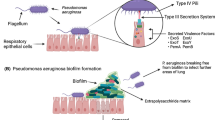
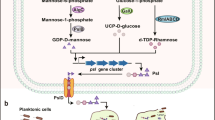
Abstract
Infectious diseases are still a worldwide important problem. This fact has led to the characterization of new biomarkers that would allow an early, fast and reliable diagnostic and targeted therapy. In this context, Pseudomonas aeruginosa can be considered one of the most threatening pathogens since it causes a wide range of infections, mainly in patients that suffer other diseases. Antibiotic treatment is not trivial given the incidence of resistance processes and the fewer new antibiotics that are placed on the market. With this scenario, relevant quorum sensing (QS) molecules that regulate the secretion of virulence factors and biofilm formation can play an important role in diagnostic and therapeutic issues. In this review, we have focused our attention on phenazines, as possible new biomarkers. They are pigmented metabolites that are produced by diverse bacteria, characterized for presenting unique redox properties. Phenazines are involved in virulence, competitive fitness and are an essential component of the bacterial QS system. Here we describe their role in bacterial pathogenesis and we revise phenazine production regulation systems. We also discuss phenazine levels previously reported in bacterial isolates and in clinical samples to evaluate them as putative good candidates to be used as P. aeruginosa infection biomarkers. Moreover we deeply go through all analytical techniques that have been used for their detection and also new approaches are discussed from a critical point.

Graphical abstract




Similar content being viewed by others
References
Jefferies JMC, Cooper T, Yam T, Clarke SC. Pseudomonas aeruginosa outbreaks in the neonatal intensive care unit – a systematic review of risk factors and environmental sources. J Med Microbiol. 2012;61(Pt 8):1052–61. https://doi.org/10.1099/jmm.0.044818-0.
Williams BJ, Dehnbostel J, Blackwell TS. Pseudomonas aeruginosa: host defence in lung diseases. Respirology (Carlton, Vic). 2010;15(7):1037–56. https://doi.org/10.1111/j.1440-1843.2010.01819.x.
Olejnickova K, Hola V, Ruzicka F. Catheter-related infections caused by Pseudomonas aeruginosa: virulence factors involved and their relationships. Pathog Dis. 2014;72(2):87–94. https://doi.org/10.1111/2049-632X.12188.
Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, et al. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis. 2001;183(3):444–52. https://doi.org/10.1086/318075.
Høiby N, Johansen HK, Moser C, Ciofu O. Clinical relevance of Pseudomonas aeruginosa: a master of adaptation and survival strategies. In Pseudomonas model organism, pathogen, cell factory. Weinheim: Wiley-VCH GmbH & Co.; 2008.
Mena A, Smith EE, Burns JL, Speert DP, Moskowitz SM, Perez JL, et al. Genetic adaptation of Pseudomonas aeruginosa to the Airways of Cystic Fibrosis Patients is Catalyzed by Hypermutation. J Bacteriol. 2008;190(24):7910–7. https://doi.org/10.1128/jb.01147-08.
Boles BR, Thoendel M, Singh PK. Genetic variation in biofilms and the insurance effects of diversity. Microbiology. 2005;151(9):2816–8. https://doi.org/10.1099/mic.0.28224-0.
Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A. 2008;105(34):12503–8. https://doi.org/10.1073/pnas.0801499105.
Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Hoiby N. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology. 2010;156(Pt 4):1108–19. https://doi.org/10.1099/mic.0.033993-0.
Rada B, Leto TL. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 2013;21(2):73–81. https://doi.org/10.1016/j.tim.2012.10.004.
Lau GW, Hassett DJ, Britigan BE. Modulation of lung epithelial functions by Pseudomonas aeruginosa. Trends Microbiol. 2005;13(8):389–97. https://doi.org/10.1016/j.tim.2005.05.011.
Faure E, Kwong K, Nguyen D. Pseudomonas aeruginosa in chronic lung infections: how to adapt within the host? Front Immunol. 2018;9:2416. https://doi.org/10.3389/fimmu.2018.02416.
Mudau M, Jacobson R, Minenza N, Kuonza L, Morris V, Engelbrecht H, et al. Outbreak of multi-drug resistant Pseudomonas aeruginosa bloodstream infection in the haematology unit of a south African academic hospital. PLoS One. 2013;8(3):e55985. https://doi.org/10.1371/journal.pone.0055985.
Muraki Y, Kitamura M, Maeda Y, Kitahara T, Mori T, Ikeue H, et al. Nationwide surveillance of antimicrobial consumption and resistance to Pseudomonas aeruginosa isolates at 203 Japanese hospitals in 2010. Infection. 2013;41(2):415–23. https://doi.org/10.1007/s15010-013-0440-0.
Rosenthal VD, Bijie H, Maki DG, Mehta Y, Apisarnthanarak A, Medeiros EA, et al. International nosocomial infection control consortium (INICC) report, data summary of 36 countries, for 2004-2009. Am J Infect Control. 2012;40(5):396–407.
Xiao M, Wang Y, Yang QW, Fan X, Brown M, Kong F, et al. Antimicrobial susceptibility of Pseudomonas aeruginosa in China: A review of two multicentre surveillance programmes, and application of revised CLSI susceptibility breakpoints. Int J Antimicrob Agents. 2012;40(5):445–9. https://doi.org/10.1016/j.ijantimicag.2012.07.002.
Marra AR, Camargo LFA, Pignatari ACC, Sukiennik T, Behar PRP, Medeiros EAS, et al. Nosocomial bloodstream infections in Brazilian hospitals: analysis of 2,563 cases from a prospective nationwide surveillance study. J Clin Microbiol. 2011;49(5):1866–71. https://doi.org/10.1128/JCM.00376-11.
Schwarzer C, Fischer H, Machen TE. Chemotaxis and binding of Pseudomonas aeruginosa to scratch-wounded human cystic fibrosis airway epithelial cells. PLoS One. 2016;11(3):e0150109. https://doi.org/10.1371/journal.pone.0150109.
Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, et al. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66(1):43–51.
Adamo R, Sokol S, Soong G, Gomez MI, Prince A. Pseudomonas aeruginosa flagella activate airway epithelial cells through asialoGM1 and toll-like receptor 2 as well as toll-like receptor 5. Am J Respir Cell Mol Biol. 2004;30(5):627–34. https://doi.org/10.1165/rcmb.2003-0260OC.
Pier GB, Grout M, Zaidi TS. Cystic fibrosis transmembrane conductance regulator is an epithelial cell receptor for clearance of Pseudomonas aeruginosa from the lung. Proc Natl Acad Sci U S A. 1997;94(22):12088–93.
Kucharska I, Liang B, Ursini N, Tamm LK. Molecular interactions of lipopolysaccharide with an outer membrane protein from Pseudomonas aeruginosa probed by solution NMR. Biochemistry. 2016;55(36):5061–72. https://doi.org/10.1021/acs.biochem.6b00630.
Pier GB. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int J Med Microbiol. 2007;297(5):277–95. https://doi.org/10.1016/j.ijmm.2007.03.012.
Hauser AR. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol. 2009;7(9):654–65. https://doi.org/10.1038/nrmicro2199.
Winstanley C, Fothergill JL. The role of quorum sensing in chronic cystic fibrosis Pseudomonas aeruginosa infections. FEMS Microbiol Lett. 2009;290(1):1–9. https://doi.org/10.1111/j.1574-6968.2008.01394.x.
Jakobsen TH, Bjarnsholt T, Jensen PO, Givskov M, Hoiby N. Targeting quorum sensing in Pseudomonas aeruginosa biofilms: current and emerging inhibitors. Future Microbiol. 2013;8(7):901–21. https://doi.org/10.2217/fmb.13.57.
Williams P, Camara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12(2):182–91. https://doi.org/10.1016/j.mib.2009.01.005.
Middleton B, Rodgers HC, Camara M, Knox AJ, Williams P, Hardman A. Direct detection of N-acylhomoserine lactones in cystic fibrosis sputum. FEMS Microbiol Lett. 2002;207(1):1–7. https://doi.org/10.1111/j.1574-6968.2002.tb11019.x.
Collier DN, Anderson L, McKnight SL, Noah TL, Knowles M, Boucher R, et al. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol Lett. 2002;215(1):41–6. https://doi.org/10.1111/j.1574-6968.2002.tb11367.x.
Morlon-Guyot J, Mere J, Bonhoure A, Beaumelle B. Processing of Pseudomonas aeruginosa exotoxin A is dispensable for cell intoxication. Infect Immun. 2009;77(7):3090–9. https://doi.org/10.1128/iai.01390-08.
Jaffar-Bandjee MC, Lazdunski A, Bally M, Carrere J, Chazalette JP, Galabert C. Production of elastase, exotoxin A, and alkaline protease in sputa during pulmonary exacerbation of cystic fibrosis in patients chronically infected by Pseudomonas aeruginosa. J Clin Microbiol. 1995;33(4):924–9.
Hall S, McDermott C, Anoopkumar-Dukie S, McFarland AJ, Forbes A, Perkins AV et al. Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins (Basel). 2016;8(8). doi:https://doi.org/10.3390/toxins8080236.
Bell SC, De Boeck K, Amaral MD. New pharmacological approaches for cystic fibrosis: promises, progress, pitfalls. Pharmacol Ther. 2015;145:19–34. https://doi.org/10.1016/j.pharmthera.2014.06.005.
Mossine VV, Waters JK, Chance DL, Mawhinney TP. Transient Proteotoxicity of bacterial virulence factor Pyocyanin in renal tubular epithelial cells induces ER-related Vacuolation and can be efficiently modulated by Iron chelators. Toxicol Sci. 2016;154(2):403–15. https://doi.org/10.1093/toxsci/kfw174.
Muller M, Li Z, Maitz PK. Pseudomonas pyocyanin inhibits wound repair by inducing premature cellular senescence: role for p38 mitogen-activated protein kinase. Burns. 2009;35(4):500–8. https://doi.org/10.1016/j.burns.2008.11.010.
Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10(12):599–606. https://doi.org/10.1016/j.molmed.2004.10.002.
Costa KC, Bergkessel M, Saunders S, Korlach J, Newman DK. Enzymatic degradation of Phenazines can generate energy and protect sensitive organisms from toxicity. mBio. 2015;6(6):e01520–15. https://doi.org/10.1128/mBio.01520-15.
Wu HJ, Wang AH, Jennings MP. Discovery of virulence factors of pathogenic bacteria. Curr Opin Chem Biol. 2008;12(1):93–101. https://doi.org/10.1016/j.cbpa.2008.01.023.
Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 2006;44:417–45. https://doi.org/10.1146/annurev.phyto.44.013106.145710.
Ramos I, Dietrich LE, Price-Whelan A, Newman DK. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol. 2010;161(3):187–91. https://doi.org/10.1016/j.resmic.2010.01.003.
Du X, Li Y, Zhou Q, Xu Y. Regulation of gene expression in Pseudomonas aeruginosa M18 by phenazine-1-carboxylic acid. Appl Microbiol Biotechnol. 2015;99(2):813–25. https://doi.org/10.1007/s00253-014-6101-0.
Dietrich LE, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321(5893):1203–6. https://doi.org/10.1126/science.1160619.
Dowling DN, O'Gara F. Metabolites of Pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol. 1994;12(4):133–41. https://doi.org/10.1016/0167-7799(94)90091-4.
Laursen JB, Nielsen J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem Rev. 2004;104(3):1663–86. https://doi.org/10.1021/cr020473j.
Pierson LS III, Pierson EA. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol. 2010;86(6):1659–70. https://doi.org/10.1007/s00253-010-2509-3.
Fordos M. Recherches sur les matieres colorantes des suppurations bleues, pyocyanine et pyoxanthose. CR Hebd Seances Acad Sci. 1863;56:1128–31.
Reyes EA, Bale MJ, Cannon WH, Matsen JM. Identification of Pseudomonas aeruginosa by pyocyanin production on tech agar. J Clin Microbiol. 1981;13(3):456–8.
Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW, Cole PJ. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect Immun. 1988;56(9):2515–7.
Penesyan A, Kumar SS, Kamath K, Shathili AM, Venkatakrishnan V, Krisp C, et al. Genetically and phenotypically distinct Pseudomonas aeruginosa cystic fibrosis isolates share a Core proteomic signature. PLoS One. 2015;10(10):e0138527. https://doi.org/10.1371/journal.pone.0138527.
Bonfield TL, Konstan MW, Berger M. Altered respiratory epithelial cell cytokine production in cystic fibrosis. J Allergy Clin Immunol. 1999;104(1):72–8.
Moura-Alves P, Fae K, Houthuys E, Dorhoi A, Kreuchwig A, Furkert J, et al. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 2014;512(7515):387–92. https://doi.org/10.1038/nature13684.
Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61(5):1308–21. https://doi.org/10.1111/j.1365-2958.2006.05306.x.
Usher LR, Lawson RA, Geary I, Taylor CJ, Bingle CD, Taylor GW, et al. Induction of neutrophil apoptosis by the Pseudomonas aeruginosa exotoxin pyocyanin: a potential mechanism of persistent infection. J Immunol. 2002;168(4):1861–8. https://doi.org/10.4049/jimmunol.168.4.1861.
Allen L, Dockrell DH, Pattery T, Lee DG, Cornelis P, Hellewell PG, et al. Pyocyanin production by Pseudomonas aeruginosa induces neutrophil apoptosis and impairs neutrophil-mediated host defenses in vivo. J Immunol. 2005;174(6):3643–9. https://doi.org/10.4049/jimmunol.174.6.3643.
Rada B, Leto TL. Redox warfare between airway epithelial cells and Pseudomonas: dual oxidase versus pyocyanin. Immunol Res. 2009;43(1–3):198–209. https://doi.org/10.1007/s12026-008-8071-8.
Liu GY, Nizet V. Color me bad: microbial pigments as virulence factors. Trends Microbiol. 2009;17(9):406–13. https://doi.org/10.1016/j.tim.2009.06.006.
Kanthakumar K, Taylor GW, Cundell DR, Dowling RB, Johnson M, Cole PJ, et al. The effect of bacterial toxins on levels of intracellular adenosine nucleotides and human ciliary beat frequency. Pulm Pharmacol. 1996;9(4):223–30. https://doi.org/10.1006/pulp.1996.0028.
Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun. 2004;72(7):4275–8. https://doi.org/10.1128/IAI.72.7.4275-4278.2004.
Shellito J, Nelson S, Sorensen RU. Effect of pyocyanine, a pigment of Pseudomonas aeruginosa, on production of reactive nitrogen intermediates by murine alveolar macrophages. Infect Immun. 1992;60(9):3913–5.
Kamath JM, Britigan BE, Cox CD, Shasby DM. Pyocyanin from Pseudomonas aeruginosa inhibits prostacyclin release from endothelial cells. Infect Immun. 1995;63(12):4921–3.
Muller M, Sorrell TC. Leukotriene B4 omega-oxidation by human polymorphonuclear leukocytes is inhibited by pyocyanin, a phenazine derivative produced by Pseudomonas aeruginosa. Infect Immun. 1992;60(6):2536–40.
Muller M, Sztelma K, Sorrell TC. Inhibition of platelet eicosanoid metabolism by the bacterial phenazine derivative pyocyanin. Ann N Y Acad Sci. 1994;744:320–2. https://doi.org/10.1111/j.1749-6632.1994.tb52752.x.
Muller M, Sorrell TC. Modulation of neutrophil superoxide response and intracellular diacylglyceride levels by the bacterial pigment pyocyanin. Infect Immun. 1997;65(6):2483–7.
Britigan BE, Railsback MA, Cox CD. The Pseudomonas aeruginosa secretory product pyocyanin inactivates alpha1 protease inhibitor: implications for the pathogenesis of cystic fibrosis lung disease. Infect Immun. 1999;67(3):1207–12.
Denning GM, Railsback MA, Rasmussen GT, Cox CD, Britigan BE. Pseudomonas pyocyanine alters calcium signaling in human airway epithelial cells. Am J Phys. 1998;274(6 Pt 1):L893–900. https://doi.org/10.1152/ajplung.1998.274.6.L893.
Denning GM, Wollenweber LA, Railsback MA, Cox CD, Stoll LL, Britigan BE. Pseudomonas pyocyanin increases interleukin-8 expression by human airway epithelial cells. Infect Immun. 1998;66(12):5777–84.
Denning GM, Iyer SS, Reszka KJ, O'Malley Y, Rasmussen GT, Britigan BE. Phenazine-1-carboxylic acid, a secondary metabolite of Pseudomonas aeruginosa, alters expression of immunomodulatory proteins by human airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2003;285(3):L584–92. https://doi.org/10.1152/ajplung.00086.2003.
Look DC, Stoll LL, Romig SA, Humlicek A, Britigan BE, Denning GM. Pyocyanin and its precursor phenazine-1-carboxylic acid increase IL-8 and intercellular adhesion molecule-1 expression in human airway epithelial cells by oxidant-dependent mechanisms. J Immunol. 2005;175(6):4017–23. https://doi.org/10.4049/jimmunol.175.6.4017.
Caldwell CC, Chen Y, Goetzmann HS, Hao Y, Borchers MT, Hassett DJ, et al. Pseudomonas aeruginosa exotoxin Pyocyanin causes cystic fibrosis airway pathogenesis. Am J Pathol. 2009;175(6):2473–88. https://doi.org/10.2353/ajpath.2009.090166.
Mavrodi DV, Parejko JA, Mavrodi OV, Kwak YS, Weller DM, Blankenfeldt W, et al. Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ Microbiol. 2013;15(3):675–86. https://doi.org/10.1111/j.1462-2920.2012.02846.x.
Haas D, Defago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3(4):307–19. https://doi.org/10.1038/nrmicro1129.
Hunter RC, Klepac-Ceraj V, Lorenzi MM, Grotzinger H, Martin TR, Newman DK. Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am J Respir Cell Mol Biol. 2012;47(6):738–45. https://doi.org/10.1165/rcmb.2012-0088OC.
Cui Q, Lv H, Qi Z, Jiang B, Xiao B, Liu L, et al. Cross-regulation between the phz1 and phz2 operons maintain a balanced level of Phenazine biosynthesis in Pseudomonas aeruginosa PAO1. PLoS One. 2016;11(1):e0144447. https://doi.org/10.1371/journal.pone.0144447.
Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, Raaijmakers JM, Lemanceau P, et al. Diversity and evolution of the phenazine biosynthesis pathway. Appl Environ Microbiol. 2010;76(3):866–79. https://doi.org/10.1128/aem.02009-09.
Sakhtah HP-WA, Dietrich LEP. Regulation of Phenazyne biosynthesis. In: Chincholkar SaT L, editor. Microbial Phenazines. Heidelberg: Springer-Verlag; 2013. p. 19–42.
Dubern JF, Diggle SP. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol BioSyst. 2008;4(9):882–8. https://doi.org/10.1039/b803796p.
Subramaniyan S, Divyasree S, Sandhia GS. Phytochemicals as effective quorum quenchers against bacterial communication. Recent Pat Biotechnol. 2016;10(2):153–66.
D'Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Deziel E, Smith EE, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64(2):512–33. https://doi.org/10.1111/j.1365-2958.2007.05678.x.
Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros. 2009;8(1):66–70. https://doi.org/10.1016/j.jcf.2008.09.006.
Fothergill J, Panagea S, Hart C, Walshaw M, Pitt T, Winstanley C. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol. 2007;7(1):45. https://doi.org/10.1186/1471-2180-7-45.
Firoved AM, Boucher JC, Deretic V. Global genomic analysis of AlgU (sigma(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J Bacteriol. 2002;184(4):1057–64. https://doi.org/10.1128/jb.184.4.1057-1064.2002.
Ryall B, Carrara M, Zlosnik JE, Behrends V, Lee X, Wong Z, et al. The mucoid switch in Pseudomonas aeruginosa represses quorum sensing systems and leads to complex changes to stationary phase virulence factor regulation. PLoS One. 2014;9(5):e96166. https://doi.org/10.1371/journal.pone.0096166.
Damron FH, Barbier M, McKenney ES, Schurr MJ, Goldberg JB. Genes required for and effects of alginate overproduction induced by growth of Pseudomonas aeruginosa on Pseudomonas isolation agar supplemented with ammonium metavanadate. J Bacteriol. 2013;195(18):4020–36. https://doi.org/10.1128/JB.00534-13.
Mowat E, Paterson S, Fothergill JL, Wright EA, Ledson MJ, Walshaw MJ, et al. Pseudomonas aeruginosa population diversity and turnover in cystic fibrosis chronic infections. Am J Resp Crit Care. 2011;183(12):1674–9. https://doi.org/10.1164/rccm.201009-1430OC.
Bohn YS, Brandes G, Rakhimova E, Horatzek S, Salunkhe P, Munder A, et al. Multiple roles of Pseudomonas aeruginosa TBCF10839 PilY1 in motility, transport and infection. Mol Microbiol. 2009;71(3):730–47. https://doi.org/10.1111/j.1365-2958.2008.06559.x.
Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103(22):8487–92. https://doi.org/10.1073/pnas.0602138103.
Cabeen MT. Stationary phase-specific virulence factor overproduction by a lasR mutant of Pseudomonas aeruginosa. PLoS One. 2014;9(2):e88743. https://doi.org/10.1371/journal.pone.0088743.
Aaron SD, Kottachchi D, Ferris WJ, Vandemheen KL, St Denis ML, Plouffe A, et al. Sputum versus bronchoscopy for diagnosis of Pseudomonas aeruginosa biofilms in cystic fibrosis. Eur Respir J. 2004;24(4):631–7. https://doi.org/10.1183/09031936.04.00049104.
Goss CH, Burns JL. Exacerbations in cystic fibrosis · 1: epidemiology and pathogenesis. Thorax. 2007;62(4):360–7. https://doi.org/10.1136/thx.2006.060889.
Suzuki S, Mano Y, Fujitani K, Furuya N. Effects of long- term, low-dose macrolide treatment on Pseudomonas aeruginosa PAO1 virulence factors in vitro. Arch Clin Microbiol. 2017;8(4):50. https://doi.org/10.4172/1989-8436.100050.
El-Fouly MZ, Sharaf AM, Shahin AAM, El-Bialy HA, Omara AMA. Biosynthesis of pyocyanin pigment by Pseudomonas aeruginosa. J Radiat Res Appl Sci. 2015;8(1):36–48. https://doi.org/10.1016/j.jrras.2014.10.007.
Silva LV, Galdino AC, Nunes AP, Dos Santos KR, Moreira BM, Cacci LC, et al. Virulence attributes in Brazilian clinical isolates of Pseudomonas aeruginosa. Int J Med Microbiol. 2014;304:990–1000. https://doi.org/10.1016/j.ijmm.2014.07.001.
Tielen P, Narten M, Rosin N, Biegler I, Haddad I, Hogardt M, et al. Genotypic and phenotypic characterization of Pseudomonas aeruginosa isolates from urinary tract infections. Int J Med Microbiol. 2011;301(4):282–92. https://doi.org/10.1016/j.ijmm.2010.10.005.
Cigana C, Melotti P, Baldan R, Pedretti E, Pintani E, Iansa P, et al. Genotypic and phenotypic relatedness of Pseudomonas aeruginosa isolates among the major cystic fibrosis patient cohort in Italy. BMC Microbiol. 2016;16(1):142. https://doi.org/10.1186/s12866-016-0760-1.
Khadim MM, Al Marjani MF. Pyocyanin and biofilm formation in Pseudomonas aeruginosa isolated from burn infections in Baghdad. Iraq Jordan J Biol Sci. 2019;12:31–5.
Wilson R, Pitt T, Taylor G, Watson D, MacDermot J, Sykes D, et al. Pyocyanin and 1-hydroxyphenazine produced by Pseudomonas aeruginosa inhibit the beating of human respiratory cilia in vitro. J Clin Invest. 1987;79(1):221–9. https://doi.org/10.1172/jci112787.
Reimer Å. Concentrations of the Pseudomonas aeruginosa toxin Pyocyanin in human ear secretions. Acta Otolaryngol. 2000;120(543):86–8. https://doi.org/10.1080/000164800454062.
Retraction: Phenazine Content in the Cystic Fibrosis Respiratory Tract Negatively Correlates with Lung Function and Microbial Complexity. Am. J. Respir. Cell Mol Biol 2019;60(1):134-. doi:https://doi.org/10.1165/rcmb.601retraction.
Reszka KJ, O'Malley Y, McCormick ML, Denning GM, Britigan BE. Oxidation of pyocyanin, a cytotoxic product from Pseudomonas aeruginosa, by microperoxidase 11 and hydrogen peroxide. Free Radical Bio Med. 2004;36(11):1448–59. https://doi.org/10.1016/j.freeradbiomed.2004.03.011.
Cheluvappa R. Standardized chemical synthesis of Pseudomonas aeruginosa pyocyanin. MethodsX. 2014;1:67–73. https://doi.org/10.1016/j.mex.2014.07.001.
El-Mowafy SA, Abd El Galil KH, El-Messery SM, Shaaban MI. Aspirin is an efficient inhibitor of quorum sensing, virulence and toxins in Pseudomonas aeruginosa. Microb Pathog. 2014;74(0):25–32. https://doi.org/10.1016/j.micpath.2014.07.008.
Essar DW, Eberly L, Hadero A, Crawford IP. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1991;172(2):884–900. https://doi.org/10.1128/jb.172.2.884-900.1990.
Kurachi M. Studies on the biosynthesis of Pyocyanine (II): isolation and determination of Pyocyanine. Bull Inst Chem Res. 1958;36(6):174–87.
Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol. 2011;193(14):3606–17. https://doi.org/10.1128/jb.00396-11.
Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183(21):6454–65. https://doi.org/10.1128/jb.183.21.6454-6465.2001.
Watson D, MacDermot J, Wilson R, Cole PJ, Taylor GW. Purification and structural analysis of pyocyanin and 1-hydroxyphenazine. Eur J Biochem. 1986;159(2):309–13.
Sharp D, Gladstone P, Smith RB, Forsythe S, Davis J. Approaching intelligent infection diagnostics: carbon fibre sensor for electrochemical pyocyanin detection. Bioelectrochemistry. 2010;77(2):114–9. https://doi.org/10.1016/j.bioelechem.2009.07.008.
Webster TA, Sismaet HJ, Goluch ED. Amperometric detection of pyocyanin in nanofluidic channels. Nano LIFE. 2013;03(01):1340011. https://doi.org/10.1142/S1793984413400114.
Webster TA, Goluch ED. Electrochemical detection of pyocyanin in nanochannels with integrated palladium hydride reference electrodes. Lab Chip. 2012;12(24):5195–201. https://doi.org/10.1039/C2LC40650K.
Alatraktchi FA, Johansen HK, Molin S, Svendsen WE. Electrochemical sensing of biomarker for diagnostics of bacteria-specific infections. Nanomedicine (Lond). 2016;11(16):2185–95. https://doi.org/10.2217/nnm-2016-0155.
Alatraktchi FA, Andersen SB, Johansen HK, Molin S, Svendsen WE. Fast Selective Detection of Pyocyanin Using Cyclic Voltammetry. Sensors (Basel). 2016;16(3). doi:https://doi.org/10.3390/s16030408.
Webster TA, Sismaet HJ, Conte JL, Chan IJ, Goluch ED. Electrochemical detection of Pseudomonas aeruginosa in human fluid samples via pyocyanin. Biosens Bioelectron. 2014;60(0):265–70. https://doi.org/10.1016/j.bios.2014.04.028.
Sismaet HJ, Banerjee A, McNish S, Choi Y, Torralba M, Lucas S, et al. Electrochemical detection of Pseudomonas in wound exudate samples from patients with chronic wounds. Wound Repair Regen. 2016;24(2):366–72. https://doi.org/10.1111/wrr.12414.
Sismaet HJ, Pinto AJ, Goluch ED. Electrochemical sensors for identifying pyocyanin production in clinical Pseudomonas aeruginosa isolates. Biosens Bioelectron. 2017;97:65–9. https://doi.org/10.1016/j.bios.2017.05.042.
Yang Y, Yu YY, Wang YZ, Zhang CL, Wang JX, Fang Z, et al. Amplification of electrochemical signal by a whole-cell redox reactivation module for ultrasensitive detection of pyocyanin. Biosens Bioelectron. 2017;98:338–44. https://doi.org/10.1016/j.bios.2017.07.008.
Alatraktchi FA, Noori JS, Tanev GP, Mortensen J, Dimaki M, Johansen HK, et al. Paper-based sensors for rapid detection of virulence factor produced by Pseudomonas aeruginosa. PLoS One. 2018;13(3):e0194157. https://doi.org/10.1371/journal.pone.0194157.
Elkhawaga AA, Khalifa MM, El-Badawy O, Hassan MA, El-Said WA. Rapid and highly sensitive detection of pyocyanin biomarker in different Pseudomonas aeruginosa infections using gold nanoparticles modified sensor. PLoS One. 2019;14(7):e0216438. https://doi.org/10.1371/journal.pone.0216438.
Rusciano G, Capriglione P, Pesce G, Abete P, Carnovale V, Sasso A. Raman spectroscopy as a new tool for early detection of bacteria in patients with cystic fibrosis. Laser Phys Lett. 2013;10(7):075603. https://doi.org/10.1088/1612-2011/10/7/075603.
Wu X, Chen J, Li X, Zhao Y, Zughaier SM. Culture-free diagnostics of Pseudomonas aeruginosa infection by silver nanorod array based SERS from clinical sputum samples. Nanomedicine. 2014. https://doi.org/10.1016/j.nano.2014.04.010.
Polisetti S, Baig NF, Morales-Soto N, Shrout JD, Bohn PW. Spatial mapping of Pyocyanin in Pseudomonas aeruginosa bacterial communities using surface enhanced Raman scattering. Appl Spectrosc. 2017;71(2):215–23. https://doi.org/10.1177/0003702816654167.
Jia F, Barber E, Turasan H, Seo S, Dai R, Liu L, et al. Detection of Pyocyanin using a new biodegradable SERS biosensor fabricated using gold coated Zein nanostructures further decorated with gold nanoparticles. J Agric Food Chem. 2019;67(16):4603–10. https://doi.org/10.1021/acs.jafc.8b07317.
Pastells C, Pascual N, Sanchez-Baeza F, Marco MP. Immunochemical determination of Pyocyanin and 1-Hydroxyphenazine as potential biomarkers of Pseudomonas aeruginosa infections. Anal Chem. 2016;88(3):1631–8. https://doi.org/10.1021/acs.analchem.5b03490.
Karatuna O, Yagci A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin Microbiol Infect. 2010;16(12):1770–5. https://doi.org/10.1111/j.1469-0691.2010.03177.x.
Acknowledgements
This work has been funded by the Spanish government through the project QS4CF (RTI2018-096278-B-C21), by Fundació La Marató de TV3 (201825-30-31) and by the program Marie Sklodowska Research Fellowship through the project New Diagnostics for Infectious Diseases (ND4ID, 675412-ND4ID-H2020-MSCA-ITN-2015). The Nb4D group (formerly Applied Molecular Receptors group, AMRg) is a consolidated research group (Grup de Recerca) of the Generalitat de Catalunya and has support from the Departament d’Universitats, Recerca i Societat de la Informació de la Generalitat de Catalunya (expedient: 2014 SGR 1484). CIBER-BBN is an initiative funded by the Spanish National Plan for Scientific and Technical Research and Innovation 2013-2016, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions are financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. The ICTS “NANBIOSIS”, and particularly the Custom Antibody Service (CAbS, IQAC-CSIC, CIBER-BBN), is acknowledged for the assistance and support related to some immunoreagents mentioned in this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Published in the topical collection featuring Female Role Models in Analytical Chemistry.
Rights and permissions
About this article
Cite this article
Vilaplana, L., Marco, MP. Phenazines as potential biomarkers of Pseudomonas aeruginosa infections: synthesis regulation, pathogenesis and analytical methods for their detection. Anal Bioanal Chem 412, 5897–5912 (2020). https://doi.org/10.1007/s00216-020-02696-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-020-02696-4