expression of QS-controlled genes
RNA-seq was then performed to assess the influence of Harmine (50 µM) on the global transcription of P. aeruginosa PA14. Compared to the untreated group, a total of 437 down-regulated genes and 575 up-regulated genes were identified in the Harmine treated group (Fig. 3a). KEGG functional annotation demonstrated significant enrichment of Quorum-sensing, biofilm formation, phenazine biosynthesis, and starch/sucrose metabolism pathways among the down-regulated genes in the treated group, and significant enrichment of ribosome, oxidative phosphorylation, and peptidoglycan biosynthesis pathways among the up-regulated genes (Fig. 3b). Additionally, the treatment of Harmine caused enrichment mainly in biological process of GO terms among the down-regulated genes such as pathogenesis and antibiotic metabolic (Fig. 3c and Fig. S2a), though not significant, and the treatment of Harmine caused significant enrichment of 94 GO terms among the up-regulated genes (Fig. S2b). These findings suggested that Harmine significantly influenced the biological metabolic processes of P. aeruginosa PA14 at the transcriptional level. We then accessed(assessed?) the down-regulation of typical virulence genes induced by QS through qPCR analysis. As shown in the results, the relative expression of lasB, rhlA, rhlR, pqsA, pqsD, pqsR, hcnA and pslA genes in P. aeruginosa PA14 all significantly decreased upon treatment of Harmine (Fig. 3d).
Harmine curbed QS system of P. aeruginosa PA14 in transcriptional level. The volcano plot (a) of differentially expressed genes of PA14 influenced by 50 µM of Harmine compared to DMSO group. The significantly changed KEGG pathways influenced by Harmine (b). The Venn diagram and the list (c) of significantly changed genes in QS-induced genes in PA14 treated with 50 µM of Harmine. Expression of typical QS-induced genes determined by quantitative PCR (d) in PA14 treated with 50 µM of Harmine. The target verification assay of Harmine with LasR (e), RhlR (f) and PqsR (g). Data shown were means ± SD of three independent replicates. Unpaired two-tailed t test was used to determine P values. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001
Harmine targets three QS regulators of P. aeruginosa
The autoinducer defect isolates PAO1-ΔlasI, PAO1-rhlI, and PAO1-ΔpqsA were utilized to assess the interactions of Harmine with LasR, RhlR, and PqsR. With the addition of exogenous autoinducers C12-HSL, C4-HSL, and PQS, the expression of downstream genes lasB, rhlA, and phzA1, which were directly controlled by three receptors, increased to about fourfold (Fig. 3e-g). Compared to the exogenous autoinducer groups, the addition of both autoinducers and Harmine significantly curbed the expression of downstream genes (Fig. 3e-g). It suggested that when competing with autoinducers, Harmine was more likely to bind with receptors and hence curb the expression of downstream genes, which aligned with the results of molecular docking. Moreover, compared to the untreated group, the addition of Harmine alone did not affect the expression of downstream genes controlled by LasR and RhlR (Fig. 3e,f) while increasing the expression of phzA1 controlled by PqsR (Fig. 3g). Results displayed that Harmine could target three QS regulators of P. aeruginosa and hence inhibit the downstream genes they controlled.
Harmine protects C. elegans and mice from PA14 infection
The C. elegans-P. aeruginosa infection model was employed to investigate in vivo activity of Harmine. In comparison to the infected groups, which were inoculated with E. coli OP50, Harmine exhibited no toxicity to nematodes (Fig. 4a). The survival proportion of Harmine treated group was 60% at hour 120 when all nematodes of the control group had died (Fig. 4a). Moreover, in mice-P. aeruginosa infection model (Fig. 4b), all mice in the control group died within 48 h while the survival rate in Harmine treated group was 33.3% higher at hour 48. Results suggested that in the fast-killing assay of C. elegans and mice model, which mimicked the conditions of an acute infection, the presence of Harmine decreased the mortality of both nematodes and mice, especially in C. elegans-P. aeruginosa infection model.
Synergistic effect of Harmine and antibiotics against P. aeruginosa
The levofloxacin-intermediate clinical isolate 3-100-1, polymyxin B-susceptible clinical isolate 7-61-28, meropenem-resistant clinical isolate PA2, kanamycin-resistant clinical isolate W8a and PA14 were utilized to assess the potential synergistic effects between Harmine and the commonly used antibiotics in clinical (Fig. 5a-p). For clinical isolates, Harmine increased the susceptibility of isolates against antibiotics, especially of 7-61-28 (Fig. 5e,f). The same results were found in PA14 against polymyxin B and kanamycin (Fig. 5h,p). However, all concentrations of Harmine decreased the susceptibility of PA14 against levofloxacin and meropenem (Fig. 5d, l). Compared to PA14, the isolates 3-100-1 and PA2 had higher MIC values of levofloxacin and meropenem, which suggested that Harmine can strengthen the inhibitory effect of antibiotics against hyper-virulence isolates.
The synergic interactions of Harmine with antibiotics against P. aeruginosa. The heat map of Harmine combined with levofloxacin against 3-100-1 (a) and PA14 (c) and the killing curve of 3-100-1 (b) and PA14 (d). The heat map of Harmine combined with polymyxin B against 7-61-28 (e) and PA14 (g) and the killing curve of 7-61-28 (f) and PA14 (h). The heat map of Harmine combined with meropenem against PA2 (i) and PA14 (k) and the killing curve of PA2 (j) and PA14 (l). The heat map of Harmine combined with kanamycin against W8a (m) and PA14 (o) and the killing curve of W8a (n) and PA14 (p). The orange-grey bar displayed the degree of synergy calculated by the Bliss independence model
Harmine curbs the pathogenicity of clinical isolates
We then assessed the inhibitory effect of Harmine on typical QS-related phenotypes of clinical isolates, and it was found to significantly inhibit the pyocyanin production and biofilm formation of all isolates (Fig. 6a,c). However, hyper-virulence clinical isolates limited Harmine’s capacity for pathogenicity inhibition, and Harmine did not curb the exocellular protease production or twitching motility of all isolates (Fig. 6b,d), displaying a stronger inhibitory effect on pyocyanin production and biofilm formation than on exocellular protease production or twitching motility across controlled PA14 experiments.
Harmine inhibited the virulence-related phenotypes of P. aeruginosa clinical isolates. The pyocyanin assay (a), exocellular protease assay (b), biofilm assay (c), and motility assay (d) of clinical isolates treated with 50 µM and 100 µM of Harmine. Each experiment included a DMSO negative control. Data shown were means ± SD of three independent replicates. One-way ANOVA test was used to determine P values. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001
Discussion
Pseudomonas aeruginosa is a Gram-negative, opportunistic pathogen that can be isolated from several nosocomial and life-threatening infections in patients with cystic fibrosis (CF), chronic obstructive pulmonary disease (COPD), urinary tract infections (UTIs), burn wounds, etc. [37] P. aeruginosa has strong abilities to develop natural and acquired drug resistance through various mechanisms. The novel development of antibiotics is rapidly decelerating as new AMR strains emerge. Moreover, due to the increasing difficulty of treating AMR strain infections, the development of anti- P. aeruginosa therapy depends on targeting resistance mechanism [38]. Recent research has displayed that Quorum Sensing, as a central regulatory mechanism, controls biofilm formation, secretion systems, efflux pumps, motility, and antibiotic resistance in a population density-dependent manner. Developments have already been made in QS inhibitors [39, 40], making it a perfect target for anti-virulence therapy [41].
The objective of this study was to find a potent QSI of P. aeruginosa with the high-throughput virtual screening approach, which can address the limitations of traditional methods [42]. Seven compounds were selected from DrugBank Database—Prednisone acetate, Dioxybenzone, Glipizide, Haloperidol, Meticrane, Irsogladine, and Harmine. All the compounds were estimated to bind with LasR receptor, but the results of phenotype experiments displayed that Harmine was the most effective QSI among all compounds. Hence, Harmine was taken for further analysis. Harmine (Fig. 1d) is a β-carboline alkaloid widely existing in natural plants and extractable from Banisteria caapi and Peganum harmala L [43]. Although Harmine shows a wide range of pharmacological activities, including anti-inflammatory, antitumor, and anti-depressant uses [44], no anti-virulence activity has been reported before.
The in vitro results show that Harmine possesses multifaceted QS inhibiting activities and no bactericidal effects in concentrations of 50 µM and 100 µM (Fig. 2a), which are consistent with the characteristics of an anti-virulence drug [45]. We then performed a series of experiments to identify the mechanism of Harmine’s targeting of QS. Interestingly, Harmine up-regulated lasR in q-PCR assay (Fig. 3d) while down-regulating lasB in LasR target verification experiment (Fig. 3e). We assumed that the overexpression of lasR might activate the negative feedback loop such as RsaL-dependent regulation. This kind of system creates balance to prevent dominant control by regulators of opposing functions [46]. Moreover, in PqsR target verification experiment, the Harmine-treated group had a higher level of phzA1 expression than untreated group (Fig. 3g), which suggested that Harmine up-regulated the expression of phzA1 through an unknown mechanism. Notably, the transcriptomic analysis results displayed that gacA, which encodes the global regulator GacA, expressed differently in Harmine treated group. GacA, the activator of LasR and RhlR [47], was enriched in an up-regulated KEGG pathway, though not significantly (Padj = 0.097). This might be one of the reasons why the expression of lasR was not curbed by Harmine in qPCR assay. Both the molecular docking and target verification assays suggested that Harmine had the potential of binding with LasR, RhlR and PqsR. The phenotypic experiments were also promising. Hence, the possibility of Harmine being a multi-targeted QSI cannot be rule out and the mechanism of it needs further exploration due to the complicity of quorum sensing network.
The non-bactericidal characteristic of QSIs makes this infection treatment a double-edged sword, as less AMR development comes with the risk of failing to clear infections, which is especially dangerous for immune-compromised patients [35]. Hence, we employed the synergistic effect assays with both Harmine and antibiotics in P. aeruginosa. For polymyxin B and kanamycin, Harmine increased the susceptibility of both PA14 and clinical isolates (Fig. 5e-h and Fig. 5m-p), especially when combined with polymyxin B against PA14. Harmine was found to significantly down-regulate genes such as PA1238 and oprG associated with multidrug efflux pumps or outer membrane proteins, which are related to the common mechanism of polymyxin B resistance in Gram-negative bacteria [48]. This might explain the robust synergistic effect of Harmine-polymyxin B combination. Notably, when Harmine was combined with levofloxacin and meropenem, it increased the susceptibility of clinical isolates while decreasing the susceptibility of PA14 (Fig. 5a-d and Fig. 5i-l). We cannot rule out the possibility that the key genes responsible for antibiotic resistance of clinical isolates 3–100-1 and PA2 may have mutations, which is worth investigating.
As a dominant global pathogen, the development of antibiotic resistance in P. aeruginosa has emerged as a profound public health challenge, compounded by the absence of viable alternative therapies [49, 50]. Quorum sensing interference has emerged as a promising anti-virulence strategy for managing antibiotic resistance [51]. Extensive research has focused on discovering quorum sensing inhibitors (QSIs) to advance anti-infective drug development, including evaluating the QS inhibitory activities of compounds derived from traditional medicinal plants. These plant-derived compounds are primarily secondary metabolites. Major groups responsible for antimicrobial activity include phenolics, phenolic acids, quinones, saponins, flavonoids, tannins, coumarins, terpenoids, and alkaloids [52]. As a naturally occurring alkaloid known for its medicinal properties, the antibacterial properties of Harmine has been reported in recent years [18]. Harmine-derived compound has demonstrated antimicrobial activity against carbapenem-resistant Acinetobacter baumannii clinical isolates [19]. In this study, we identified Harmine through virtual screening based on three QS receptors of P. aeruginosa and explored its effects on QS-controlled and pathogenicity-related phenotypes. We also assessed Harmine's protective effect in C. elegans and mice infection models. The results suggested that Harmine acts as an anti-virulence compound against QS targets, decreasing the virulence and antibiotic resistance of P. aeruginosa.
The present study has some limitations, mainly due to the complexity of QS system of P. aeruginosa. Harmine was proven to interact with QS receptors LasR, RhlR, and PqsR, but we cannot rule out the possibility that Harmine may curb the QS system through other pathways. Since mutations are frequently observed in isolates from clinical patients, it was also difficult to determine the mechanism of the synergistic effect of Harmine with antibiotics.
Conclusion
In conclusion, this study reveals QS inhibitory activity of Harmine on P. aeruginosa. Harmine inhibits the production of pyocyanin and exocellular protease, biofilm formation and twitching motility, which are all directly or indirectly controlled by QS. Moreover, Harmine showed significant protective activity in C. elegans infection model and reduced the mortality rate of the mice infection model. For synergy assay, Harmine enhanced the susceptibility of P. aeruginosa clinical isolates and PA14 toward polymyxin B and kanamycin treatments. Results highlight the QS activity of Harmine, its potential for development as an anti-virulence drug, and the value of more molecules obtained by structural modification of Harmine.
Availability of data and materials
The datasets generated and analyzed during the current study are available in the NCBI BioProject repository and the registration number PRJNA1059290.
Abbreviations
- AMR:
-
Antimicrobial resistance
- C. elegans :
-
Caenorhabditis elegans
- DMSO:
-
Dissolved in dimethyl sulfoxide
- ESKAPE:
-
Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter sp
- MHB:
-
Mueller–Hinton broth
- LB:
-
Lysogeny broth
- QS:
-
Quorum sensing
- P. aeruginosa :
-
Pseudomonas aeruginosa
References
Moriel DG, Piccioli D, Raso MM, Pizza M. The overlooked bacterial pandemic. Semin Immunopathol. 2024;45:481–91.
Venter H. Reversing resistance to counter antimicrobial resistance in the World Health Organisation’s critical priority of most dangerous pathogens. Biosci Rep. 2019;29:1–12.
Rolff J, Bonhoeffer S, Kloft C, Leistner R, Regoes R, Hochberg ME. Forecasting antimicrobial resistance evolution. Trends Microbiol. 2024;18,S0966-842X(23)00361-X. https://doi.org/10.1016/j.tim.2023.12.009.
Huemer M, Mairpady Shambat S, Brugger SD, Zinkernagel AS. Antibiotic resistance and persistence—implications for human health and treatment perspectives. EMBO Rep. 2020;21:1–24.
Calvert MB, Jumde VR, Titz A. Pathoblockers or antivirulence drugs as a new option for the treatment of bacterial infections. Beilstein J Org Chem. 2018;14:2607–17.
Scoffone VC, Barbieri G, Irudal S, Trespidi G, Buroni S. New antimicrobial strategies to treat multi-drug resistant infections caused by gram-negatives in cystic fibrosis. Antibiotics. 2024;13:1–33.
Defoirdt T. Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 2018;26:313–28.
Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12:300–8.
Zhao X, Yu Z, Ding T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms. 2020;8:1–21.
Venkateswaran P, Vasudevan S, David H, Shaktivel A, Shanmugam K, Neelakantan P, et al. Revisiting ESKAPE pathogens: virulence, resistance, and combating strategies focusing on quorum sensing. Front Cell Infect Microbiol. 2023;13:1–30.
Cruz RL, Asfahl KL, Van den Bossche S, Coenye T, Crabbé A, Dandekar AA. RhlR-regulated acyl-homoserine lactone quorum sensing in a cystic fibrosis isolate of Pseudomonas aeruginosa. mBio. 2020;11:1–14.
Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6:26–41.
Sánchez-Jiménez A, Llamas MA, Marcos-Torres FJ. Transcriptional regulators controlling virulence in Pseudomonas aeruginosa. Int J Mol Sci. 2023;24:11895.
Jakobsen TH, Bjarnsholt T, Jensen PØ, Givskov M, Høiby N. Targeting quorum sensing in Pseudomonas aeruginosa biofilms: current and emerging inhibitors. Future Microbiol. 2013;8:901–21.
Hibbert TM, Whiteley M, Renshaw SA, Neill DR, Fothergill JL. Emerging strategies to target virulence in Pseudomonas aeruginosa respiratory infections. Crit Rev Microbiol. 2023;24:1–16.
Sanya DRA, Onésime D, Vizzarro G, Jacquier N. Recent advances in therapeutic targets identification and development of treatment strategies towards Pseudomonas aeruginosa infections. BMC Microbiol. 2023;23:1–18.
Oselusi SO, Dube P, Odugbemi AI, Akinyede KA, Ilori TL, Egieyeh E, et al. The role and potential of computer-aided drug discovery strategies in the discovery of novel antimicrobials. Comput Biol Med. 2024;169:107927.
Qin S, Xiao W, Zhou C, Pu Q, Deng X, Lan L, Liang H, Song X, Wu M. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther. 2022;7:199.
Breine A, Van Gysel M, Elsocht M, et al. Antimicrobial activity of a repurposed harmine-derived compound on Carbapenem-Resistant Acinetobacter baumannii clinical isolates. Front Cell Infect Microbiol. 2021;11:789672.
Zhao K, Yang X, Zeng Q, Zhang Y, Li H, Huang T, et al. Evolution of lasR mutants in polymorphic Pseudomonas aeruginosa populations facilitates chronic infection of the lung. Nat Commun. 2023;14:5976.
Wen F, Wu Y, Yuan Y, Yang X, Ran Q, Gan X, et al. Discovery of psoralen as a quorum sensing inhibitor suppresses Pseudomonas aeruginosa virulence. Appl Microbiol Biotechnol. 2024;108:1–15.
Du L, Geng C, Zeng Q, Huang T, Tang J, Chu Y, et al. Dockey: a modern integrated tool for large-scale molecular docking and virtual screening. Brief Bioinform. 2023;24:bbad047.
Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:W443-7.
Toussaint JP, Farrell-Sherman A, Feldman TP, Smalley NE, Schaefer AL, Greenberg EP, Dandekar AA. Gene duplication in Pseudomonas aeruginosa improves growth on adenosine. J Bacteriol. 2017;199(21):e00261–17.
Vilaplana L, Marco MP. Phenazines as potential biomarkers of Pseudomonas aeruginosa infections: synthesis regulation, pathogenesis and analytical methods for their detection. Anal Bioanal Chem. 2020;412:5897–912.
Thabit AK, Eljaaly K, Zawawi A, Ibrahim TS, Eissa AG, Elbaramawi SS, et al. Muting bacterial communication: evaluation of prazosin anti-quorum sensing activities against gram-negative bacteria Pseudomonas aeruginosa, Proteus mirabilis, and Serratia marcescens. Biology (Basel). 2022;11:1349.
Wang H, Chu W, Ye C, Gaeta B, Tao H, Wang M, et al. Chlorogenic acid attenuates virulence factors and pathogenicity of Pseudomonas aeruginosa by regulating quorum sensing. Appl Microbiol Biotechnol. 2019;103:903–15.
Turnbull L, Whitchurch CB. Motility assay: twitching motility. Methods Mol Biol. 2014;1149:73–86.
Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–79.
Kirienko NV, Cezairliyan BO, Ausubel FM, Powell JR. Pseudomonas aeruginosa PA14 pathogenesis in Caenorhabditis elegans. Methods Mol Biol. 2014;1149:653–69.
Yuan Y, Yang X, Zeng Q, Li H, Fu R, Du L, et al. Repurposing Dimetridazole and Ribavirin to disarm Pseudomonas aeruginosa virulence by targeting the quorum sensing system. Front Microbiol. 2022;13:1–15.
Utari PD, Setroikromo R, Melgert BN, Quax WJ. PvdQ quorum quenching acylase attenuates Pseudomonas aeruginosa virulence in a mouse model of pulmonary infection. Front Cell Infect Microbiol. 2018;8:119.
Bellio P, Fagnani L, Nazzicone L, Celenza G. New and simplified method for drug combination studies by checkerboard assay. MethodsX. 2021;8:101543.
Huang T, Zeng M, Fu H, Zhao K, Song T, Guo Y, et al. A novel antibiotic combination of linezolid and polymyxin B octapeptide PBOP against clinical Pseudomonas aeruginosa strains. Ann Clin Microbiol Antimicrob. 2022;21:1–12.
Rezzoagli C, Archetti M, Mignot I, Baumgartner M, Kümmerli R. Combining antibiotics with antivirulence compounds can have synergistic effects and reverse selection for antibiotic resistance in Pseudomonas aeruginosa. PLoS Biol. 2020;18:1–27.
Heurlier K, Haenni M, Guy L, Krishnapillai V, Haas D. Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J Bacteriol. 2005;187:4875–83.
Azam MW, Khan AU. Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov Today. 2019;24:350–9.
Qin S, Xiao W, Zhou C, Pu Q, Deng X, Lan L, et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther. 2022;7:1–27.
Ogawara H. Possible drugs for the treatment of bacterial infections in the future: anti-virulence drugs. J Antibiot. 2021;74:24–41.
Rather MA, Saha D, Bhuyan S, Jha AN, Mandal M. Quorum quenching: a drug discovery approach against Pseudomonas aeruginosa. Microbiol Res. 2022;264:127173.
Botelho J, Grosso F, Peixe L. Antibiotic resistance in Pseudomonas aeruginosa – mechanisms, epidemiology and evolution. Drug Resist Updates. 2019;44:100640.
Sadiq S, Rana NF, Zahid MA, Zargaham MK, Tanweer T, Batool A, et al. Virtual Screening of FDA-approved drugs against LasR of Pseudomonas aeruginosa for antibiofilm potential. Molecules. 2020;25:3723.
Zhang L, Li D, Yu S. Pharmacological effects of harmine and its derivatives: a review. Arch Pharm Res. 2020;43:1259–75.
Huang J, Liu Y, Chen JX, Lu XY, Zhu WJ, Qin L, et al. Harmine is an effective therapeutic small molecule for the treatment of cardiac hypertrophy. Acta Pharmacol Sin. 2022;43:50–63.
Kalia VC, Patel SKS, Kang YC, Lee JK. Quorum sensing inhibitors as antipathogens: biotechnological applications. Biotechnol Adv. 2019;37:68–90.
Schinner S, Preusse M, Kesthely C, Häussler S. Analysis of the organization and expression patterns of the convergent Pseudomonas aeruginosa lasR/rsaL gene pair uncovers mutual influence. Mol Microbiol. 2021;115:643–57.
Moghaddam MM, Khodi S, Mirhosseini A. Quorum sensing in bacteria and a glance on Pseudomonas aeruginosa. Clin Microbial. 2014;3:156.
Kim BO, Jang HJ, Chung IY, Bae HW, Kim ES, Cho YH. Nitrate respiration promotes polymyxin B resistance in Pseudomonas aeruginosa. Antioxid Redox Signal. 2021;34:442–51.
Behzadi P, Ambrosi C, Scribano D, Zanetti S, Sarshar M, Gajdács M, Donadu MG. Editorial: current perspectives on Pseudomonas aeruginosa: epidemiology, virulence and contemporary strategies to combat multidrug-resistant (MDR) pathogens. Front Microbiol. 2022;13:975616.
Behzadi P, Gajdács M, Pallós P, Ónodi B, Stájer A, Matusovits D, Kárpáti K, Burián K, Battah B, Ferrari M, Doria C, Caggiari G, Khusro A, Zanetti S, Donadu MG. Relationship between biofilm-formation, phenotypic virulence factors and antibiotic resistance in environmental Pseudomonas aeruginosa. Pathogens. 2022;11:1015.
Asfour HZ. Anti-quorum sensing natural compounds. J Microsc Ultrastruct. 2018;6:1–10.
Guzzo F, Scognamiglio M, Fiorentino A, Buommino E, D’Abrosca B. Plant derived natural products against Pseudomonas aeruginosa and Staphylococcus aureus: antibiofilm activity and molecular mechanisms. Molecules. 2020;25:5024.
Acknowledgements
Not applicable.
Funding
This research was funded by the National Natural Science Foundation of China (32200525), and in part by the Antibiotics Research and Re-evaluation Key Laboratory of Sichuan Province (ARRLKF21-06).
Author information
Authors and Affiliations
Contributions
QZ and LD designed the research. PC, JQ and QZ performed the experiments. JQ and LD performed the bioinformatic analyses. LD and HKS performed the docking prediction. HKS performed English editing. PC, JQ and QL analyzed data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Clinical isolates were obtained from the patients hospitalized in the Affiliated Hospital of Chengdu University (Chengdu, China), which approved by the Ethics Committee of the Affiliated Hospital of Chengdu University (PJ2020-021-03). Animal experiments were carried out the performed at the State Key Laboratory of Biotherapy, Sichuan University, which approved by the Ethics Committee of the State Key Laboratory of Biotherapy (2021559A).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
12879_2024_9639_MOESM1_ESM.tiff
Supplementary Material 1: Figure S1. The structures of screening compounds and phenotypic assays. The structure of Prednisone acetate, Dioxybenzone, Glipizide, Haloperidol, Meticrane, and Irsogladine. The pyocyanin assay, exocellular protease assay, biofilm assay, and motility assayof PA14 treated with 50 µM and 100 µM of compounds. Data shown were means ± SD of three independent replicates. One-way ANOVA test was used to determine P values. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
12879_2024_9639_MOESM2_ESM.tiff
Supplementary Material 2: Figure S2. The secondary classification diagramof down-regulated GO terms influenced by 50 µM of Harmine compared to DMSO group. The bubble diagramof significantly up-regulated GO terms influenced by 50 µM of Harmine, compared to DMSO group.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, P., Qin, J., Su, H.K. et al. Harmine acts as a quorum sensing inhibitor decreasing the virulence and antibiotic resistance of Pseudomonas aeruginosa. BMC Infect Dis 24, 760 (2024). https://doi.org/10.1186/s12879-024-09639-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-024-09639-9