Abstract
Background
Enterococcus species continues to be an important cause of hospital-acquired infection worldwide. This study was designed to determine the antibiotic resistance profiles, virulence genes and molecular characteristics of Enterococcus faecium strains isolated from an Iranian children hospital in a four-years period.
Results
A total 189 Enterococcus strains, comprising 108 (57%) E. faecium, 67 (35%) E. faecalis and 14 (7%) isolates of other spp. were isolated during the collection period. More than 92% of E. faecium isolates were resistant to ampicillin (92.5%), ciprofloxacin (96%), erythromycin (100%) and clindamycin (96%). A high frequency of resistance to clindamycin (100%), erythromycin (98.5%) and ciprofloxacin (80.5%) was observed among E. faecalis isolates, while resistance to ampicillin (7%) was less frequent. The prevalence of vanA gene among vancomycin resistant E. faecium and vancomycin resistant E. faecalis was 95 and 50%, respectively. The analysis of 108 E. faecium isolates revealed 34 variable number tandem repeat (VNTR) patterns and 27 Multi Locus VNTR Analysis (MLVA) types (MTs).
Conclusions
The results show a shift from E. faecalis to E. faecium as the dominant enterococcal species among patients at the children Hospital. Our data revealed that the majority of E. faecium isolates (66%) belonged to three common MTs and these types were isolated from different wards in children hospital.
Similar content being viewed by others
Background
Enterococcus continues to be an important cause of hospital-acquired infection worldwide [1]. Two species (Enterococcus faecalis and Enterococcus faecium) are responsible for the majority of enterococcal infections in humans and these species have become resistant to multiple antimicrobial agents such as vancomycin (vancomycin resistant enterococci; VRE), aminoglycosides (the high-level gentamicin resistant; HLGR), macrolides, and tetracyclines [2, 3]. The glycopeptide resistance in enterococci is mediated by nine (vanA, vanB, vanC, vanD, vanE, vanG, vanL, vanM, vanN) mobile gene clusters [4]. Among them, vanA genotype is the most common type of enterococcal vancomycin resistance in several countries [5]. The presence of aac (6′)-Ie-aph(2″)-Ia gene, which is carried on transposon is the main cause of HLGR emergence [6, 7]. In addition to the increasing antibiotic resistance, some virulence determinants described to be associated with pathogenesis in E. faecium including, collagen-binding adhesin of E. faecium (Acm), aggregation substance (Asa1), cytolysin (CylA), enterococcal surface protein (Esp), gelatinase (GelE) [4, 8]. Recently, several reports have described Multilocus variable-number of tandem repeat analysis based on PCR-amplification of variable number tandem repeat (VNTR) located on chromosome, is a suitable tool for learning the genetic relationships of important bacterial pathogens, including E. faecium [3, 9]. Despite the high incidence rate of resistant enterococci in Iran, especially VRE and HLGR [10, 11], there is limited information on enterococcal strains isolated from children infections. This study was designed to determine the antibiotic resistance profiles, virulence genes and the prevalence of different VNTR patterns among E. faecium strains isolated from an Iranian children hospital in a four-years period.
Results
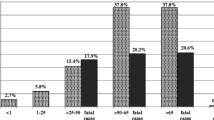
A total 189 Enterococcus strains, comprising 108 (57%) E. faecium, 67 (35%) E. faecalis and 14 (7%) isolates of other spp. were isolated during the collection period. Distribution of E. faecium and E. faecalis isolates based on isolation time (Fig. 1) was showed that during 2015, the prevalence of E. faecium were significantly higher than E. faecalis (P = 0.0001). Most of the E. faecium strains (74%) were isolated from urine, followed by blood (11%), body fluids (7%) and wound (2%). The majority proportion of E. faecium isolates were obtained from urology hospitalized patients (14%) and outpatients (13%).
Antimicrobial susceptibility
More than 92% of E. faecium isolates were resistant to ampicillin (92.5%), ciprofloxacin (96%), erythromycin (100%) and clindamycin (96%). A high frequency of resistance to clindamycin (100%), erythromycin (98.5%) and ciprofloxacin (80.5%) was observed among E. faecalis isolates, while resistance to ampicillin (7%) was less frequent. HLGR was found in 75 and 49% of E. faecium and E. faecalis strains‚ respectively. Inducible resistance to clindamycin was 7% among E. faecium isolates, but not in E. faecalis strains. Vancomycin resistance were detected in 70% of E. faecium and 9% of E. faecalis isolates. The MIC values of Vancomycin Resistant E. faecium (VREfm) and Vancomycin Resistant E. faecalis (VREfs) were ≥ 128 μg̸ml and ≥ 128 μg̸ml respectively. The prevalence of vanA gene among VREfm and VREfs isolates was 95 and 50%, respectively. The presence of aac(6′)-Ie-aph(2″)-Ia gene among HLGR isolates of E. faecium and E. faecalis was 48 and 67%, respectively.
Prevalence of virulence genes among E. faecium isolates
The acm was the most commonly detected gene (81%), followed by esp (17.5%), gelE (16%), and ace (6%). Only two (2%) isolates carried asa1 gene and cylA was not seen in any of the isolates. The presence of the esp gene was significantly higher (P = 0.011) among VREfm isolates than vancomycin sensitive E. faecium isolates.
Molecular analysis of E. faecium
The results of MLVA typing of E. faecium isolates are presented in Table 1. The analysis of 108 E. faecium strains revealed 34 VNTR patterns and 27 MTs. Forty-three isolates (40%) were identified as MT1, 15 (13.8%) as MT2 and 14 (12.9%) as MT3. MT1 was isolated from different wards of the hospital, while MT2 and MT3 were not found in outpatients who were referred to this center. By comparing antibiotic resistance genes in three common types (MT1- MT3), aac(6′)-Ie-aph(2“)-Ia was significantly higher in MT3 than MT1 (P = 0.0046). Also, virulence gene esp had more frequency in MT3 than MT1 (P = 0.0003). The most prevalent pattern of antibiotic resistance in common types (MT1- MT3) was related to pattern gentamicin, ampicillin, ciprofloxacin, erythromycin, and clindamycin. Moreover, the results of the antibiotic resistance genes pattern in common types indicated that pattern vanA+ aac(6’)-Ie-aph(2”)-Ia in MT3 was significantly more frequent than MT1 (P = 0.013).
Discussion
In the current study, the majority (57%) of the isolates was E. faecium. This observation is similar to reports from other countries in which the distribution of Enterococcal species derived from clinical samples (blood, urine, pleural fluid, cerebrospinal fluid, sputum, ascites and hydrothorax) was changed in the favour of E. faecium [3, 4, 12, 13]. The increase in the prevalence of E. faecium species may be due to common resistance of this bacteria to anti-enterococcal drugs, such as ampicillin, aminoglycosides and glycopeptides [3]. In our study, resistance to vancomycin in E. faecium and E. faecalis isolates was 70 and 9%, relatively. The occurrence of VRE varies in different countries, with a high frequency described in VRE in the US, the UK, Ireland, Saudi Arabia and Turkey [13,14,15,16], whereas a low percentage is specific for some European countries such as France and Italy [17, 18]. In spite of past studies in Iran, which showed that all VRE were vanA genotype, in our study, this gene was observed in 95 and 50% of VREfm and VREfs [5, 19, 20]. A possible explanation for this variation is probably related to the presence of other resistance gene such as vanB or presence of other resistance mechanism including thicker cell wall production [21,22,23]. Similar to previous finding in Iran, 75% of E. faecium and 49% of E. faecalis isolates were HLGR [11, 24]. In the current study, 48 and 61% of HLGR in E. faecium and E. faecalis strains carried the aac(6′)-Ie-aph(2“)-Ia gene. This finding was similar with previous studies in which have been shown that the aac(6’)-Ie-aph(2”)-Ia gene is the predominant gene responsible for HLGR. [5, 11, 25,26,27]. In this study, inducible resistance to clindamycin was observed in only 7% of E. faecium isolates. Since the resistance to erythromycin and clindamycin antibiotics depends on the use of these agents in the clinic, inducible resistance to clindamycin between E. faecium strains might be attributed to the wide intake of these antibiotics in our study center [28]. Our result showed that the acm gene was most prevalent virulence gene in E. faecium strains. Similar findings were observed in other studies [8, 25, 29]. It seems that the acm gene have a role in the improved ability of members of the hospital-associated E. faecium to cause disease [30]. Similar to previous report, the prevalence of ace and gelE genes was 6 and 16% [25]. The cylA gene was not detected in any of the 108 E. faecium isolates which is in line with the results stated by other investigators who also tested E. faecium strains for the presence of cylA or more of virulence genes [27, 31]. The rates of esp and asa1 genes were 17.5 and 2%. In some studies, these genes were reported in higher prevalence but in our study and some other studies these genes were identified in lower prevalence among E. faecium isolates [32, 33]. Similar to former studies, the presence of the esp gene was significant among VRE isolates [34, 35]. Recently, a variant of esp gene in VREfm clones has been reported. Also, esp gene has been found to be more common in clinical isolates than fecal isolates, which shows the role of esp gene in pathogens of enterococci [34, 36]. The MLVA typing of 108 E. faecium isolates produced 34 VNTR patterns and 27 MTs. In a study conducted by Top et al. MLVA of 392 E. faecim isolates revealed 127 different MTs [9]. In a study piloted by Gawryszewska et al. MLVA of 112 invasive E. faecium isolates showed 12 different MTs [3]. Unlike MT1 strains that were isolated from all wards in the 4 years period; two MT2 and MT3 were only found in hospitalized patients in the 4 years of study. Differences in the number of types between the present study and previous studies are probably due to different naming patterns for MTs and the term “VNTR pattern” in the present study is equivalent to MT in two other studies. Three common types (MT1, MT2 and MT3) were resistant to gentamicin, ampicillin, ciprofloxacin, erythromycin, clindamycin and had acm and ampicillin resistance, which is more prevalent in nosocomial strains [2], had high frequency in isolates of three common types. This probably indicates the presence of a multi-drug resistant clone that is compatible with the treatment center and the infection control strategies appear to be ineffective so the organism is stable and spreading to patients in different departments and outpatients referring to this center. On the other hand, MT2 and MT3 strains were likely to mutate in order to adapt to the hospital setting. For example the resistance gene pattern vanA+ aac(6′)-Ie-aph(2“)-Ia and esp virulence gene in MT3 were significantly more abundant than MT1. Since the esp gene in isolates of E. faecium is a marker of a pathogenic island that can be transmitted through conjugation to other isolates and vanA and aac(6’)-Ie-aph(2”)-Ia genes are often found on plasmids [7, 37], identification of these isolates is necessary in order to review the infection control strategies to prevent the release of resistance genes, vanA and aac(6′)-Ie-aph(2″)-Ia, and the virulance gene, esp, to other cells.
Conclusions
This study has demonstrated changes over time in species distribution in enterococci isolated from an Iranian children’s hospital. The results show a shift from E. faecalis to E. faecium as the dominant enterococcal species among patients at the children Hospital. Our data revealed that the majority of E. faecium isolates (66%) belonged to three common MTs and these types were isolated from different wards in children hospital. Moreover, the results of this study shows that there is a significant difference in the prevalence rate of antimicrobial resistance and virulence genes among common MTs.
Methods
Bacterial isolates
One hundred and eighty-nine non-repetitive isolates of Enterococcus spp. were collected during December 2011 to November 2015 from various clinical samples of children admitted to a children hospital in Tehran, Iran. Enterococcal isolates were initially re-identified in the microbiology laboratory of Tehran university of Medical Science based on a series of conventional microbiological tests [38]. To confirm the identity of isolate as E. faecium and E. faecalis, the ddl gene was amplified by a Polymerase Chain reaction (PCR)-based method as described previously [39]. Isolates identified as E. faecium were studied further. The study was approved by the Ethics Committee of Tehran University of Medical Sciences.
Antimicrobial susceptibility testing
Antibiotic susceptibility testing was performed by disc diffusion method according to the Clinical Laboratory Standards Institute (CLSI) guidelines [40] with the following antimicrobial disks (Mast Group Ltd., Merseyside, UK.): ampicillin (10 μg), ciprofloxacin (5 μg), erythromycin (15 μg), clindamycin (2 μg). HLGR isolates were also determined by disk diffusion method by using 120 μg gentamicin disk. Inducible clindamycin resistance was determined by D-test [40]. The minimum inhibitory concentrations (MICs) of vancomycin was determined by the agar dilution method. E. faecalis ATCC29212 and S. aureus ATCC25923 were used as controls [40].
Antimicrobial resistance and virulence genes detection
Bacterial genomic DNA was extracted from overnight grown colonies by boiling method [19]. The genes encoding resistance to vancomycin (vanA) and aminoglycoside (aac(6′)-Ie-aph(2″)-Ia) among E. faecium and E. faecalis isolates and virulence factor genes (cylA, gelE, esp, acm, ace, asa1) among E. faecium were detected by a series of PCR assays [5, 25, 34, 41].
Molecular analysis
To examine the genotypic diversity of E. faecium isolates, MLVA was carried out by a modified Top method [9], as previously described, Briefly, 5 VNTR loci (VNTR-1, VNTR-7, VNTR-8, VNTR-9, VNTR-10) were targeted by PCR using the following steps: an initial denaturation at 95 °C for 5 min and final extension at 72 °C for 5 min. For VNTR-1, 30 cycles of 95 °C for 50 s, 54 °C for 50 s and 72 °C for 80 s were performed. For VNTR-7 a touchdown PCR was done that involved 30 cycles, comprising of 30 s at 95 °C, 30 s at 65 °C down to 55 °C and 1 min at 72 °C. For VNTR-8, VNTR-9, and VNTR-10, 50 s at 95 °C, 45 s at 59 °C and 1 min at 72 °C was prepared. Amplified products were separated by electrophoresis in 2% agarose gels with 0.5X TBE (Tris/Borate/EDTA) buffer. The amplicon bands were visualized with UV illumination after staining with KBC power load dye (GelRed Nucleic Acid Gel Stain, 10,000× in water, Kawsar Biotech Co., Tehran, Iran). MLVA type (MT) were assigned on the basis of one or more loci differences, congruence with a similarity index of approximately 80%. Therefore, MTs were defined as isolates sharing 80% or higher similarity.
Statistical analysis
The Fisher’s test was used to compare the frequency of antibiotic resistance, virulence factors and resistance genes in common MTs (95% confidence intervals and P value ≤ 0.05 considered significant). All results were rounded down if they were < 0.5, were presented as whole numbers if they were > 0.5 and were regarded 0.5 itself if they were = 0.5.
Availability of data and materials
Please contact author for data requests.
Abbreviations
- Acm:
-
Collagen-binding adhesin
- AP:
-
Ampicillin
- Asa1:
-
Aggregation substance
- CD:
-
Clindamycin
- CICU:
-
Coronary Intensive Care Unit
- CIP:
-
Ciprofloxacin
- CLSI:
-
Clinical and Laboratory Standards Institute
- CSF:
-
Cerebrospinal fluid
- CylA:
-
Cytolysin
- E:
-
Erythromycin
- Esp:
-
Enterococcal surface protein
- GelE:
-
Gelatinase
- GM:
-
Gentamicin
- HLGR:
-
High-level gentamicin resistant
- m/y:
-
month/year
- MIC:
-
Minimal inhibitory concentration
- MTfm :
-
MLVA Type E. faecium
- NICU:
-
Neonatal Intensive Care Unit
- PCR:
-
Polymerase Chain reaction
- PICU:
-
Paediatric Intensive Care Unit
- VNTR:
-
Variable number tandem repeat
- VRE:
-
Vancomycin resistant enterococci
- VREfm:
-
Vancomycin resistant enterococci E. faecium
- VREfs:
-
Vancomycin resistant enterococci E. faecalis
References
Graham PL 3rd. Staphylococcal and enterococcal infections in the neonatal intensive care unit. Semin Perinatol. 2002;26(5):322–31.
Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10(4):266–78.
Gawryszewska I, Zabicka D, Bojarska K, Malinowska K, Hryniewicz W, Sadowy E. Invasive enterococcal infections in Poland: the current epidemiological situation. Eur J Clin Microbiol Infect Dis. 2016;35(5):847–56.
Yang JX, Li T, Ning YZ, Shao DH, Liu J, Wang SQ, Liang GW. Molecular characterization of resistance, virulence and clonality in vancomycin-resistant Enterococcus faecium and Enterococcus faecalis: a hospital-based study in Beijing, China. Infect Genet Evol. 2015;33:253–60.
Emaneini M, Aligholi M, Aminshahi M. Characterization of glycopeptides, aminoglycosides and macrolide resistance among Enterococcus faecalis and Enterococcus faecium isolates from hospitals in Tehran. Pol J Microbiol. 2008;57(2):173–8.
Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti-Infect Ther. 2014;12(10):1221–36.
Rosvoll TC, Lindstad BL, Lunde TM, Hegstad K, Aasnaes B, Hammerum AM, Lester CH, Simonsen GS, Sundsfjord A, Pedersen T. Increased high-level gentamicin resistance in invasive Enterococcus faecium is associated with aac(6′)Ie-aph(2″)Ia-encoding transferable megaplasmids hosted by major hospital-adapted lineages. FEMS Immunol Med Microbiol. 2012;66(2):166–76.
Camargo IL, Gilmore MS, Darini AL. Multilocus sequence typing and analysis of putative virulence factors in vancomycin-resistant and vancomycin-sensitive Enterococcus faecium isolates from Brazil. Clin Microbiol Infect. 2006;12(11):1123–30.
Top J, Schouls LM, Bonten MJ, Willems RJ. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J Clin Microbiol. 2004;42(10):4503–11.
Emaneini M, Hosseinkhani F, Jabalameli F, Nasiri MJ, Dadashi M, Pouriran R, Beigverdi R. Prevalence of vancomycin-resistant Enterococcus in Iran: a systematic review and meta-analysis. Eur J Clin Microbiol Infect Dis. 2016;35(9):1387–92.
Emaneini M, Khoramian B, Jabalameli F, Beigverdi R, Asadollahi K, Taherikalani M, Lari AR. Prevalence of high-level gentamicin-resistant Enterococcus faecalis and Enterococcus faecium in an Iranian hospital. J Prev Med Hyg. 2016;57(4):E197–e200.
Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, Fridkin SK. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect Control Hosp Epidemiol. 2008;29(11):996–1011.
Niu H, Yu H, Hu T, Tian G, Zhang L, Guo X, Hu H, Wang Z. The prevalence of aminoglycoside-modifying enzyme and virulence genes among enterococci with high-level aminoglycoside resistance in Inner Mongolia, China. Braz J Microbiol. 2016;47(3):691–6.
Brown DF, Hope R, Livermore DM, Brick G, Broughton K, George RC, Reynolds R. Non-susceptibility trends among enterococci and non-pneumococcal streptococci from bacteraemias in the UK and Ireland, 2001-06. J Antimicrob Chemother. 2008;62(Suppl 2):ii75–85.
Saba Copur S, Sahin F, Gocmen JS. Determination of virulence and multidrug resistance genes with polymerase chain reaction method in vancomycin-sensitive and -resistant enterococci isolated from clinical samples. Turk J Med Sci. 2016;46(3):877–91.
Alotaibi FE, Bukhari EE. Emergence of vancomycin-resistant enterococci at a teaching hospital, Saudi Arabia. Chin Med J. 2017;130(3):340–6.
Peta M, Carretto E, Barbarini D, Zamperoni A, Carnevale L, Perversi L, Pagani M, Bonora MG, Fontana R, Marone P, et al. Outbreak of vancomycin-resistant Enterococcus spp. in an Italian general intensive care unit. Clin Microbiol Infect. 2006;12(2):163–9.
Ozsoy S, Ilki A. Detection of vancomycin-resistant enterococci (VRE) in stool specimens submitted for Clostridium difficile toxin testing. Braz J Microbiol. 2017;48(3):489–92.
Fatholahzadeh B, Hashemi FB, Emaneini M, Aligholi M, Nakhjavani FA, Kazemi B. Detection of Vancomycin Resistant Enterococci (VRE) isolated from Urinary Tract Infections (UTI) in Tehran, Iran. DARU. 2006;14(3):141–5.
Shokouhi S, Darazam IA, Javadi A, Rouhani M, Ghasemnejad M. Genotypic characterization of vancomycin-resistant enterococcus spp. in tertiary center, Iran. Infect Disord Drug Targets. 2017;17(2):90–4.
Emaneini M, Hashemi FB, Aligholi M, Fatholahzadeh B, Kazemi B, Sadeghi F. Detection of vanB genotype enterococci in Iran. Int J Antimicrob Agents. 2005;26(1):98–9.
Sharifi Y, Hasani A, Ghotaslou R, Varshochi M, Hasani A, Aghazadeh M, Milani M. Survey of virulence determinants among vancomycin resistant Enterococcus faecalis and Enterococcus faecium isolated from clinical specimens of hospitalized patients of north west of Iran. Open Microbiol J. 2012;6:34–9.
Kafil HS, Asgharzadeh M. Vancomycin-resistant enteroccus faecium and enterococcus faecalis isolated from education hospital of Iran. Maedica (Buchar). 2014;9(4):323–7.
Zarrilli R, Tripodi MF, Di Popolo A, Fortunato R, Bagattini M, Crispino M, Florio A, Triassi M, Utili R. Molecular epidemiology of high-level aminoglycoside-resistant enterococci isolated from patients in a university hospital in southern Italy. J Antimicrob Chemother. 2005;56(5):827–35.
Yu J, Shi J, Zhao R, Han Q, Qian X, Gu G, Zhang X, Xu J. Molecular characterization and resistant spectrum of enterococci isolated from a haematology unit in China. J Clin Diagn Res. 2015;9(6):Dc04–7.
Celik S, Cakirlar FK, Torun MM. Presence of vancomycin, aminoglycosides, and erythromycin resistance genes in enterococci isolated from clinical samples in Turkey. Clin Lab. 2014;60(11):1801–6.
Heidari H, Emaneini M, Dabiri H, Jabalameli F. Virulence factors, antimicrobial resistance pattern and molecular analysis of Enterococcal strains isolated from burn patients. Microb Pathog. 2016;90:93–7.
Lim JA, Kwon AR, Kim SK, Chong Y, Lee K, Choi EC. Prevalence of resistance to macrolide, lincosamide and streptogramin antibiotics in gram-positive cocci isolated in a korean hospital. J Antimicrob Chemother. 2002;49(3):489–95.
Kang M, Xie Y, He C, Chen ZX, Guo L, Yang Q, Liu JY, Du Y, Ou QS, Wang LL. Molecular characteristics of vancomycin-resistant Enterococcus faecium from a tertiary care hospital in Chengdu, China: molecular characteristics of VRE in China. Eur J Clin Microbiol Infect Dis. 2014;33(6):933–9.
Nallapareddy SR, Singh KV, Okhuysen PC, Murray BE. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect Immun. 2008;76(9):4110–9.
Semedo T, Almeida Santos M, Martins P, Silva Lopes MF, Figueiredo Marques JJ, Tenreiro R, Barreto Crespo MT. Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J Clin Microbiol. 2003;41(6):2569–76.
Enayati M, Sadeghi J, Nahaei MR, Aghazadeh M, Pourshafie MR, Talebi M. Virulence and antimicrobial resistance of Enterococcus faecium isolated from water samples. Lett Appl Microbiol. 2015;61(4):339–45.
Strateva T, Atanasova D, Savov E, Petrova G, Mitov I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz J Infect Dis. 2016;20(2):127–33.
Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42(10):4473–9.
Biswas PP, Dey S, Sen A, Adhikari L. Molecular characterization of virulence genes in vancomycin-resistant and vancomycin-sensitive enterococci. J Glob Infect Dis. 2016;8(1):16–24.
Mundy LM, Sahm DF, Gilmore M. Relationships between enterococcal virulence and antimicrobial resistance. Clin Microbiol Rev. 2000;13(4):513–22.
Top J, Willems R, van der Velden S, Asbroek M, Bonten M. Emergence of clonal complex 17 Enterococcus faecium in The Netherlands. J Clin Microbiol. 2008;46(1):214–9.
Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27(4):731–4.
Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33(5):1434.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Twenty-fifth informational supplement; 2015. p. M100–S25.
Emaneini M, Bigverdi R, Kalantar D, Soroush S, Jabalameli F, Noorazar Khoshgnab B, Asadollahi P, Taherikalani M. Distribution of genes encoding tetracycline resistance and aminoglycoside modifying enzymes in Staphylococcus aureus strains isolated from a burn center. Ann Burns Fire Disasters. 2013;26(2):76–80.
Acknowledgements
Not applicable.
Funding
This research has been supported by Tehran University of Medical Sciences and Health Services. Study grant no 95–02–30-32393.
Author information
Authors and Affiliations
Contributions
ME and FJ designed the experiments. AS and NNF conducted the experiments, AS drafted the manuscript. ME and RB revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Tehran University of Medical Sciences. Consent to participate is not applicable for this study because the isolates included in the study were obtained from existing clinical collections routinely assembled as part of laboratory practices of university hospitals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sattari-Maraji, A., Jabalameli, F., Node Farahani, N. et al. Antimicrobial resistance pattern, virulence determinants and molecular analysis of Enterococcus faecium isolated from children infections in Iran. BMC Microbiol 19, 156 (2019). https://doi.org/10.1186/s12866-019-1539-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-019-1539-y