Abstract
Introduction
Chronic lung disease is a major cause of morbidity in African children with HIV infection; however, the microbial determinants of HIV-associated chronic lung disease (HCLD) remain poorly understood. We conducted a case–control study to investigate the prevalence and densities of respiratory microbes among pneumococcal conjugate vaccine (PCV)-naive children with (HCLD +) and without HCLD (HCLD-) established on antiretroviral treatment (ART).
Methods
Nasopharyngeal swabs collected from HCLD + (defined as forced-expiratory-volume/second < -1.0 without reversibility postbronchodilation) and age-, site-, and duration-of-ART-matched HCLD- participants aged between 6–19 years enrolled in Zimbabwe and Malawi (BREATHE trial-NCT02426112) were tested for 94 pneumococcal serotypes together with twelve bacteria, including Streptococcus pneumoniae (SP), Staphylococcus aureus (SA), Haemophilus influenzae (HI), Moraxella catarrhalis (MC), and eight viruses, including human rhinovirus (HRV), respiratory syncytial virus A or B, and human metapneumovirus, using nanofluidic qPCR (Standard BioTools formerly known as Fluidigm). Fisher's exact test and logistic regression analysis were used for between-group comparisons and risk factors associated with common respiratory microbes, respectively.
Results
A total of 345 participants (287 HCLD + , 58 HCLD-; median age, 15.5 years [IQR = 12.8–18], females, 52%) were included in the final analysis. The prevalence of SP (40%[116/287] vs. 21%[12/58], p = 0.005) and HRV (7%[21/287] vs. 0%[0/58], p = 0.032) were higher in HCLD + participants compared to HCLD- participants. Of the participants positive for SP (116 HCLD + & 12 HCLD-), 66% [85/128] had non-PCV-13 serotypes detected. Overall, PCV-13 serotypes (4, 19A, 19F: 16% [7/43] each) and NVT 13 and 21 (9% [8/85] each) predominated. The densities of HI (2 × 104 genomic equivalents [GE/ml] vs. 3 × 102 GE/ml, p = 0.006) and MC (1 × 104 GE/ml vs. 1 × 103 GE/ml, p = 0.031) were higher in HCLD + compared to HCLD-. Bacterial codetection (≥ any 2 bacteria) was higher in the HCLD + group (36% [114/287] vs. (19% [11/58]), (p = 0.014), with SP and HI codetection (HCLD + : 30% [86/287] vs. HCLD-: 12% [7/58], p = 0.005) predominating. Viruses (predominantly HRV) were detected only in HCLD + participants. Lastly, participants with a history of previous tuberculosis treatment were more likely to carry SP (adjusted odds ratio (aOR): 1.9 [1.1 -3.2], p = 0.021) or HI (aOR: 2.0 [1.2 – 3.3], p = 0.011), while those who used ART for ≥ 2 years were less likely to carry HI (aOR: 0.3 [0.1 – 0.8], p = 0.005) and MC (aOR: 0.4 [0.1 – 0.9], p = 0.039).
Conclusion
Children with HCLD + were more likely to be colonized by SP and HRV and had higher HI and MC bacterial loads in their nasopharynx. The role of SP, HI, and HRV in the pathogenesis of CLD, including how they influence the risk of acute exacerbations, should be studied further.
Trial registration
The BREATHE trial (ClinicalTrials.gov Identifier: NCT02426112, registered date: 24 April 2015).
Similar content being viewed by others
Introduction
In 2019, over 2.8 million children and adolescents were living with HIV globally, 90% in sub-Saharan Africa [1]. Respiratory infections remain the most common manifestation of HIV among these children and adolescents [2, 3]. The scale-up of antiretroviral therapy (ART) has increased survival so that growing numbers of children are entering adulthood. In addition, ART has resulted in a reduction in the rate of respiratory disorders, including tuberculosis and lymphocytic interstitial pneumonitis [4,5,6,7]. However, studies in sub-Saharan Africa revealed that approximately 30% of HIV-infected older children experience chronic respiratory symptoms, including chronic cough and reduced tolerance to exercise, which often leads to presumptive tuberculosis treatment [8]. The clinical and radiological picture of this chronic lung disease is consistent with small airway disease, predominantly constrictive obliterative bronchiolitis [9].
The pathogenesis of this condition is incompletely understood. It is speculated that HIV-induced chronic inflammation and dysregulated immune activation may play a role [10,11,12]. A previous study of older children with HIV-associated chronic lung disease (HCLD) conducted by our group demonstrated that there was increased inflammatory activation in children with HCLD (HCLD +) compared to their HIV-infected counterparts without HCLD (HCLD-) [13]. In the same cohort, there was an association between the carriage of specific bacteria in the nasopharynx and HCLD [14]. Specifically, we observed that older children with HCLD were more likely to be colonized with Streptococcus pneumoniae (SP) and Moraxella catarrhalis (MC) than their HCLD- counterparts [14]. The study utilized bacterial culture, which is limited by viability and a narrow spectrum of culturable bacterial species. Although we observed that SP was associated with HCLD, we did not investigate the specific serotypes that may be involved in this condition, which is important to inform pneumococcal immunization. Furthermore, the prevalence of respiratory viruses was also not studied.
Viruses facilitate bacterial infections in the host through various mechanisms, including damaging the respiratory epithelium, modifying the immune response, and altering cell membranes [15]. Coinfection of viruses and bacteria leads to increased bacterial load, thus making individuals more susceptible to complications related to upper respiratory tract infections [16]. Prior to COVID-19, respiratory syncytial virus, influenza virus and human rhinovirus (HRV) were the most common causative agents of upper respiratory infection and have been linked to exacerbations of COPD [17, 18], asthma development [17], and severe bronchiolitis in children [19,20,21].
To overcome these limitations, we investigated the prevalence of respiratory pathogens in both HCLD + and HCLD- participants using real-time quantitative polymerase chain reaction (qPCR) to detect and quantify a large number of bacterial and viral targets and elucidate common SP serotypes (94 serotypes). We also assessed clinical and sociodemographic factors associated with microbial carriage and density.
Materials and methods
Study design, population, and setting
This case–control study was nested within the BREATHE trial (ClinicalTrials.gov Identifier: NCT02426112, registered date: 24 April 2015) investigating whether azithromycin therapy could improve lung function and reduce the risk of exacerbations among children with HCLD [22]. BREATHE was a two-site, double-blinded, placebo-controlled, individually randomized trial conducted in Harare (Zimbabwe) and Blantyre (Malawi). The study setting, population, and trial procedures are described elsewhere [22,23,24]. Briefly, we enrolled perinatally HIV-infected participants aged 6 – 19 years with HCLD. HCLD was defined as a forced expiratory volume in 1 s (FEV1) z score < -1, with no reversibility (< 12% improvement in FEV1 after salbutamol 200 µg inhaled using a spacer) [22]. A group of perinatally HIV-infected children without HCLD (FEV1 z score > 0) was also recruited at the same time as the enrollment of trial participants using frequency matching for site, sex, age, and duration of ART to serve as a comparison group for pathogenesis studies. Both groups were on ART for at least six months. All participants were most likely not vaccinated due to the introduction of PCV13 in 2012 in Zimbabwe [25] and in Malawi in 2011 [26], making them ineligible for vaccination at that time because of their older age. Swabs were collected between June 1, 2016 and September 31, 2019. Sociodemographic data and clinical history were recorded through an interviewer-administered questionnaire.
Nasopharyngeal swab collection
Nasopharyngeal swabs were collected at baseline from all participants using sterile flocked flexible nylon swabs (Copan Italia, Brescia, Italy). Swabs were immediately immersed in 1 mL PrimeStore® Molecular Transport Medium (MTM) (Longhorn Vaccines & Diagnostics LLC, Bethesda, USA), transported on ice and stored at -80 °C at the diagnostic laboratory at each site. PrimeStore® MTM was used because it is a medium optimized for transporting and storing samples for molecular analyses; it also inactivates potential pathogens and stabilizes nucleic acids [27]. The samples were batched and transported on dry ice to Cape Town, South Africa, where they were stored at -80 °C until further processing.
Total nucleic acid extraction
Total nucleic acid (TNA) extraction for microbial identification was conducted on NP swabs stored in Primestore® MTM. Briefly, the samples were thawed and vortexed for 10 s, and 400 µl aliquots were transferred into ZR BashingBead™ Lysis Tubes containing 0.5 mm beads (catalog no. ZR S6002–50, Zymo Research Corp., Irvine, CA, United States) for the mechanical lysis steps. Lysis was conducted on a Qiagen Tissue lyser LTTM (Qiagen, FRITSCH GmbH, Idar-Oberstein, Germany) for 5 min at 50 Hz, followed by centrifugation (Eppendorf F-45–30-11, Merck KgaA, Darmstadt, Germany) for 1 min at 10,000 rpm (10,640 g). The supernatants (250 µl) were extracted using the QIAsymphony® DSP Virus/Pathogen Kit (Qiagen GmbH, Hilden, Germany) on the QIAsymphony SP/AS instrument (Qiagen GmbH, Hilden, Germany) following the manufacturer’s instructions. The total nucleic acid was eluted in 70 µl DNA elution buffer into the Elution Microtube (Qiagen GmbH, Hilden, Germany) and immediately stored at -80 °C until further analysis.
Real-time qPCR using the biomark HD system (Fluidigm assay)
Nanofluidic qPCR testing was performed at the WITS-VIDA Research Unit, Witwatersrand University, Johannesburg, South Africa as previously described [28, 29]. Briefly, all extracts were tested for 94 SP serotypes together with 12 bacterial species (SP, HI, MC, Staphylococcus aureus [SA], Neisseria lactamica, Neisseria meningitidis, Streptococcus pyogenes, Bordetella pertussis, Bordetella holmesii, Klebsiella pneumoniae, Acinetobacter baumanii and Streptococcus oralis), 6 HI serotypes and 8 viruses (respiratory syncytial virus A and B, human rhinovirus, influenza A and B, human parainfluenza 1 and 3, and human metapneumovirus). Furthermore, all samples were previously cultured for SP, HI, MC and SA as described elsewhere [14]. These microbial targets (Table S1) included on the nanofluidic panel might be associated with HCLD and are the most frequent pathobionts in the nasopharynx. A detailed list of these microbial targets can be found in the supplementary material (Table S1). For SP, positive samples were defined as those with a Cycle of quantification (Cq) value ≤ 36 for each serotype-specific qPCR target and positive for both LytA and PiaB. Negative samples were defined as those with Cq values ≥ 36 for each target.
The bacterial or serotype densities were determined following the method outlined by Downs et al. [28]. Briefly, culture controls and synthetic double-stranded DNA (dsDNA) template gene fragments (gBlocks) were included in the assay as external calibrators, reported as copy numbers or gene equivalents, respectively. A DNA library was prepared for the targeted pneumococcal serotypes or other bacterial species at an average concentration ranging from 103 to 104 CFU/ml. For assay-sets meeting the defined efficiency criteria (90–110%), the relative quantification of bacterial density was determined by extrapolating using the linear equation derived from standard curves of the calibrators (control strains and gBlocks with known densities), employing the equation and reported as log 10 genomic equivalents/ml:
Data management and statistical analysis
Clinical and sociodemographic data were electronically captured using Google NexusTM tablets (Google, Mountain View, CA, USA) running OpenDataKit software, managed on Microsoft Access databases (Microsoft, Redmond, WA, USA) and analyzed using Stata ((StataCorp, College Station, TX). Comparisons between groups were performed with the Student T test or Mann–Whitney U test for continuous data and chi-squared or Fisher’s exact tests for categorical data where appropriate with no further adjustment of multiplexity. Multivariate logistic regression, adjusting for age category, duration of ART, site, sex, height-for-age, HIV viral suppression, history of TB treatment, Medical Research Council dyspnea score and ART regimen, was used to investigate the factors associated with microbial carriage and density. The following were excluded from the multivariate model because of colinearity: Enrollment BMI-for-age z score, weight-for-age z score, and CD4 count. A p value of less than 0.05 was considered statistically significant.
Results
Clinical and sociodemographic characteristics
The study included 345 participants, HCLD + (n = 287) and HCLD- (n = 58), with a median age [IQR] of 15.5 (12.8 – 18.0) years and 52% (180/345)] female (Table 1). The median BMI-for-age-z score for the HCLD + group was lower compared to the HCLD- group (-1.1 vs -0.4), p < 0.001. A higher proportion of the participants from the HCLD + group were previously treated for tuberculosis (31% vs. 12%, p = 0.001), stunted (49% vs. 29%, p = 0.009) and underweight (52% vs. 14%, p < 0.001) compared to the HCLD- group. A higher number of HCLD + participants were on a second-line ART (protease inhibitor-based) regimen (25% vs. 10%, p = 0.01) compared to HCLD-. Ten percent of HCLD + participants compared to HLCD- (2%) participants had an MRC dyspnea score of 3 or above. None of the participants reported smoking.
Prevalence and densities of selected nasopharyngeal microbes in participants with and without HCLD
The prevalence and median densities of selected microbes detected in the nasopharynx of HCLD + and HCLD- participants are summarized in Table 2. The prevalence of SP colonization was significantly higher in the HCLD + group (40%, 116/278) compared to the HCLD- group (21%, 12/58; p = 0.005). However, there were no statistically significant differences between the two groups in colonization prevalence of pneumococcal serotypes covered by the 13-valent pneumococcal conjugate vaccine (PCV13 vaccine types, VT) or those not covered (non-vaccine types, NVT).
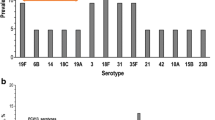
Of the 128 participants colonized with SP (116 HCLD + , 12 HCLD-), 66% (85/128) carried NVT serotypes, while 34% (43/128) carried PCV13 VT serotypes (Fig. 1). A total of 150 pneumococcal serotypes was detected in the 128 participants colonized with SP including 134 serotypes from the HCLD + group and 16 from the HCLD- group (Fig. 1). Multiple serotypes were detected in 14% (16/116) of HCLD + participants and 17% (2/12) of the HCLD- participants colonized with SP.
Pneumococcal serotypes recovered from nasopharyngeal swabs of HCLD + and HCLD- participants. Abbreviations: PCV, polysaccharide-conjugated vaccine; n, number of swabs serotyped using the fluidigm assay from the HCLD + group (n = 116) and HCLD- group (n = 12). Denominator for prevalence is the total number of SP serotypes grouped into PCV 13 and non-PCV 13 serotypes. Others* HCLD + group: PCV13 serotype [3 (0.7%), 5 (0.7%), 6B (0.7%), 9AV (0.7%), 4 (5.2%), Others**: HCLD + group non-PCV 13 serotype [18B (0.7%), 19 atypical (0.7%), 20 (0.7%), 23B (1.5%), 27 (0.7%), 25AF/38 (0.7%), 45 (0.7%), 29 (0.7%), 31 (0.7%), 33C (1.5%), 38 (0.7%)]; HCLD- group non-PCV13 serotype [10A (6.3%), 10B (6.3%), 22A (6.3%), 33B (6.3%)]. 15: HCLD + group non-PCV 13 serotype [15AF (3.7%), 15BC (2.2%), 15like (1.5%)]; HCLD- group non-PCV13 serotype [15like (6.3%)]. 11: HCLD + group non-PCV 13 serotype [11AD [3.7%), 11E (1.5%)]; HCLD- group non-PCV13 serotype [11E (6.3%)]
NVT serotypes predominated over PCV13 VTs in both groups, accounting for 69% (93/134) in HCLD + and 81% (13/16) in HCLD- (Fig. 1). While not statistically significant (p = 0.398), the prevalence of PCV13 VT serotypes trended higher in HCLD + (31%, 41/134) compared to HCLD- (19%, 3/16).
The most common PCV13 VT serotypes in both groups were 4 (16%, 7/44), 19F (16%, 7/44), 19A (16%, 7/44), and 18C (14%, 6/44). The predominant NVTs were 13 and 21 (8% each, 8/106). There were no statistically significant differences in serotype-specific colonization prevalence between HCLD + and HCLD- groups. Likewise, the median densities of the composite NVT and PCV13 VT serotypes did not differ significantly between the two groups (Figure S1). The overall median serotype density across all samples was 8.8 genomic equivalents/ml.
There were no significant differences in the colonization prevalence of any other bacteria tested. Despite there being no difference in the colonization prevalence for both HI and MC, the mean log density was higher in the HCLD + ( 2 × 104−gene equivalents [GE]/ml & 1 × 104 GE/ml) compared to the HCLD- (3 × 102 GE/ml; p = 0.006 & 0.5 × 103 GE/ml; p = 0.031,) groups, respectively (Table 2). There was no significant difference in the mean log densities between the groups for the other tested bacteria. There was a low prevalence of the viruses detected amongst the group, with HRV (7% [21/287] vs. 0% [0/58], p = 0.032) detected in the HCLD + group only. The bacterial species Klebsiella pneumoniae, Neisseria meningitidis, Actinobacter baumanii, Bordetella pertussis/holmesii, and viruses influenza A, influenza B, human parainfluenza type 1 & 3, and human metapneumovirus were not detected in any participants.
Nasopharyngeal bacterial and viral co-colonization in participants with and without HCLD
Bacterial and viral co-colonization detected in HIV-infected participants with or without HCLD is summarized in Table 3 and Table S2. Bacterial detection (any) was significantly higher in HCLD + (61% [175/287]) than in HCLD- (43.1% [25/58]) (p = 0.013). Moreover, the concurrent carriage of multiple bacterial species was higher in the HCLD + group (35.9% [103/287]) than in the HCLD- group (19% [11/58]) (p = 0.014). The most frequent bacteria detected concurrently with SP included SP were HI (HCLD + : 30% [87/287]) vs. HCLD-: 12.1% [7/58], p = 0.013) and MC (HCLD + : 23.3% [67/287] vs. HCLD-: 12.1% [7/58], p = 0.078). Viruses were detected only in the HCLD + group (8% [23/287]), with viral and bacterial co-colonization reported in 6.6% (19/287) of HCLD + participants. To determine whether the concurrent detection of bacteria in HCLD + participants was due to a true interaction or simply by chance, we compared the observed and expected values based on marginal probabilities. The results showed that the co-colonization of SP with HI (Observed [86/287] vs Expected [50.1/287], p < 0.001); SP with MC (Observed [60/287] vs Expected [30.3/287]], p < 0.001) and MC with HI (Observed [54/287] vs Expected [32.4/287], p < 0.001]) was a result of true interactions with HCLD (Table S3).
Factors associated with carriage of selected bacteria at baseline in participants with HCLD
The results of the univariate and multivariate analyses of the clinical and sociodemographic factors associated with the carriage of SP and SA are displayed in Table 4, while those for HI and MC are shown in Table 5. On multivariate analysis, participants previously treated for TB (adjusted odds ratio were more likely to carry SP (aOR): 1.9 [1.1 -3.2], p = 0.021) or HI (aOR: 2.0 [1.2 – 3.3], p = 0.011). Participants on ART for ≥ 2 years (aOR: 0.3 [0.1 – 0.8], p = 0.005) and living in Zimbabwe (aOR: 0.5 [0.3 – 0.9], p = 0.026) were less likely to carry HI (Table 5). Similarly, MC carriage was less likely in participants who had been on ART for ≥ 2 years (aOR: 0.4 [0.1 – 0.9], p = 0.039) (Table 5). Participants who were attending school were more likely to carry MC (aOR: 2.5 [1.0 -6.4], p = 0.050) (Table 5).
Discussion
In this study, we used quantitative PCR to determine the prevalence and density of bacterial and viral carriage in HIV-infected African children. As previously shown [14], microbial colonization was more frequently detected in HCLD + than HCLD- participants, with the former more likely to carry SP or HRV. Strikingly, viruses (predominantly HRV) were detected only in HCLD + children. Moreover, we observed that HCLD + participants had a higher HI and MC density than their HCLD- counterparts. The prevalence and densities of all SP serotypes tested were similar between the two groups, with more of the recovered SP serotypes (79%) being non-PCV 13. Study participants with a history of previous tuberculosis treatment were more likely to carry SP or HI, while those who used ART for ≥ 2 years were less likely to carry HI and MC. Furthermore, those living in Zimbabwe were less likely to carry HI.
The prevalence of HI in the current study in both HCLD + (43%) and HCLD- (33%) participants was higher than that observed in our previous study of the same cohort by Abotsi et al. [14] (12% and 5%, respectively). Similar studies conducted in India [30] and Zambia [31] in HIV-infected children observed similar prevalence (26% and 29%, respectively) to our current study (33%). The discrepancy in results could be attributed to the more sensitive PCR detection method used in our study compared to the culture method employed in previous studies as well as age and pathological differences.
Furthermore, HCLD + participants showed a higher density of HI than their counterparts. Previous studies have associated HI carriage in participants with other lung diseases, including asthma [32], bronchiectasis [33, 34] and chronic obstructive pulmonary disease [35,36,37]. HI has been identified as a biomarker for predicting the response to azithromycin treatment in adults with persistent uncontrolled asthma [32]. It has also been associated with negative outcomes in children suffering from respiratory viral infections [38], including hospitalization among RSV-positive children [39]. The bacterium’s ability to invade host epithelial cells, evade host defense mechanisms, form biofilms and survive as an intracellular pathogen contributes to its pathogenic nature [40], which may suggest an important role that it may play in HCLD + pathogenesis. However, further studies are needed to further elucidate this observation.
The prevalence of carriage of SA in this study (HCLD + [6%] and HCLD- [5%]) was markedly lower than that observed using bacterial culture in the same cohort (HCLD + [23%] and HCLD- [19%]) [14]. This difference in prevalence may be related to the efficiency of the nucleic acid extraction method used. The extraction of nucleic acids requires extended and vigorous lysis steps for some bacterial species (gram-positive such as SA) compared to others (gram-negative such as HI and MC) [41]; however, for this study, although we used a rigorous extraction protocol incorporating bead-beating, the inherent complexity and resilience of the SA bacterial cell wall may have contributed to the low yield. This underscores the need for tailored approaches and ongoing refinement of extraction methods to include enzymatic lysis alternatives such as lysostaphin or lysozyme in NP swabs DNA extraction for enhanced yield. Additionally, carriage may have been influenced by the efficiency of the annealing of PCR primers [42].
High SP, HI and MC density in the nasopharynx has been associated with respiratory infections in children [43, 44]. This is consistent with our study, where a higher HI and MC density was observed in HCLD + participants than in their HCLD- counterparts. The HCLD + participants may have a chronic lung infection, as evidenced by the isolation of bacteria from their sputum in our previous study [14]. Microbiota dominated by Haemophilus, Moraxella or Neisseria species are associated with chronic lung diseases, including chronic obstructive pulmonary disease and asthma [45,46,47,48]. Bhadriraju et al. [40] observed that HIV-infected children with a sputum bacteriome dominated by Haemophilus, Moraxella or Neisseria species were 1.5 times more likely to have HCLD than those with Streptococcus or Prevotella spp. [40]. These bacterial genera were also associated with enhanced inflammatory effects [40]. Interestingly, we detected Neisseria species (N. lactamica) in HCLD + participants only. Taken together, these findings support the important role of HI and MC in HCLD.
Our observation of a higher SP carriage in the HCLD + group than in the HCLD- group is consistent with our culture-based study of the same cohort [14]. Furthermore, SP carriage in the HCLD- participants (21%) is comparable to studies of HIV-infected children in South Africa (22.2%) [49] and Cambodia (17.6%) [42, 50]. Nevertheless, the prevalence is higher than that observed in children living with HIV in Ethiopia (10.3%) [51] and lower than that in studies from Ghana (27.1%) and Tanzania (81%) [52]. The differences in SP prevalence observed between studies can again be related to differences in the age of participants—younger children have a higher carriage prevalence[31, 52], socioeconomic factors [53] and the geographical location of the participants.
The prevalence of PCV 13 serotypes and densities in both study groups (HCLD + : 30.4% and HCLD-: 16.7%) did not differ significantly. The most prevalent PCV-13 serotypes were serotypes 4 (15.9%), 19F (15.9%), 19A (15.9%) and 18C (14%). A study of HIV-infected children in Malawi [54] reports 19F and 6A among the most predominant serotypes. The relatively high prevalence of serotype 19A in PCV-vaccinated children has been suggested by Kamng’ona et al. [54] to occur due to an inversion in the rmlD gene at the CPS locus. This may downregulate the rmlD gene on the CPS locus, causing an altered 19A capsule [55] that is not recognized by the PCV vaccine.
There was a higher prevalence of non-PCV13 serotypes in both the HCLD + (69% [93/134]) and HCLD- (81% [13/16]) compared to PCV13 serotypes. Similar findings were reported in Malawi [54], Nigeria [56] and Ghana [57]. We assume that community herd protection from vaccinated siblings, neighbors, and playmates may be responsible for the low prevalence of vaccine-type serotypes in our cohort. Continued surveillance of SP and its non-PCV 13 serotypes is warranted to inform future vaccine formulation and roll-out strategies, especially in this vulnerable population.
There is evidence suggesting a relationship between SP and other pathogens co-colonizing the nasal and pharyngeal mucosae [58]. Our analysis, based on expected values, revealed a positive positive association between SP with HI and MC carriage in participants with CLD, which is consistent with previous reports by Madhi et al. [59] in HIV-infected South African children. A similar positive association between SP and HI was observed in a study of HIV-infected children in India (median age was 6.5 years, IQR [4.5 – 9]) [30]. HI modulates the expression of SP genes in biofilms primarily by upregulating the type IV pilus structural protein, which is essential for adhesion and stability [60, 61]. Polymicrobial infections involving these microbes and others have been demonstrated to exacerbate higher disease severity and increased tolerance to antimicrobials [62, 63]. Further studies are warranted to comprehensively understand the mechanisms underlying these interactions and their implications for CLD + pathogenesis and treatment strategies.
The major risk factors associated with the development of pneumococcal disease are demographic (age and sex) and immune status (CD4 count and HIV viral load) [64]. We observed no association between these common factors and most bacterial species, including SP. This is supported by previous studies that reported a lack of association between CD4 count and the prevalence of pneumococcal carriage [31, 65, 66]. A longer period on an ART regimen (two years or more) was associated with reduced carriage of MC and HI. Similar results were obtained from a study among HIV-infected adults in Brazil [67]. ART therapy could help reduce the risk of infection and carriage through immune reconstitution [67].
The presence of viruses increases bacterial adherence, and the difference in the prevalence of viruses in HCLD + vs HCLD- children may partially explain the increased HI and MC densities we observed. Our findings are consistent with a study by Binks et al. [44], who reported an increased SP and HI density during coinfection with respiratory viruses within the nasopharynx of Australian children with otitis media. However, no significant difference in the bacterial load was detected in SP from the HCLD + and HCLD- groups. Viruses expose the host to bacterial infection through various mechanisms, including the destruction of the respiratory epithelium, modulation of innate defenses and alteration of cell membranes, which facilitates bacterial adherence [15]. Ishizuka et al. [68], in their in vitro studies, observed increased SP adherence to epithelial cells after infection with HRV. They suggested that this observation may explain why pneumonia develops following an HRV infection [68]. Interestingly, we found no association between any virus (HRV, RSVA and RSVB) and the prevalence or density of carriage of SP or other bacterial species tested. This contrasts with several in vitro and in vivo studies that have suggested that respiratory virus infection increases bacterial adherence and subsequent bacterial superinfection within the nasopharynx [68,69,70]. This discrepancy may be explained by the few viruses we detected due to the limited sample size, especially in the HCLD- group.
HRV is responsible for most upper respiratory tract infections and their complications, including bronchitis [15]. In a study of HIV-infected children in India [71], HRV was the most prevalent virus in these participants when asymptomatic. The GABRIEL multicenter case‒control study in Africa and Asia also found HRV in healthy control groups of pneumonia childhood studies [72]. In contrast, our study detected HRV (7%) in only HCLD + participants. Notably, RSV infection was uncommon, consistent with previous studies conducted in Africa and Asia (PERCH case‒control studies [73] and DCHS case‒control studies [74]), which showed its infrequency except during acute respiratory infections.
In conclusion, our study findings indicate that HCLD + participants were more commonly colonized by any of the bacteria tested compared to HCLD- participants. Specifically, the HCLD + group had a higher prevalence of carriage of SP bacteria, as well as a higher density of MC and HI bacteria. Interestingly, viruses, particularly HRV, were detected only in the HCLD + group. Moreover, our research revealed that previous treatment for tuberculosis was positively associated with the carriage of HI or SP bacteria among study participants. On the other hand, being a female participant was found to be less likely to be associated with SA carriage. Additionally, longer periods on the ART regimen were associated with reduced carriage of HI or MC bacteria. Our study sheds light on the quantitative information on microbial carriage and nasopharyngeal carriage of viruses and serotypes of HI and SP in children with HCLD + . The limitations of our study included a small sample size of HCLD participants, potentially impacting the statistical power and generalizability of our findings. Additionally, our statistical analysis did not correct for multiplexity. While one approach to address multiplexity is adjusting the p-value threshold to α = 0.05 divided by the number of tests conducted, this method may result in a considerable reduction in the number of statistically significant findings. Therefore, there is a need for more comprehensive studies in this population to further investigate the role of SP, HI, MC, and HRV in the pathogenesis of CLD and the underlying mechanisms behind these bacterial associations.
Availability of data and materials
The datasets used and analyzed during the current study are available from Felix Dube (sizwe.dube@uct.ac.za) on reasonable request and ethical approval.
Abbreviations
- ART:
-
Antiretroviral therapy
- AZM:
-
Azithromycin
- HCLD:
-
HIV-associated chronic lung disease
- HCLD + :
-
HIV-infected participants with HIV-associated chronic lung disease
- HCLD-:
-
HIV-infected participants without HIV-associated chronic lung disease
- NP:
-
Nasopharyngeal
- HI:
-
Haemophilus influenza
- MC:
-
Moraxella catarrhalis
- SA:
-
Staphylococcus aureus
- SP:
-
Streptococcus pneumoniae
- HRV:
-
Human rhinovirus
- RSV:
-
Respiratory syncytial virus
- NVT:
-
Non PCV 13 vaccine serotypes
- VT:
-
PCV 13 vaccine serotypes
References
Global and regional trends HIV – UNICEF DATA [https://data.unicef.org/topic/hivaids/global-regional-trends/].
Weber HC, Gie RP, Cotton MF. The challenge of chronic lung disease in HIV-infected children and adolescents. J Int AIDS Soc. 2013;16(1):18633.
Githinji LN, Gray DM, Hlengwa S, Myer L, Zar HJ. Lung Function in South African Adolescents Infected Perinatally with HIV and Treated Long Term with Antiretroviral Therapy. Ann Am Thorac Soc. 2017;14(72R):729.
Jeena PM, Coovadia HM, Thula SA, Blythe D, Buckels NJ, Chetty R. Persistent and chronic lung disease in HIV-1 infected and uninfected African children. AIDS. 1998;12(10):1185–93.
Sharland M, Gibb DM, Holland F. Respiratory morbidity from lymphocytic interstitial pneumonitis (LIP) in vertically acquired HIV infection. Arch Dis Child. 1997;76(4):334–6.
Pitcher RD, Lombard CJ, Cotton MF, Beningfield SJ, Workman L, Zar HJ. Chest radiographic abnormalities in HIV-infected African children: a longitudinal study. Thorax. 2015;70(9):840–6.
Peacock-Villada E, Richardson BA, John-Stewart GC. Post-HAART outcomes in pediatric populations: comparison of resource-limited and developed countries. Pediatrics. 2011;127(2):e423–41.
Rylance J, McHugh G, Metcalfe J, Mujuru H, Nathoo K, Wilmore S, Rowland-Jones S, Majonga E, Kranzer K, Ferrand RA. Chronic lung disease in HIV-infected children established on antiretroviral therapy. AIDS. 2016;30(18):2795–803.
Ferrand RA, Desai SR, Hopkins C, Elston CM, Copley SJ, Nathoo K, Ndhlovu CE, Munyati S, Barker RD, Miller RF, Bandason T, Wells AU, Corbett EL. Chronic lung disease in adolescents with delayed diagnosis of vertically acquired HIV infection. Clin Infect Dis. 2012;55(1):145–52.
Collini P, Morris A. Maintaining lung health with longstanding HIV. Curr Opin Infect Dis. 2016;29(1):31–8.
French MA, King MS, Tschampa JM, da Silva BA, Landay AL. Serum Immune Activation Markers Are Persistently Increased in Patients with HIV Infection after 6 Years of Antiretroviral Therapy despite Suppression of Viral Replication and Reconstitution of CD4+ T Cells. J Infect Dis. 2009;200(8):1212–5.
Zar HJ. Chronic lung disease in human immunodeficiency virus (HIV) infected children. Pediatr Pulmonol. 2008;43(1):1–10.
Hameiri-Bowen D, Sovershaeva E, Flaegstad T, Gutteberg TJ, Ngwira LG, Simms V, Rehman AM, McHugh G, Bandason T, Ferrand RA, Rowland-Jones S, Yindom LM. Soluble biomarkers associated with chronic lung disease in older children and adolescents with perinatal HIV infection. AIDS. 2021;35(11):1743–51.
Abotsi RE, Nicol MP, McHugh G, Simms V, Rehman AM, Barthus C, Mbhele S, Moyo BW, Ngwira LG, Mujuru H, Makamure B, Mayini J, Odland JØ, Ferrand RA, Dube FS. Prevalence and antimicrobial resistance profiles of respiratory microbial flora in African children with HIV-associated chronic lung disease. In: BMC Infectious Diseases. vol. 21. 2021. p. 216.
Sajjan U, Wang Q, Zhao Y, Gruenert DC, Hershenson MB. Rhinovirus disrupts the barrier function of polarized airway epithelial cells. Am J Respir Crit Care Med. 2008;178(12):1271–81.
DeMuri GP, Gern JE, Eickhoff JC, Lynch SV, Wald ER. Dynamics of Bacterial Colonization With Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis During Symptomatic and Asymptomatic Viral Upper Respiratory Tract Infection. Clin Infect Dis. 2017;66(7):1045–53.
Tan WC, Xiang X, Qiu D, Ng TP, Lam SF, Hegele RG. Epidemiology of respiratory viruses in patients hospitalized with near-fatal asthma, acute exacerbations of asthma, or chronic obstructive pulmonary disease. Am J Med. 2003;115(4):272–7.
Baillie VL, Moore DP, Mathunjwa A, Baggett HC, Brooks A, Feikin DR, Hammitt LL, Howie SRC, Knoll MD, Kotloff KL, Levine OS, O’Brien KL, Scott AG, Thea DM, Antonio M, Awori JO, Driscoll AJ, Fancourt NSS, Higdon MM, Karron RA, et al. Epidemiology of the Rhinovirus (RV) in African and Southeast Asian Children: A Case-Control Pneumonia Etiology Study. Viruses. 2021;13(7):1249.
Henquell C, Mirand A, Deusebis A-L, Regagnon C, Archimbaud C, Chambon M, Bailly J-L, Gourdon F, Hermet E, Dauphin J-B. Prospective genotyping of human rhinoviruses in children and adults during the winter of 2009–2010. J Clin Virol. 2012;53(4):280–4.
Jacobs SE, Lamson DM, St George K, Walsh TJ. Human rhinoviruses. Clin Microbiol Rev. 2013;26(1):135–62.
Louie JK, Roy-Burman A, Guardia-LaBar L, Boston EJ, Kiang D, Padilla T, Yagi S, Messenger S, Petru AM, Glaser CA, Schnurr DP. Rhinovirus Associated With Severe Lower Respiratory Tract Infections in Children. Pediatr Infect Dis J. 2009;28(4):337–9.
Gonzalez-Martinez C, Kranzer K, McHugh G, Corbett EL, Mujuru H, Nicol MP, Rowland-Jones S, Rehman AM, Gutteberg TJ, Flaegstad T, Odland JO, Ferrand RA. Azithromycin versus placebo for the treatment of HIV-associated chronic lung disease in children and adolescents (BREATHE trial): study protocol for a randomised controlled trial. Trials. 2017;18(1):622.
McHugh G, Rehman AM, Simms V, Gonzalez-Martinez C, Bandason T, Dauya E, Moyo B, Mujuru H, Rylance J, Sovershaeva E, Weiss HA, Kranzer K, Odland J, Ferrand RA, Team tBCT. Chronic lung disease in children and adolescents with HIV: a case–control study. Tropical Med Int Health. 2020;25(5):590–9.
Rehman AM, Ferrand R, Allen E, Simms V, McHugh G, Weiss HA. Exclusion of enrolled participants in randomised controlled trials: what to do with ineligible participants? BMJ Open. 2020;10(12): e039546.
Dondo V, Mujuru H, Nathoo K, Jacha V, Tapfumanei O, Chirisa P, Manangazira P, Macharaga J, de Gouveia L, Mwenda JM, Katsande R, Weldegebriel G, Pondo T, Matanock A, Lessa FC. Pneumococcal Conjugate Vaccine Impact on Meningitis and Pneumonia Among Children Aged <5 Years-Zimbabwe, 2010–2016. Clin Infect Dis. 2019;69(Suppl 2):S72-s80.
Heinsbroek E, Tafatatha T, Phiri A, Swarthout TD, Alaerts M, Crampin AC, Chisambo C, Mwiba O, Read JM, French N. Pneumococcal carriage in households in Karonga District, Malawi, before and after introduction of 13-valent pneumococcal conjugate vaccination. Vaccine. 2018;36(48):7369–76.
Daum L, Worthy SA, Yim K, Nogueras M, Schuman R, Choi YW, Fischer G. A clinical specimen collection and transport medium for molecular diagnostic and genomic applications. Epidemiol Infect. 2010;139:1764–73.
Downs SL, Madhi SA, van der Merwe L, Nunes MC, Olwagen CP. Optimization of a high-throughput nanofluidic real-time PCR to detect and quantify of 15 bacterial species and 92 Streptococcus pneumoniae serotypes. Sci Rep. 2023;13(1):4588.
Olwagen CP, Adrian PV, Madhi SA. Performance of the Biomark HD real-time qPCR System (Fluidigm) for the detection of nasopharyngeal bacterial pathogens and Streptococcus pneumoniae typing. Sci Rep. 2019;9(1):1–11.
Bhattacharya SD, Niyogi SK, Bhattacharyya S, Arya BK, Chauhan N, Mandal S. Associations between Potential Bacterial Pathogens in the Nasopharynx of HIV Infected Children. Indian J Pediatr. 2012;79(11):1447-53.
Mwenya DM, Charalambous BM, Phillips PPJ, Mwansa JCL, Batt SL, Nunn AJ, Walker S, Gibb DM, Gillespie SH. Impact of cotrimoxazole on carriage and antibiotic resistance of Streptococcus pneumoniae and Haemophilus influenzae in HIV-infected children in Zambia. Antimicrob Agents Chemother. 2010;54(9):3756–62.
Taylor SL, Ivey KL, Gibson PG, Simpson JL, Rogers GB. AMAZES Study Research Group. Airway abundance of Haemophilus influenzae predicts response to azithromycin in adults with persistent uncontrolled asthma. Eur Respir J. 2020;56(4):2000194. https://doi.org/10.1183/13993003.00194-2020.
Kapur N, Grimwood K, Masters IB, Morris PS, Chang AB. Lower airway microbiology and cellularity in children with newly diagnosed non-CF bronchiectasis. Pediatr Pulmonol. 2012;47(3):300–7.
Hare KM, Grimwood K, Leach AJ, Smith-Vaughan H, Torzillo PJ, Morris PS, Chang AB. Respiratory bacterial pathogens in the nasopharynx and lower airways of Australian indigenous children with bronchiectasis. J Pediatr. 2010;157(6):1001–5.
Murphy T, Brauer A, Grant B, Sethi S. Moraxella catarrhalis in Chronic Obstructive Pulmonary Disease: Burden of Disease and Immune Response. Am J Respir Crit Care Med. 2005;172:195–9.
Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med. 2008;359(22):2355–65.
Sethi S, Evans N, Grant BJ, Murphy TF. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N Engl J Med. 2002;347(7):465–71.
Rosas-Salazar C, Shilts MH, Tovchigrechko A, Chappell JD, Larkin EK, Nelson KE, Moore ML, Anderson LJ, Das SR, Hartert TV. Nasopharyngeal Microbiome in Respiratory Syncytial Virus Resembles Profile Associated with Increased Childhood Asthma Risk. Am J Respir Crit Care Med. 2016;193(10):1180–3.
de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, Chaussabel D, Cohen DM, Sanders EA, Ramilo O, Bogaert D, Mejias A. Nasopharyngeal Microbiota, Host Transcriptome, and Disease Severity in Children with Respiratory Syncytial Virus Infection. Am J Respir Crit Care Med. 2016;194(9):1104–15.
Bhadriraju S, Fadrosh DW, Shenoy MK, Lin DL, Lynch KV, McCauley K, Ferrand RA, Majonga ED, McHugh G, Huang L, Lynch SV, Metcalfe JZ. Distinct lung microbiota associate with HIV-associated chronic lung disease in children. Sci Rep. 2020;10(1):16186.
Rantakokko-Jalava K, Jalava J. Optimal DNA isolation method for detection of bacteria in clinical specimens by broad-range PCR. J Clin Microbiol. 2002;40(11):4211–7.
Rogers GB, Shaw D, Marsh RL, Carroll MP, Serisier DJ, Bruce KD. Respiratory microbiota: addressing clinical questions, informing clinical practice. Thorax. 2015;70(1):74–81. https://doi.org/10.1136/thoraxjnl-2014-205826.
Vu HTT, Yoshida LM, Suzuki M, Nguyen HAT, Nguyen CDL, Nguyen ATT, Oishi K, Yamamoto T, Watanabe K, Vu TD. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J. 2011;30(1):11–8.
Binks MJ, Cheng AC, Smith-Vaughan H, Sloots T, Nissen M, Whiley D, McDonnell J, Leach AJ. Viral-bacterial co-infection in Australian Indigenous children with acute otitis media. BMC Infect Dis. 2011;11(1):161.
Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, Boushey H. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol. 2015;136(4):874–84.
Durack J, Boushey HA, Lynch SV. Airway Microbiota and the Implications of Dysbiosis in Asthma. Curr Allergy Asthma Rep. 2016;16(8):52.
Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, Dyer AM, Israel E, Kraft M, Martin RJ, Mauger DT, Rosenberg SR, Sharp-King T, White SR, Woodruff PG, Avila PC, Denlinger LC, Holguin F, Lazarus SC, Lugogo N, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. 2017;140(1):63–75.
Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WOC. Disordered Microbial Communities in Asthmatic Airways. PLoS ONE. 2010;5(1): e8578.
Cotton MF, Wasserman E, Smit J, Whitelaw A, Zar HJ. High incidence of antimicrobial resistant organisms including extended spectrum beta-lactamase producing Enterobacteriaceae and methicillin-resistant Staphylococcus aureus in nasopharyngeal and blood isolates of HIV-infected children from Cape Town, South Africa. BMC Infect Dis. 2008;8(1):40.
Krcmery V, Sokolova J, Kulkova N, Liskova A, Shahum A, Benca G. Nasopharyngeal bacterial colonisation in HIV-positive children in Cambodia. Tropical Med Int Health. 2013;18(10):1267–8.
Mulu W, Yizengaw E, Alemu M, Mekonnen D, Hailu D, Ketemaw K, Abera B, Kibret M. Pharyngeal colonization and drug resistance profiles of Morraxella catarrrhalis, Streptococcus pneumoniae, Staphylococcus aureus, and Haemophilus influenzae among HIV infected children attending ART Clinic of Felegehiwot Referral Hospital, Ethiopia. PLoS ONE. 2018;13(5): e0196722.
Abdullahi O, Karani A, Tigoi CC, Mugo D, Kungu S, Wanjiru E, Jomo J, Musyimi R, Lipsitch M, Scott JAG. The Prevalence and Risk Factors for Pneumococcal Colonization of the Nasopharynx among Children in Kilifi District, Kenya. PLoS ONE. 2012;7(2): e30787.
Adegbola RA, DeAntonio R, Hill PC, Roca A, Usuf E, Hoet B, Greenwood BM. Carriage of Streptococcus pneumoniae and other respiratory bacterial pathogens in low and lower-middle income countries: a systematic review and meta-analysis. PLoS ONE. 2014;9(8): e103293.
Kamng’ona AW, Hinds J, Bar-Zeev N, Gould KA, Chaguza C, Msefula C, Cornick JE, Kulohoma BW, Gray K, Bentley SD, French N, Heyderman RS, Everett DB. High multiple carriage and emergence of Streptococcus pneumoniae vaccine serotype variants in Malawian children. BMC Infect Dis. 2015;15(1):234.
Naseeb S, Delneri D. Impact of chromosomal inversions on the yeast DAL cluster. PLoS ONE. 2012;7(8): e42022.
Adetifa IM, Antonio M, Okoromah CA, Ebruke C, Inem V, Nsekpong D, Bojang A, Adegbola RA. Pre-vaccination nasopharyngeal pneumococcal carriage in a Nigerian population: epidemiology and population biology. PLoS ONE. 2012;7(1): e30548.
Dayie NT, Baffuor-Asare M, Labi AK, Obeng-Nkrumah N, Olayemi E, Lartey M, Slotved HC, Donkor ES. Epidemiology of Pneumococcal Carriage among HIV-Infected Individuals in the Conjugate Vaccine Era: A Study in Southern Ghana. Biomed Res Int. 2019;2019:3427174.
Margolis E, Yates A, Levin BR. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host’s immune response. BMC Microbiol. 2010;10:59.
Madhi SA, Adrian P, Kuwanda L, Cutland C, Albrich WC, Klugman KP. Long-Term Effect of Pneumococcal Conjugate Vaccine on Nasopharyngeal Colonization by Streptococcus pneumoniae—and Associated Interactions with Staphylococcus aureus and Haemophilus influenzae Colonization—in HIV-Infected and HIV-Uninfected Children. J Infect Dis. 2007;196(11):1662–6.
Bakaletz LO. Bacterial biofilms in otitis media: evidence and relevance. Pediatr Infect Dis J. 2007;26(10):S17–9.
Cope EK, Goldstein-Daruech N, Kofonow JM, Christensen L, McDermott B, Monroy F, Palmer JN, Chiu AG, Shirtliff ME, Cohen NA, Leid JG. Regulation of virulence gene expression resulting from Streptococcus pneumoniae and nontypeable Haemophilus influenzae interactions in chronic disease. PLoS ONE. 2011;6(12): e28523.
Frisan T. Co- and polymicrobial infections in the gut mucosa: The host-microbiota-pathogen perspective. Cell Microbiol. 2021;23(2): e13279.
Perez A, Murphy T. Do we need a vaccine against Moraxella catarrhalis in chronic lung disease? What are the options and opportunities? Vaccine. 2017;37:5551-8.
Licciardi PV, Tan EL, Li P, Ng OT. Pneumococcal vaccination for HIV-infected individuals in Singapore. Proceedings of Singapore Healthcare. 2019;28:55–60.
Anthony L, Meehan A, Amos B, Mtove G, Mjema J, Malahiyo R, Yin JK, Oftadeh S, Gilbert GL, Shingadia D, Reyburn H, Deen J, Richmond PC, Booy R. Nasopharyngeal carriage of Streptococcus pneumoniae: prevalence and risk factors in HIV-positive children in Tanzania. Int J Infect Dis. 2012;16(10):e753-757.
Pemba L, Charalambous S, von Gottberg A, Magadla B, Moloi V, Seabi O, Wasas A, Klugman KP, Chaisson RE, Fielding K, Churchyard GJ, Grant AD. Impact of cotrimoxazole on non-susceptibility to antibiotics in Streptococcus pneumoniae carriage isolates among HIV-infected mineworkers in South Africa. J Infect. 2008;56(3):171–8.
Nicoletti C, Brandileone MC, Guerra ML, Levin AS. Prevalence, serotypes, and risk factors for pneumococcal carriage among HIV-infected adults. Diagn Microbiol Infect Dis. 2007;57(3):259–65.
Ishizuka S, Yamaya M, Suzuki T, Takahashi H, Ida S, Sasaki T, Inoue D, Sekizawa K, Nishimura H, Sasaki H. Effects of rhinovirus infection on the adherence of Streptococcus pneumoniae to cultured human airway epithelial cells. J Infect Dis. 2003;188(12):1928–39.
Bakaletz LO. Viral potentiation of bacterial superinfection of the respiratory tract. Trends Microbiol. 1995;3(3):110–4.
Rodrigues F, Foster D, Nicoli E, Trotter C, Vipond B, Muir P, Gonçalves G, Januário L, Finn A. Relationships between rhinitis symptoms, respiratory viral infections and nasopharyngeal colonization with Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in children attending daycare. Pediatr Infect Dis J. 2013;32(3):227–32.
Khan T, Das RS, Chaudhary A, Chatterjee J, Bhattacharya SD. Association of nasopharyngeal viruses and pathogenic bacteria in children and their parents with and without HIV. Pneumonia. 2021;13(1):8.
Bénet T, Sánchez Picot V, Messaoudi M, Chou M, Eap T, Wang J, Shen K, Pape J-W, Rouzier V, Awasthi S, Pandey N, Bavdekar A, Sanghavi S, Robinson A, Rakoto-Andrianarivelo M, Sylla M, Diallo S, Nymadawa P, Naranbat N, Russomando G, et al. Microorganisms Associated With Pneumonia in Children <5 Years of Age in Developing and Emerging Countries: The GABRIEL Pneumonia Multicenter, Prospective. Case-Control Study Clinical Infectious Diseases. 2017;65(4):604–12.
O’Brien KL, Baggett HC, Brooks WA, Feikin DR, Hammitt LL, Higdon MM, Howie SRC, Deloria Knoll M, Kotloff KL, Levine OS, Madhi SA, Murdoch DR, Prosperi C, Scott JAG, Shi Q, Thea DM, Wu Z, Zeger SL, Adrian PV, Akarasewi P, et al. Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. Lancet. 2019;394(10200):757–79.
Zar HJ, Nduru P, Stadler JAM, Gray D, Barnett W, Lesosky M, Myer L, Nicol MP. Early-life respiratory syncytial virus lower respiratory tract infection in a South African birth cohort: epidemiology and effect on lung health. Lancet Glob Health. 2020;8(10):e1316–25.
Acknowledgements
We would like to acknowledge the BREATHE trial participants, their families, and the study team. We would also like to thank the staff of the Division of Medical Microbiology and the Department of Molecular and Cell Biology, particularly members of Dube Lab and the UCT community, for providing all the resources required for the project and training whenever needed. We are grateful to Lara Van Der Merwe of the WITS-VIDA research team for assistance with the Fluidigm assay and data analysis.
BREATHE Study Team Members: Prince K. Mushunje1, Msc; Felix S. Dube1,2, PhD; Jon Ø Odland5,6,7, PhD; Rashida A Ferrand8,9, PhD; Mark P. Nicol10, PhD; Regina E. Abotsi1,11, Tsitsi Bandason8, MSc; Ethel Dauya8, MPH; Tafadzwa Madanhire8, PhD; Elizabeth L. Corbett9, 17, PhD; Katharina Kranzer8,9, PhD; Edith D. Majonga8, PhD; Victoria Simms8,12; PhD, Andrea M Rehman8,12; PhD; Helen A.Weiss12, PhD; Hilda Mujuru13, MSc; Dan Bowen14, MSc; Louis-Marie Yindom14, PhD; Sarah L. Rowland-Jones14, DM; Trond Flaegstad15, PhD; Tore J. Gutteberg15, PhD; Jorunn Pauline Cavanagh15, PhD; Trym Thune Flygel 15, MD; Evegeniya Sovarashaeva15, PhD; Jessica Chikwana16, MBBS; Gugulethu Newton Mapurisa16, MBBS; Carmen Gonzalez-Martinez16, 17, MSc; Robina Semphere16, MBBS; Brewster Wisdom Moyo17, MSc; Lucky Gift Ngwira17, MPH; 18Slindile Mbhele, MSc
Affiliations: 1Department of Molecular and Cell Biology & Institute of Infectious Diseases and Molecular Medicine, University of Cape Town, Cape Town, South Africa; 2School of Medicine, University of Lusaka, Lusaka, Zambia; 5Nord University, Faculty of Biosciences and Aquaculture, Bodø, Norway; 6International Research Laboratory for Reproductive Ecotoxicology (IL RET), The National Research University Higher School of Economics, Moscow, Russia; 7School of Health Systems and Public Health, Faculty of Health Sciences, University of Pretoria, Pretoria, South Africa; 8Biomedical Research and Training Institute, Harare, Zimbabwe; 9Clinical Research Department, London School of Hygiene and Tropical Medicine, London, United Kingdom; 10Marshall Centre, Division of Infection and Immunity, School of Biomedical Sciences, Faculty of Health and Medical Sciences, University of Western Australia, Perth, Australia; 11Department of Pharmaceutical Microbiology, School of Pharmacy, University of Health and Allied Sciences, Ho, Ghana; 12MRC International Statistics and Epidemiology Group, Department of Infectious Disease Epidemiology, London School of Hygiene and Tropical Medicine, London, United Kingdom; 13Department of Paediatrics, University of Zimbabwe, Harare, Zimbabwe; 14Nuffield Department of Medicine, University of Oxford, Oxford, UK; 15Faculty of Health Sciences, UiT, The Arctic University of Norway, Tromsø, Norway; and Department of Paediatrics, University Hospital of North Norway, Tromsø, Norway); 16Department of Paediatrics and Child Health, University of Malawi College of Medicine, Blantyre, Malawi; 17Malawi-Liverpool-Wellcome Trust Clinical Research Programme, Blantyre, Malawi; 18Division of Medical Microbiology, Faculty of Health Sciences, University of Cape Town, Cape Town, South Africa
Funding
This parent study was funded by the Global Health and Vaccination Research (GLOBVAC) Programme of the Medical Research Council of Norway. This substudy was funded by the Royal Society through the Future Leaders African Independent Research award and the National Institute for Health Research (NIHR) Global Health Research Unit on Mucosal Pathogens using UK aid from the UK Government (Project number 16/136/46). The views expressed are those of the authors and not necessarily those of the NIHR, the Department of Health and Social Care. PM received funding from the UCT Postgraduate Funding, UCT's Building Research Active Academic Staff (B.R.A.A.S.) award, the Molecular and Cell Biology_Equity Development Programme scholarship, and the Dube-lab scholarship. REA acknowledges the financial support of the Swedish International Development Cooperation Agency (SIDA) through the Organization of Women in Science for the Developing World (OWSD) PhD Fellowship, Margaret McNamara Education Grants and L'Oréal UNESCO For Women in Science PhD Fellowship. MN is supported by an Australian National Health and Medical Research Council Investigator Grant [APP1174455]. FSD is supported by the National Research Foundation of South Africa (112160), Future Leaders – African Independent Research (FLAIR) Fellowship, the National Institute for Health Research (NIHR) Global Health Research Unit on Mucosal Pathogens using UK aid from the UK Government, the University of Cape Town and the Allergy Society of South Africa (ALLSA). RAF is funded by the Wellcome Trust (206316_Z_17_Z). The funders had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Consortia
Contributions
FSD conceived the study. PKM conducted the laboratory experiments and data analysis and wrote the first draft of the manuscript supervised by REA and FSD. CO and SM supervised the Fluidigm assay. JOO, MN, and RAF conceived and led the parent BREATHE study. JOO secured funding from GLOBVAC on behalf of the consortium. All authors contributed to, read, and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The parent study (BREATHE) was approved by the Human Research and Ethics Committee of the University of Cape Town—UCT HREC (HREC/REF:754/2015), the London School of Hygiene and Tropical Medicine Ethics Committee (reference 8818), the Harare Central Hospital Ethics Committee and Medical Research Council of Zimbabwe (reference MRCZ/A/1946), the College of Medicine Research Ethics Committee Malawi (reference P.04/15/1719) and the Regional Committee for Medical and Health Research Ethics, Northern Norway (reference 2015/1650). The University of Oxford waived approval. Additional ethical approval was received for this substudy from the UCT HREC (HREC/REF: 092/2019). No additional data were collected other than those approved in the parent study. Written informed consent and assent were given by guardians and participants, respectively. Participants who were 18 years old and above consented independently at the time of enrollment. All data obtained and generated during the study were kept confidential. This research was conducted in accordance with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mushunje, P.K., Dube, F.S., Olwagen, C. et al. Characterization of bacterial and viral pathogens in the respiratory tract of children with HIV-associated chronic lung disease: a case–control study. BMC Infect Dis 24, 637 (2024). https://doi.org/10.1186/s12879-024-09540-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-024-09540-5