Abstract
Background
Enterobacter cloacae complex (ECC) including different species are isolated from different human clinical samples. ECC is armed by many different virulence genes (VGs) and they were also classified among ESKAPE group by WHO recently. The present study was designed to find probable association between VGs and antibiotic susceptibility in different ECC species.
Methods
Forty-five Enterobacter isolates that were harvested from different clinical samples were classified in four different species. Seven VGs were screened by PCR technique and antibiotic susceptibility assessment was performed by disk-diffusion assay.
Result
Four Enterobacter species; Enterobacter cloacae (33.3%), Enterobacter hormaechei (55.6%), Enterobacter kobei (6.7%) and Enterobacter roggenkampii (4.4%) were detected. Minimum antibiotic resistance was against carbapenem agents and amikacin even in MDR isolates. 33.3% and 13.3% of isolates were MDR and XDR respectively. The rpoS (97.8%) and csgD (11.1%) showed maximum and minimum frequency respectively. Blood sample isolated were highly virulent but less resistant in comparison to the other sample isolates. The csgA, csgD and iutA genes were associated with cefepime sensitivity.
Conclusion
The fepA showed a predictory role for differentiating of E. hormaechei from other species. More evolved iron acquisition system in E. hormaechei was hypothesized. The fepA gene introduced as a suitable target for designing novel anti-virulence/antibiotic agents against E. hormaechei. Complementary studies on other VGs and ARGs and with bigger study population is recommended.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enterobacter genus is a facultative anaerobic, Gram-negative rod-shape bacterium which categorized in the family Enterobacteriaceae. The taxonomy of Enterobacter is complicated and modified continuously, many members of Enterobacter genus were moved to the other genus recently [1, 2]. The species that collectively classified as Enterobacter cloacae complex (ECC) are the most reported pathogens form human infections [3, 4]. Recently, whole-genome sequencing investigations suggested possible reclassification of Enterobacter aerogenes as Klebsiella aerogenes or Klebsiella mobilis. However, there are many morphological and biochemical differences between two genera [1, 5]. Several species were reported from ECC till now. Enterobacter cloacae, Enterobacter asburiae, Enterobacter hormaechei, Enterobacter kobei, Enterobacter ludwigii, and Enterobacter nimipressuralis were introduced as most common isolates from human clinical samples [6].
But, taxonomy of Enterobacter is updating continuously and precise identification of Enterobacter species has a multistep challenging, expensive and time-consuming protocol. The ECC members identified primarily through Matrix-assisted laser desorption/ionization -Time of flight (MALDI-TOF) mass spectrometry (MS) technique and it usually will be completed by some molecular methods [3, 7]. Recently Yang et al., introduced a one-step multiplex PCR method for differential identification of four member of ECC, including; E cloacae, E hormaechei, E kobei and E roggenkampii [7].
Numerous virulence genes were detected in Enterobacter species including gene encoding different adhesins, biofilm related genes, iron acquisition system genes, stress response genes, and various secretion system attributed genes [8,9,10].
Type I fimbriae was detected in different Gram-negative uropathogens. This adhesin is responsible for binding of bacterium to mannosylated receptor molecules on the uroepithelium. It is encoded by fimACDFIHZ locus and FimH is the fimbriae tip located molecule [11]. Another major adhesin of Enterobacter is curli fimbriae. The major construction subunit of curli is CsgA that encoded by csgBAC operon and another homologous operon csgDEFG encode transcription activator CsgD and two incorporated chaperons. Curli fimbriae were reported from other genus of Enterobacteriaceae, it is involved in adhesion and biofilm formation [12,13,14]. Several Enterobacter species were well known for their inhibitory actions against plant pathogens, that is attributed to the competition on iron uptake from soil due to production of different siderophores [15]. Enterobactin, aerobactin and yersiniabactin were reported from Enterobacter [16]. Ferric enterobactin receptor (FepA) is a multifunctional outer membrane protein that is detected in different genera of Enterobacteriaceae. It is the receptor for colicins, enterobactin iron complex and also it could be used by some bacteriophages for entry to bacterial cell [17, 18]. The iutA is encoding gene of Enterobactin receptor that delivered by type 3 secretion system (T3SS) into the host cells [19]. Flagellum specific ATP-synthetase is encoded by fliI gene that plays a critical role in stress adaptation and pathogenesis of E. cloacae [20]. Another gene that plays role in stress responses of Enterobacter species is rpoS gene. It is RNA polymerase sigma factor encoding gene that has some different homologues genes in different bacteria [21].
In 2017 Enterobacter species were placed in ESKAPE group (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) by WHO, that they are the most critical antibiotic resistance bacteria and were represented as very high level threats for human kind [4]. Different types of extended spectrum β-lactamases (ESBLs) and carbapenemases were detected from Enterobacter species that cause intrinsic resistance to wide range of cell wall synthesis active antibiotics. Many strains of Enterobacter are regarded as multidrug-resistant (MDR). The colistin was used as the one of the few effective antibiotic agents against these MDR strains, but recently colistin resistant isolates were reported repeatedly worldwide [22,23,24]. Therefore, continuous research in order to finding new alternatives is a constant need. Some of the studies reported that anti-virulence agents could be suitable candidates in combating against antibiotic resistant infections [25,26,27,28]. In the present study we tried to investigate antibiotic susceptibility profile and also a list of virulence genes of Enterobacter species. We also tried to discover the possible logical relationships between antibiotic resistance and virulence-related genes in order to introduce new targets for planning future anti-virulence strategies.
Material and methods
Specimen collection and bacteria isolation
In this study, a total of 53 laboratory isolates with initial diagnosis of Enterobacter were collected from different samples of hospitalized patients referred to three medical centers affiliated to Kerman University of Medical Sciences in Kerman. All bacterial isolates were cultivated during routine diagnosis and treatment protocols, not by means of research. We only collect the grown media unknown. Finally, 45 out of 53 isolates were confirmed as a distinct Enterobacter species in subsequent steps and 8 unknown species were omitted from the study.
The isolates were obtained from different samples (urine, burning wound, surgical wound, upper respiratory tract and blood). Confirmatory identification and also species determination took placed in the bacteriology laboratory of Afzalipour school of medicine.
Species identification
The bacterial isolates were primarily identified by standard biochemical tests. Swarming on blood agar, monosaccharide fermentation pattern, H2S production in triple sugar iron agar, Indole test, Methyl Red-Voges Proskauer test, citrate utilization test, urease test, lysine decarboxylase test and ornithine decarboxylase test were used in identification of genus and also species differentiation. Species identification take placed through PCR amplification by species-specific primers [7]. Finally, four Enterobacter species(E cloacae, E hormaechei, E kobei and E roggenkampii) were tracked by specific primers through PCR technique. The confirmed isolates were inoculated into the tryptic soy broth enriched with 20% glycerol and stored at -70 °C for next steps experiments.
Antimicrobial susceptibility assessment
Disk diffusion method was used to determine antibiotic susceptibility of the isolates according to the guidelines of the Institute of Clinical and Laboratory Standards (CLSI) and supplementary reviews [29, 30].
Antibiotic discs were provided from Padtan-Teb.Co (Iran) and were as following: Amoxicillin (25 µg), Ceftazidime (30 µg), Ceftriaxone (30 µg), Cefotaxime (30 µg), Aztreonam (30 µg), Imipenem (10 µg), Meropenem (10 µg), Tobramycin (10 µg), Norfloxacin (10 µg), Gentamicin (10 µg), Cefalexin (30 µg), Trimethoprim/Sulfamethoxazole (1.23 /25.75 µg), Cefoxitin(30 µg), Amikacin (30 µg), Cefepime (30 µg), Cefuroxime (30 µg), Tetracycline (30 µg).
Multidrug-resistance (MDR) and extremely drug resistant (XDR) isolates were detected by CLSI guideline [30].
Bacterial genome lysate preparation
Lysate preparation was performed by boiling method. In summary, a loopful of an overnight culture of bacterial isolates were inoculated into the microtubes including of 500 µl of distilled water. The microtubes were places in water bath at 100 °C for 10 min. In the next step, the sample was centrifuged (12,000 rpm for 5 min). After centrifugation, the supernatant was separated and kept in -70 °C.
Polymerase chain reaction conditions and primers list for detection of virulence genes (VGs)
The presence of some virulence genes was investigated by polymerized by chain reaction method through Biometra thermocycler (Germany) and specific primers (Table 1).
PCR products were separated by gel electrophoresis using 1.5% agarose gel with 100 bp DNA Ladder.
Statistical analysis
Data analysis was performed using SPSS 19 statistical software. Chi-square and Fisher’s exact test were used to analysis differences in antibiotic resistance between four studied Enterobacter species and also the association between antibiotic resistance against different antibiotic agents with other studied variables such as; virulence genes, multidrug-resistance. Binary logistic regression analysis was used to evaluate the predictory role of virulence genes for resistance to different antibiotic agents. The p ≤ 0.05 was considered as significant.
Results
Distribution of bacterial isolates in different samples
Totally 45 Enterobacter species were identified in this study. Fifteen (33.3%) Enterobacter cloacae, 25 (55.6%) Enterobacter hormaechei, 3 (6.7%) Enterobacter kobei and 2 (4.4%) Enterobacter roggenkampii were confirmed. These known isolates were used in the all experiments. Thirteen isolates were obtained from surgical wound (28.9%), 10 isolates (22.2%) were harvested from upper respiratory tract (bronchoalveolar lavage fluid and throat swabs). Eleven (24.4%), 7 (15.6%) and 4 (8.9%) of isolates were harvested from blood culture, urine and burning wounds respectively. Frequency of different species was not significantly different among various samples (Pearson Chi-square, p = 0.189). Data are presented in Table 2.
Frequency of antibiotic resistance among the identified Enterobacter species
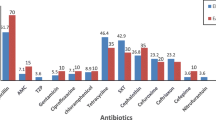
Data collected from disk diffusion test showed different frequency of resistance against various antibiotic agents (Fig. 1) and also antibiotic resistance was different among four studied Enterobacter species (Table 3). But, by means of reach a trustable finding the frequency of antibiotic resistant isolates for each species was compared with same frequency in collection of other species separately through Pearson Chi-square analysis. Results showed that, ceftriaxone resistance was significantly more prevalent among E. cloacae isolates (93.8% vs 70.3%, p = 0.049) and also less prevalent among E. hormaechei isolates (60% vs 92.9%, p = 0.004) in comparison to the other isolates. Significant less frequency of cefotaxime resistance (68% vs 96.4%, p = 0.007), aztreonam resistance (52% vs 78.6%, p = 0.041) and tobramycin resistance (24% vs 53.6%, p = 0.028) also were detected in E. hormaechei isolates in comparison to the other isolates. Binary logistic regression analysis also proposed a positive predictory expression of ceftriaxone resistance in E. cloacae isolates ( p = 0.016. odds ration = 2.278). Resistance against other antibiotic agents was not significantly different in four studied Enterobacter species.
Cumulative resistance against ceftriaxone, gentamicin and ciprofloxacin were regarded as multidrug-resistant and XDR isolates had extra resistant to meropenem in comparison to MDR isolates. Fifteen (33.3%) isolates were MDR and XDR was detected in 6 (13.3%) isolates. Pearson chi-square analysis (p ≤ 0.05) showed higher resistance against cefotaxime, ceftazidime, cefepime, trimethoprim/sulfamethoxazole (cotrimoxazole), imipenem, meropenem, tobramycin, amikacin, norfloxacin, aztreonam and tetracycline in MDR isolates in comparison to the non-MDRs (Table 4).
Pearson chi-square analysis also showed, significant higher frequency of resistance against ceftazidime (100% vs 46.2%, p = 0.014), cefepime (100% vs 43.6%, p = 0.010), imipenem (83.3% vs 10.3%, p = 0.000), aztreonam (100% vs 56.4%, p = 0.040) amikacin (100% vs 10.3%, p = 0.000) and tobramycin (100% vs 25.6%, p = 0.000) among XDR isolates in comparison to the other isolates.
Pearson chi-square test analysis showed that, frequency of resistance against ciprofloxacin was significantly different (p ≤ 0.05) in the isolates which were recovered from different sources, URT samples (33.3%), urine (18.5%) and surgical wound (29.6%), blood (18.5%) and burning wound (0%). There were not significant differences in the frequency of resistance against other antibiotics among samples taken from different wards and different sources.
Frequency of different VGs in the studied isolates
All of the virulence genes were detected in studied isolates. The rpoS and csgD were most frequent and less common genes respectively. The rpoS was detected in 44 (97.8%) out of 45 isolates and csgD was detected only in 5 (11.1%) isolates. The frequency of other virulence genes were as follow; csgA 7 (15.6%), fliI 40 (88.9%), fimH 21 (46.7%), fepA 35 (77.8%) and iutA 40 (88.9%).
Pearson Chi-square analysis revealed that prevalence of iutA and fepA genes was significantly different among four Enterobacter species and they were more frequent in E. hormaechei isolates in comparison to the other isolates (Table 5). Binary logistic regression analysis also proposed a positive predictory role of fepA gene ( p = 0.020. odds ration = 2.216) for differentiation of E. hormaechei from other species and also a negative predictory role of csgD gene ( p = 0.026. odds ration = -2.686) for differentiation of E. hormaechei from other species. Three (6.7%) isolates (two E. cloacae and one E. hormaechei)were armed by all studied virulence genes. Statistical analysis showed that all of these highly virulent isolates were non-MDR and they were susceptible to majority of studied antibiotic agents including; cefepime, imipenem, meropenem, tobramycin, gentamicin, amikacin, trimethoprim/sulfamethoxazole and tetracycline.
Frequency of different virulence genes regarding to the antibiotic resistance in the studied isolates
Results obtained from Chi-square analysis showed different frequency of csgA, csgD, and iutA genes among antibiotic resistant isolates in comparison to antibiotic sensitive isolates (Fig. 2). While, the frequency of the other studied virulence genes was not significantly different between these two groups (data not shown).
The Fischer’s exact test analysis showed that csgA gene was more prevalent among cefepime sensitive isolates in comparison to the cefepime resistant isolates (27.3% vs 4.3%, p = 0.042). Similar finding was also observed for csgD (22.7% vs 0%, p = 0.022) and iutA (100% vs 78.3%, p = 0.028) genes. The csgD gene also had higher frequency among the isolates that were sensitive against trimethoprim/sulfamethoxazole combination (19.2% vs 0%, p = 0.043) and tetracycline (19.2% vs 0%, p = 0.043) in comparison to the resistant isolates (Fig. 2).
Discussion
Resistance against third generation cephalosporins and carbapenem antibiotics among Enterobacter species were recently categorized as priority 1 or critical by WHO experts, which had to considered in prescription of antibacterial agents [31]. Therefore, continuous screening of antibiotic resistance pattern and also finding new targets for development of new drugs with antibacterial or anti-virulence properties is mandatory.
Maximum resistance was detected against penicillin antibiotics and also first and second generation cephalosporins in the present study. High level of resistance against third generation cephalosporins also was observed (Fig. 1). Expression of chromosomal ampC gene as intrinsic mechanism could be responsible of resistance against penicillin antibiotics and also first and second generation cephalosporins in the studied isolates. Continuous hyperproduction of AmpC and also plasmid encoded ESBLs were probable major factors behind resistance against third generation cephalosporins and aztreonam as it reported recently [32].
Data obtained from the present study showed that minimum antibiotic resistant was against carbapenem antibiotics and aminoglycosides specially amikacin. Similar results were reported by other researchers [10, 31, 33, 34]. A previously published review from Iran reported a raising pattern of antibiotic resistance rate science 1999 to June 2021in Enterobacter clinical isolates [33]. Comparison the results reported in this review with our finding also confirmed increasing rate of antibiotic resistance concerning different group of antibiotics including; 3rd generation cephalosporins (5.4% to 27.3%), cefepime (7.5%), ciprofloxacin (24.5%), imipenem (3.4%) and aztreonam (21.3%), from June 2021 till now.
Data analyzing showed, meanwhile, some antibiotic agents such as cefepime, carbapenems and different aminoglycoside antibiotics are still effective against non-MDR E. cloacae complex. High frequency of resistance was detected against majority of antibiotic agents in MDR isolates. Minimum antibiotic resistance in MDR isolates was detected against imipenem, meropenem and also amikacin (Table 4). In the recent decade, carbapenems like imipenem was reported as the most effective antibiotic agent against MDR E. cloacae complex and amikacin also mentioned as the most effective aminoglycoside against them [5, 6, 34, 35]. Thus, carbapenem antibiotics and amikacin could introduced as the most effective antibiotics against MDR E. cloacae complex. Even though, carbapenem resistance in E. cloacae complex also were reported repeatedly in the recent years [32].
The E. hormaechei was most prevalent species among studied Enterobacter species in the present study. Some other reports also have supported this finding [9, 36,37,38]. E. cloacae was the most frequent isolate after E. hormaechei in the present study. Some reports also introduced E. hormaechei and E. cloacae as the most common Enterobacter isolates from human clinical samples [6]. Statistical analysis also revealed that E. hormaechei isolates were significantly more susceptible to ceftriaxone and aztreonam in comparison to E. cloacae. It could be interpreted as higher ESBLs production or AmpC over production among E. cloacae isolates in comparison to the E. hormaechei isolates.
Antibiotic susceptibility findings were not significant different among various studied species. Therefore, similar susceptibility pattern in studied Enterobacter species could be concluded and similar antibiotic therapy regiment will be probably effective against infections caused by different species. However, supporting results have reported recently [6]. But our study population was very limited and only few reports existed in this respect. Thus, it is expected that it had to further investigated in the future to reach a clear conclusion.
The rpos gene had maximum prevalence among investigated virulence genes in the present study and similar results was reported also by Ghanavati et al., recently [8]. Data analysis showed that, blood cultivated isolates (11 isolates, 24.4%) were more virulent in comparison to other sample isolates and all virulence genes except fepA were more prevalent in blood isolates(data not showed). These isolates were categorized as E. hormaechei (6 isolates, 54.5%) and E. cloacae (5 isolates, 45.5%). The percentage increase calculators including; 23.8% for csgA, 106.8% for csgD, 17.2% for fliI, 23.6% for fimH, 3% for rpoS and 3.1% for iutA were detected for these isolates. Three of these 11 isolates were armed by all virulence genes and they were categorized as highly virulent isolates (two E. hormaechei and one E. cloacae). Regarding to these results, it could be concluded that E. hormaechei and E. cloacae are more virulent in comparison to E. kobei and E. roggenkampii. Ganbold et al., also reported same conclusion recently [9].
They were non-MDR and also were susceptible to majority of antibiotics except of beta-lactam agents (antibiotics with intrinsic resistance). This could be reasonable because, such blood colonizing isolates that are targeting directly by immune cell activities continuously, preferred to acquire many virulence properties that enable them to survive challenging conditions of surrounding harsh environment. Instead, they may acquire other genes such as antibiotic resistance genes with comparatively lower frequency or even tolerate some gene reductions specially regarding ARGs. Such fitness-cost phenomenon also was reported in Pseudomonas aeruginosa recently [39].
Based on the results, the iutA and fepA genes were significantly more frequent in E. hormaechei in comparison to other specie (Table 5). The iutA and fepA genes categorized as enterobactin and aerobactin receptor genes respectively and they are superior to other siderophore receptors such as yersiniabactin or salmochelin receptors specially in an iron depleted environment [40]. Therefore, it could be estimated that E. hormaechei evolved to acquire iron more efficiently specially in a depleted iron environment and subsequently outcompete other species. We thought, it may explain higher prevalence of E. hormaechei in the human clinical samples in comparison to other species. Key role of aerobactin in virulence, biofilm and stress resistance of other Enterobacteriaceae member Yersinia pseudotuberculosis was also suggested recently [41]. Using iron acquisition systems as a carrier for antibiotic agents was hypothesized recently [42]. Daoud et al., reported that siderophore receptor fepA involved in delivering some drugs such as catechol-cephalosporins in to the bacterial cell recently [43]. Therefore, fepA gene could be a suitable target for designing novel anti-virulence/antibiotic agents against E. hormaechei infections.
The positive predictory role of fepA and negative predictory role of csgD for E. hormaechei against other studied species also was confirmed by logistic regression analysis. Therefore, condition expressing presence of fepA gene and absence of csgD gene could be regarded as a diagnostic key point for differentiation of E. hormaechei from other species.
Data analysis showed that csgA, csgD and iutA genes were associated with cefepime sensitivity (Fig. 2). The csgD gene was detected only in cefepime sensitive isolates. The csgA gene also was detected in very limited number (1 from 23, 4.3%) of cefepime resistant isolates. Both csgA and csgD genes involved production and control of curli fimbria and also, they promoting bacterial cells to shift from planktonic condition to biofilm former [13, 44]. Therefore, cefepime could introduced as a suitable antibiotic agent against biofilm associated infections caused by Enterobacter spp. However, it has to further investigated in bigger bacterial groups.
Conclusion
Despite continuous increasing of antibiotic resistance in Enterobacter cloacae complex, the present study finding showed that carbapenem antibiotics and amikacin could be effective even against MDR isolates. Blood isolates were significantly more virulent but less resistant against many antibiotic agents in comparison to the other sample isolates. Our findings hypothesized that hyper evolution of some virulence properties such as iron acquisition systems in E. hormaechei could explain higher prevalence of this species in human clinical samples in comparison to the other species. Some virulence genes such as csgA, csgD were associated with cefepime susceptibility in ECC specially biofilm former isolates. For the first time, differential diagnostic role of some virulence genes such as fepA and csgD proposed and the fepA gene introduced as a suitable target for designing novel anti-virulence/antibiotic agents against E. hormaechei isolates. Collectively, we believe that in spite to many investigations some interaction between different genes may remained unknown. In addition to the environmental stresses, some genes may affect the expression of others and also acquisition rate of other genes directly or indirectly. But these interactions have to revealed during future studies and our study proposed some preliminary idea about these ambiguous interactions that may lead other investigations in a even direction that finally results to novel and clear conclusions and introducing suitable target for designing novel anti-virulence agents or new therapeutic regiments including combination of anti-virulence agents and antibiotics.
Availability of data and materials
The datasets used and /or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ECC:
-
Enterobacter cloacae Complex
- PCR:
-
Polymerase Chain Reaction
- MDR:
-
Multidrug-Resistant
- XDR:
-
Extremely Drug Resistant
- ESBL:
-
Extended Spectrum Beta-Lactamase
- CLSI:
-
Clinical & Laboratory Standards Institute
- SPSS:
-
Statistical Package for the Social Sciences
- VGs:
-
Virulence Genes
- ARGs:
-
Antibiotic Resistance Genes
- MOLDI-TOF:
-
Matrix-assisted laser desorption/ionization -time-of-flame
- URT:
-
Upper Respiratory Tract
- WHO:
-
World Health Organization
References
Tindall, BJ, G Sutton, and GM Garrity, Enterobacter aerogenes Hormaeche and Edwards 1960 (Approved Lists 1980) and Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980) share the same nomenclatural type (ATCC 13048) on the Approved Lists and are homotypic synonyms, with consequences for the name Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980). Int J Syst Evol Microbiol, 2017. 67(2):502–504.https://doi.org/10.1099/ijsem.0.001572.
Wu, W., Y. Feng, and Z. Zong, Precise species identification for enterobacter: A genome sequence-based study with reporting of two novel species, enterobacter quasiroggenkampii sp. nov. and Enterobacter quasimori sp. nov. mSystems, 2020. 5(4): 10.1128/msystems.00527–20.https://doi.org/10.1128/mSystems.00527-20.
Davin-Regli A, Lavigne J-P, Pages J-M. Enterobacter spp.: Update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev. 2019;32(4):e00002-19. https://doi.org/10.1128/CMR.00002-19.
Mancuso G, et al. Bacterial antibiotic resistance: The most critical pathogens. Pathogens. 2021;10(10):1310. https://doi.org/10.3390/pathogens10101310.
Davin-Regli A, Pages JM. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol. 2015;6:392. https://doi.org/10.3389/fmicb.2015.00392.
Mezzatesta ML, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012;7(7):887–902. https://doi.org/10.2217/fmb.12.61.
Ji Y, et al. Development of a one-step multiplex PCR assay for differential detection of four species (Enterobacter cloacae, Enterobacter hormaechei, Enterobacter roggenkampii, and Enterobacter kobei) belonging to enterobacter cloacae complex with clinical significance. Front Cell Infect Microbiol. 2021;11:677089. https://doi.org/10.3389/fcimb.2021.677089.
Ghanavati R, et al. Clonal relation and antimicrobial resistance pattern of extended-spectrum β-lactamase-and AmpC β-lactamase-producing Enterobacter spp. isolated from different clinical samples in Tehran, Iran. Rev Soc Bras Med Tro. 2018;51:88–93. https://doi.org/10.1590/0037-8682-0227-2017.
Ganbold M, et al. Species identification, antibiotic resistance, and virulence in Enterobacter cloacae complex clinical isolates from South Korea. Front Microbiol. 2023;14:1122691. https://doi.org/10.3389/fmicb.2023.1122691.
Liu S, et al. Cluster differences in antibiotic resistance, biofilm formation, mobility, and virulence of clinical enterobacter cloacae complex. Front Microbiol. 2022;13:814831. https://doi.org/10.3389/fmicb.2022.814831.
Bolourchi N, et al. Comparative resistome and virulome analysis of clinical NDM-1–producing carbapenem-resistant Enterobacter cloacae complex. J Glob Ant Res. 2022;28:254–63. https://doi.org/10.1016/j.jgar.2022.01.021.
Akbari M, et al. Detection of curli biogenesis genes among Enterobacter cloacae isolated from blood cultures. Int J Enteric Pathog. 2015;3(4):1–3.
Kim SM, et al. Involvement of curli fimbriae in the biofilm formation of Enterobacter cloacae. J Microbiol. 2012;50(1):175–8. https://doi.org/10.1007/s12275-012-2044-2.
Zogaj X, et al. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun. 2003;71(7):4151–8. https://doi.org/10.1128/IAI.71.7.4151-4158.2003.
Jha CK, et al. Enterobacter: Role in plant growth promotion. in: bacteria in agrobiology: Plant growth responses. Berlin, Heidelberg: Springer; 2011. p. 159–82.
Mokracka J, Koczura R, Kaznowski A. Yersiniabactin and other siderophores produced by clinical isolates of Enterobacter spp. and Citrobacter spp. FEMS Immunol Med Microbiol. 2004;40(1):51–5. https://doi.org/10.1016/S0928-8244(03)00276-1.
Nemati Zargaran F, et al. Detecting the dominant T and B epitopes of Klebsiella pneumoniae ferric enterobactin protein (FepA) and introducing a single epitopic peptide as vaccine candidate. Int J Pept Res Ther. 2021;27(4):2209–21. https://doi.org/10.1007/s10989-021-10247-3.
Rabsch W, et al. FepA-and TonB-dependent bacteriophage H8: receptor binding and genomic sequence. J Bacteriol. 2007;189(15):5658–74. https://doi.org/10.1128/jb.00437-07.
Compain F, et al. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol. 2014;52(12):4377–80. https://doi.org/10.1128/JCM.02316-14.
Guérin F, et al. Landscape of in vivo fitness-associated genes of Enterobacter cloacae complex. Front Microbiol. 2020;11:1609. https://doi.org/10.3389/fmicb.2020.01609.
Robbe-Saule V, et al. Identification of a non-haem catalase in Salmonella and its regulation by RpoS (σS). Mol Microbiol. 2001;39(6):1533–45. https://doi.org/10.1046/j.1365-2958.2001.02340.x.
Zong Z, Feng Y, McNally A. Carbapenem and colistin resistance in enterobacter: determinants and clones. Trends Microbiol. 2021;29(6):473–6. https://doi.org/10.1016/j.tim.2020.12.009.
Liao W, et al. High prevalence of colistin resistance and mcr-9/10 genes in Enterobacter spp. in a tertiary hospital over a decade. Int J Antimicrob Agents. 2022;59(5):106573. https://doi.org/10.1016/j.ijantimicag.2022.106573.
Smith RD, et al. A Novel Lipid-Based MALDI-TOF Assay for the rapid detection of colistin-resistant enterobacter species. Microbiol Spectr. 2022;10(1):e0144521. https://doi.org/10.1128/spectrum.01445-21.
Brannon, J.R. and M. Hadjifrangiskou, The arsenal of pathogens and antivirulence therapeutic strategies for disarming them. Drug Des Devel Ther, 2016:1795–1806.https://doi.org/10.2147/DDDT.S98939.
Cegelski L, et al. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6(1):17–27. https://doi.org/10.1038/nrmicro1818.
Defoirdt T. Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 2018;26(4):313–28. https://doi.org/10.1016/j.tim.2017.10.005.
Sarshar M, et al. FimH and anti-adhesive therapeutics: A disarming strategy against uropathogens. Antibiotics (Basel). 2020;9(7):397. https://doi.org/10.3390/antibiotics9070397.
Humphries R, et al. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st Edition. J Clin Microbiol. 2021;59(12):e0021321. https://doi.org/10.1128/JCM.00213-21.
CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed.CLSI supplement M100. Wayne: Clinical and Laboratory Standards Institute; 2020.
Markovska R, et al. Quinolone resistance mechanisms among third-generation cephalosporin resistant isolates of Enterobacter spp. in a Bulgarian university hospital. Infect Drug Resist. 2019;12:1445. https://doi.org/10.2147/IDR.S204199.
Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. Multidrug-resistant enterobacter cloacae complex emerging as a global diversifying threat. Front Microbiol. 2019;10:44. https://doi.org/10.3389/fmicb.2019.00044.
Khademi F, et al. Prevalence of ESBL-producing enterobacter species resistant to carbapenems in Iran: A systematic review and meta-analysis. Int J Microbiol. 2022;2022:8367365. https://doi.org/10.1155/2022/8367365.
Dong X, et al. Whole-genome sequencing-based species classification, multilocus sequence typing, and antimicrobial resistance mechanism analysis of the enterobacter cloacae complex in Southern China. Microbiol Spectr. 2022;10(6):e0216022. https://doi.org/10.1128/spectrum.02160-22.
Intra J, et al. Antimicrobial resistance patterns of enterobacter cloacae and klebsiella aerogenes strains isolated from clinical specimens: A twenty-year surveillance study. Antibiotics (Basel). 2023;12(4):775. https://doi.org/10.3390/antibiotics12040775.
Chavda, KD, et al. Comprehensive genome analysis of carbapenemase-producing enterobacter spp.: New insights into phylogeny, population structure, and resistance mechanisms. mBio, 2016. 7(6):10.1128/mbio.02093–16.https://doi.org/10.1128/mBio.02093-16.
Kremer A, Hoffmann H. Prevalences of the Enterobacter cloacae complex and its phylogenetic derivatives in the nosocomial environment. Eur J Clin Microbiol Infect Dis. 2012;31(11):2951–5. https://doi.org/10.1007/s10096-012-1646-2.
Akbari M, Bakhshi B, Najar Peerayeh S. Particular distribution of enterobacter cloacae Strains Isolated from urinary tract infection within clonal complexes. Iran Biomed J. 2016;20(1):49–55. https://doi.org/10.7508/ibj.2016.01.007.
Morgan SJ, et al. Bacterial fitness in chronic wounds appears to be mediated by the capacity for high-density growth, not virulence or biofilm functions. PLoS pathog. 2019;15(3):e1007511. https://doi.org/10.1371/journal.ppat.1007511.
Watts RE, et al. Contribution of siderophore systems to growth and urinary tract colonization of asymptomatic bacteriuria Escherichia coli. Infect Immun. 2012;80(1):333–44. https://doi.org/10.1128/IAI.05594-11.
Li C, et al. Aerobactin-mediated iron acquisition enhances biofilm formation, oxidative stress resistance, and virulence of yersinia pseudotuberculosis. Front Microbiol. 2021;12:699913. https://doi.org/10.3389/fmicb.2021.699913.
Braun V. Iron uptake by Escherichia coli. Front Biosci. 2003;8:s1409–21. https://doi.org/10.2741/1232.
Daoud L, et al. Elucidating the effect of iron acquisition systems in Klebsiella pneumoniae on susceptibility to the novel siderophore-cephalosporin cefiderocol. PLoS ONE. 2022;17(12):e0277946. https://doi.org/10.1371/journal.pone.0277946.
Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol. 2011;193(10):2587–97. https://doi.org/10.1128/jb.01468-10.
Acknowledgements
The authors are grateful for the kindly cooperation of bacteriology laboratory staff of the Afzalipour school of medicine, Kerman University of medical sciences.
Funding
This study was approved by Vice-Chancellor of research and technology of Kerman University of Medical Sciences, but no fund was provided to purpose of publication.
Author information
Authors and Affiliations
Contributions
OT introduced primary idea. OT and FS were involved in designing the study. Resources were provided by OT. Bacterial isolates were collected by FM and MV. OT, FS, MV and FM had equal roles in designing and implementation of actual steps of methods. Molecular screening through PCR technique was performed by MV and FS. Statistical analysis and interpretation were done by OT. The primary version of manuscript was prepared by OT but it was revised and completed by all authors. The submitted version of the manuscript was read and accepted by all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by ethics committee of Kerman university of medical sciences. Ethic’s code: 1399–189. Obtaining consent to participate is not applicable for this study because our isolates were collected from the patients as a part of laboratory practices and treatment protocol(we received unnamed bacteria on agar media) and human or animals were not participated as sample source.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mosaffa, F., Saffari, F., Veisi, M. et al. Some virulence genes are associated with antibiotic susceptibility in Enterobacter cloacae complex. BMC Infect Dis 24, 711 (2024). https://doi.org/10.1186/s12879-024-09608-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-024-09608-2