Abstract
Background
Pulmonary infections caused by non-diphtheriae corynebacteria are increasing. However, rapid identification of Corynebacterium species poses a challenge due to the low genetic variation within the genus.
Methods
Three reference strains and 99 clinical isolates were used in this study. A qPCR followed by high-resolution melting (HRM) targeting ssrA was performed to simultaneously identify C. striatum, C. propinquum and C. simulans. To further evaluate this assay’s performance, 88 clinical sputum samples were tested by HRM and the detection results were compared with those of the traditional culture method and multiple cross-displacement amplification (MCDA) assay.
Results
The melting curve produced by a pair of universal primers generated species-specific HRM curve profiles and could distinguish the three target species from other related bacteria. The limit of detection of HRM assay for DNA from the three purified Corynebacterium species was 100 fg. Compared with the culture method, HRM detected 22 additional positive specimens, representing a 23.9% relative increase in detection rate. The HRM assay had 98.4% (95% confidence interval [CI], 90.5–99.9%) sensitivity and 100% (95% CI, 82.8–100%) specificity. Additionally, 95.5% concordance between HRM and MCDA (κ = 0.89 [95% CI, 0.79–0.99]) was noted.
Conclusions
The HRM assay was a simple, rapid, sensitive, and specific diagnostic tool for detecting C. striatum, C. propinquum, and C. simulans, with the potential to contribute to early diagnosis, epidemiological surveillance, and rapid response to outbreak.
Similar content being viewed by others
Background
The genus Corynebacterium is composed of aerobic, non–spore-forming, pleomorphic, Gram-positive bacilli with worldwide distribution. The most well-established and well-described pathogen in this genus, C. diphtheria, is the main causative agent of diphtheria, the incidence of which has dropped due to effective vaccination programs [1]. However, in recent years, there has been a considerable increase in reports of non-diphtheriae Corynebacterium species, which have been linked to multiple hospital outbreaks and nosocomial infections [2,3,4]. Although these microorganisms are common components of the skin microbiota and mucous membranes, their clinical significance as emerging respiratory pathogens has been demonstrated by various studies [4,5,6]. Of note, recent reports show that multidrug-resistant strains of the species C. striatum are emerging rapidly [7,8,9,10]. Early detection and identification of Corynebacterium species are essential to intervention and infection treatment efforts.
Traditionally, these microorganisms have been routinely identified by biochemical tests using the API Coryne system (bioMérieux, Craponne, France) or the RapID CB PLUS system (Thermo Fisher Scientific, Waltham, MA, USA) in clinical microbiology laboratories [11, 12]. However, these methods have low sensitivity, and are time-consuming and unreliable for species identification, especially in the case of C. simulans, due to its similarities with C. striatum [13, 14]. Identification by 16S ribosomal ribonucleic acid (rRNA) and rpoB gene sequencing produces more reliable results, but is slow and cost prohibitive in developing countries [15, 16]. Matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) has been applied to perform accurate species-level identification of Corynebacterium spp. clinical isolates, but this technology is not yet fully accessible to clinical microbiology laboratories in resource-limited settings [17]. Also, MALDI-TOF requires pure cultures as starting material, which precludes rapid diagnosis. Therefore, suitable detection assays that are rapid, reliable, and cost-effective are always in demand for effective control and treatment strategies against infections caused by emerging Corynebacterium species.
The high-resolution melting (HRM) assay, a recently developed technique based on quantitative real-time polymerase chain reaction (qPCR) that detects genetic variation in deoxyribonucleic acid (DNA) sequences, provides a good alternative for molecular diagnosis [18]. Before HRM analysis can be performed, the region of interest is amplified using PCR in the presence of a fluorescent dye (EvaGreen) that is homogenously intercalated into the double-stranded (ds) DNA. After PCR, the amplicon is gradually heated at increasing temperatures, and the ds PCR product denatures into two single strands, releasing the binding dye and leading to a decrease in fluorescence level. The rate of dissociation of the amplicon mainly depends on GC-content, sequence length, complementarity, and nearest-neighbor thermodynamics [19]. A specific and characteristic melting profile can be produced for the amplicon by monitoring changes in fluorescence intensity. The HRM assay is an easily implemented, closed-tube method that can simultaneously detect closely related species within approximately 2 h. In addition, it is not restricted to cultured material, but can detect DNA extracted directly from clinical specimens. The HRM technique has been successfully employed in concurrently identifying and differentiating between several pathogens, such as bacteria, viruses, and fungi [20,21,22,23].
In this study, we report the development of a qPCR–based HRM assay capable of detecting C. striatum, C. propinquum, and C. simulans, as well as distinguishing between them in pure cultures and clinical specimens with increased specificity and sensitivity.
Methods
Bacterial strains
Three reference strains (C. striatum ATCC 6940, C. propinquum DSM 44285, and C. simulans DSM 44415) and 99 clinical isolates were used in this study (Additional file 1). All clinical isolates were recovered from pulmonary specimens (80 sputum samples, 14 tracheal aspirates, and 5 bronchoalveolar lavage fluid samples) of patients clinically suspected of having respiratory tract infections, and were collected from 2016 to 2018 in the Department of Clinical Laboratory Medicine, Peking University People’s Hospital, Beijing, China. The organisms were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using the Vitek MS (bioMérieux, France) system and the results were confirmed by 16S rRNA gene sequencing.
Genomic DNA extraction
We extracted genomic DNA from reference strains and clinical isolates using QIAamp DNA Mini Kits (QIAGEN, Hilden, Germany) in accordance with the manufacturer’s instructions. DNA concentration and purity were measured using a spectrophotometer (NanoDrop ND-1000; Thermo Fisher) at A260/280. Purified DNA were kept at − 20 °C for subsequent experiments.
Genomic target selection and primer design for HRM
As the ssrA gene, which encodes a transfer-messenger RNA (tmRNA), is highly conserved and phylogenetically informative [24], it was selected as the target. The sequences of ssrA loci for three reference strains were obtained from published genomes in GenBank (National Center for Biotechnology Information [NCBI], Bethesda, MA, USA; https://www.ncbi.nlm.nih.gov/genbank/) and aligned using SeqMan software (version 7.1.0; Fig. 1). In order to identify the best primer pair, 20 pairs of primers were designed based on the ssrA gene in the conserved region using CmSuite software version 8.0 (https://www.scied.com/pr_cmpro.htm). C. striatum, C. propinquum, and C. simulans were tested by PCR using 20 primer sets. The primers ssrA-Fwd (5′-TCAGCGTGACTACGCCCTC-3′) and ssrA-Rev (5′-RCYTCGCCAGGGCTTCTC-3′) displayed positive amplification and were selected for qPCR and HRM assays (data not shown).
qPCR and HRM assays
A qPCR assay was set up containing the following components per reaction:
15 μl 2× Taqman PCR Master Mix (Thermo Fisher); 0.9 μl Primer F (10 μΜ); 0.9 μl Primer R (10 μΜ); 0.3 μl Rox Reference Dye II (100×); 1.5 μl EvaGreen, 20× in water (Biotium, Fremont, CA, USA); 1 ng DNA sample; and nuclease-free water to a total reaction volume of 30 μl. The qPCR assay was performed and validated using the ABI QuantStudio 6 Flex system (Thermo Fisher Scientific, USA) with the following run conditions: 1 cycle of 95 °C for 10 min, followed by 35 cycles of 95 °C for 15 s and 63 °C for 45 s. Next, HRM analysis was initiated by raising the temperature to 95 °C for 15 s and decreasing it to 60 °C for 1 min. Then, the melting curves were generated by increasing the temperature from 60 °C to 95 °C in increments of 0.025 °C/s. All of the amplicons were tested in triplicate to detect technical errors. The HRM data were analyzed using QuantStudio™ Real-Time PCR Software version 1.3. All of the experiments were performed in triplicate.
Evaluation of the sensitivity of the HRM assay
To assess the sensitivity of the HRM assay for identification of Corynebacterium species, the genomic DNA templates of three reference strains were serially diluted with distilled water (10 ng, 100 pg, 10 pg, 1 pg, 100 fg, and 10 fg per μl) to define the limit of detection. DNA templates of Nocardia farcinica were used as the negative control, and distilled water was the blank control. Each DNA concentration was assayed in duplicate by HRM.
Evaluation of the specificity of the HRM assay
To assess the specificity of the HRM assay, the PCR-HRM reactions were performed under the conditions described above with purely genomic DNA templates (10 ng/reaction) from 60 C. striatum, 3 C. propinquum, 6 C. simulans, and 30 non-target clinical samples (Additional file 1). All of the strains were cultured overnight on brain–heart infusion ager at 37 °C. Bacterial genomic DNA from all of the cultured strains were extracted using QIAamp DNA Mini Kits (Qiagen, Germany) in accordance with the manufacturer’s instructions. Analysis of each sample was carried out twice independently.
Practical application of HRM in clinical sputum samples
In total, 88 human sputum samples were recovered from hospitalized patients with pulmonary infections. These samples were previously identified using the culture method and stored at − 70 °C until use in the HRM assay. To evaluate the practical application of the novel HRM method, we employed this assay in routine detection for 88 clinical sputum samples, and compared the results with those of the traditional culture method and the multiple cross displacement amplification (MCDA) assay developed in a previous study [25]. The DNA templates from the sputum samples were extracted using a Wizard® Genomic DNA Purification Kit (Promega, Fitchburg, WI, USA) in accordance with the manufacturer’s instructions. The extracted genomic DNA were used for HRM and MCDA tests. This experiment was carried out in duplicate independently.
DNA sequencing
To further confirm the reliability of the HRM assay, all of the HRM-positive/culture-negative amplicons from qPCR-HRM were sent to Sangon Biotech (Shanghai, China) to undergo direct sequencing in both strands. In brief, the PCR amplicons produced from ssrA were purified with a QIAquick PCR purification kit (QIAGEN, Germany) and subjected to sequencing using the primers ssrA-Fwd and ssrA-Rev on the ABI PRISM 7500 Sequence Detection System (Applied Biosystems, USA). The sequencing results were analyzed using SeqMan software, and were further compared with the reference samples using the Basic Local Alignment Search Tool (BLAST; NCBI; https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analysis
To analyze assay sensitivity and specificity, we used the conventional culture method as the gold standard for the culture-positive specimens, while using consensus results from both the HRM and MCDA assays as a reference standard for the remaining specimens. The sensitivities of the HRM and MCDA assays were determined by the following equation:
Specificity was determined by the following equation [26]:
Additionally, 95% confidence intervals (CIs) were calculated for sensitivity and specificity using the Wilson score method. We used McNemar’s chi-square test to assess whether the performances of these tests were different. A P < 0.05 was considered to indicate statistical significance. The agreement levels between HRM and MCDA results were measured using Cohen’s kappa test, which was calculated as described previously [27].
Results
HRM analysis for identification of C. striatum, C. propinquum, and C. simulans
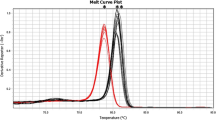
A pair of universal primers (ssrA-Fwd and ssrA-Rev), designed to detect C. striatum, C. propinquum, and C. simulans in HRM analysis, amplified fragments of 233, 240, and 233 bp, respectively, for these species. In the HRM analysis, C. striatum and C. propinquum each had a single peak, while C. simulans produced two melting peaks (Fig. 2a). A distinct HRM peak was observed in the normalized melting curves, allowing them easy to distinguish from each other. The melting temperatures for C. striatum, C. propinquum, and C. simulans using HRM primers were 88.91 °C, 88.44 °C, and 87.86 °C, respectively. Specifically, a high-resolution difference plot was constructed using the curved shape of C. striatum as a baseline, which indicated that these three species were indeed different through the HRM curves (Fig. 2b).
Limit of detection of the HRM assay
To assess the sensitivity of the HRM assay, serial dilutions (10 ng–10 fg) of total genomic DNA extracted from the three reference strains were subjected to a HRM assay. The decreasing concentrations of genomic DNA are presented from left to right in Fig. 3. The limit of detection of the HRM assay was 100 fg of genomic DNA per reaction for C. striatum, C. propinquum, and C. simulans.
Analytical specificity of the HRM assay
To determine the specificity of the HRM assay, the purely genomic DNA extracted from 60 C. striatum, 3 C. propinquum, 6 C. simulans, and 30 non-target strains were tested using the HRM assay under the standard conditions described above. As expected, all 69 cultured samples belonging to C. striatum, C. propinquum, and C. simulans were amplified and formed unique and reproducible melting curves (Additional file 2). In contrast, the 30 non-target strains showed no melting curves. These results suggested that the HRM assay targeting ssrA identified C. striatum, C. propinquum, and C. simulans with 100% specificity.
Evaluation of the HRM assay using clinical sputum samples
To further assess the suitability and usefulness of the HRM assay as a tool for Corynebacterium spp. detection, a total of 88 clinical human sputum samples were analyzed using the traditional culture method, a MCDA assay, and a HRM assay. The results are summarized in Table 1. In the 88 sputum samples, the culture produced 42 positives (47.7%), the MCDA produced 59 (67.0%), and the HRM assay produced 63 (71.6%).
Specimens were defined as true positives if: (i) C. striatum, C. propinquum, or C. simulans were recovered from the culture; or (ii) both MCDA and HRM results were positive for a culture-negative specimen [28,29,30]. Among the 42 culture-positive samples, the MCDA assay produced five false-negative results, while the HRM assay produced one (Table 2).
Ultimately, 22 culture-negative specimens were found to be positive by the MCDA and HRM assays, results that were further confirmed by nucleic acid sequencing. These 22 specimens were considered true positives. Sensitivity was calculated to be 65.6% (95% CI, 52.6–76.7%) for the culture method, 92.2% (95% CI, 82.0–97.1%) for the MCDA assay, and 98.4% (95% CI, 90.5–99.9%) for the HRM assay; specificities were 100% (95% CI, 82.8–100%) for all three assays (Table 3). No significant differences in sensitivity or specificity were found between the MCDA and HRM assays (McNemar’s chi-square test, P > 0.5). However, the HRM assay was more sensitive than the culture method (P < 0.001), and 95.5% concordance between the MCDA assay and our HRM assay (κ = 0.89 [95% CI, 0.79–0.99]) was noted.
Discussion
Due to being common components of the skin microbiota, non-diphtheriae Corynebacterium species are usually thought of as colonizers or contaminants [31, 32]. Because of this, as well as challenges in identification, they have not received a great deal of attention [33]. In recent years, the clinical relevance of these microorganisms has been recognized, particularly as a cause of respiratory-tract infections [5, 6, 34, 35]. A rapid, simple, specific, and sensitive molecular technique for identifying and differentiating between these species is essential for outbreak detection, epidemiological surveillance, and direct patient treatment, as most strains of C. striatum are resistant to multiple antimicrobials.
However, differentiation between these species remains difficult due to the low genetic variation between them. C. striatum, C. propinquum, and C. simulans have similar colony morphologies and cultural characteristics. C. simulans and C. striatum share high genetic homology, and their biochemical reactions are very similar [36]. Misidentification of C. simulans or C. propinquum as C. striatum by VITEK MS MALDI-TOF MS was observed in our investigation (data not shown), and this has also been reported previously [8]. Currently, few commercially available assays can rapidly differentiate between these species, and published assays rely on the use of pure cultures.
In this study, we developed a HRM assay that could differentiate between C. striatum, C. propinquum, and C. simulans in cultured samples and clinical specimens within approximately 2 h after DNA extraction. Analysis of the normalized melt curve produced with the universal primers generated species-specific HRM curve profiles. The three species were clearly differentiated by the melting temperature of the dissociation curves, with melting peaks at 88.91 °C for C. striatum, 88.44 °C for C. propinquum, and 87.86 °C for C. simulans. Furthermore, these three strains were successfully identified based on their different plots, which were mutually distinct.
We also evaluated the limit of detection of the newly established HRM assay. This assay could detect quantities as low as 100 fg using DNA from pure cultures as templates. Assay specificity was further evaluated by testing the genomic DNA of 69 corynebacteria clinical isolates and 30 non-target strains. Results showed that the high discriminatory power of the HRM assay developed for C. striatum, C. propinquum, and C. simulans also gave it the ability to specifically distinguish these three species from other related bacteria.
Furthermore, we showed that HRM could be applied to direct tests of clinical sputum samples. Among the 42 culture-positive samples, all were identified by HRM, while one false negative result occurred. This one sample was also found to be negative by the MCDA assay. This result suggested that DNA might have been lost or degraded during the preparation, processing, and storage of the sputum samples.
Furthermore, 22 samples were identified as negative by the culture method, but identified as positive by HRM and MCDA assays. In our study, the culture method was likely to miss one quarter (22/88) of Corynebacterium spp.-positive samples. Putative sensitivity increased from 65.6% using the culture method to 98.4% using the HRM assay. Compared with the culture method, the use of the HRM assay to detect the three Corynebacterium spp. could potentially improve overall sensitivity and reduce turnaround times.
Our HRM assay was also comparable to the MCDA method, showing a high concordance rate (95.5%) [25]. MCDA technology has revolutionized the detection of pathogens, but the established MCDA method can identify only one Corynebacterium species, C. striatum. In addition, this method requires the use of five pairs of primers, which can generate a complex mixture of various DNA products, and it can be difficult to distinguish between specific and non-specific products [37]. Compared with the MCDA assay, the HRM method we developed based on a pair of universal primers could specifically identify three Corynebacterium spp. in only one test.
In the present study, 95.3% of pulmonary infections were caused by C. striatum, 3.1% by C. propinquum, and 1.6% by C. simulans, as identified by HRM. Among members of Corynebacterium spp., C. striatum and C. propinquum have been recognized as pathogens of the respiratory tract, and have been found in cases of pneumonia and chronic obstructive pulmonary disease (COPD) in hospital settings [4, 38,39,40]. A comparative study noted that C. striatum was the most prevalent non-diphtheriae Corynebacterium found in sputum specimens, and found that it caused a large proportion of respiratory infections [1]. The high incidence of nosocomial outbreaks caused by C. striatum highlights the importance of routine species-level identification to avoid further spread and outbreaks [41].
Unlike C. striatum and C. propinquum, C. simulans has not been described as a cause of respiratory infection. Only a few well-documented infections have been reported, including one case of acute pyogenic spondylitis, one prosthetic joint infection, and one case of endocarditis [42,43,44]. The recovery of C. simulans from a respiratory specimen here appears to be novel and has clinical relevance, emphasizing its potential role as a causative pathogen of respiratory tract infections.
There were several limitations in our study. i) Sample sizes of C. propinquum and C. simulans clinical isolates were small. This was because, despite our efforts, we could not find additional isolates. ii) C. pseudodiphtheriticum and C. amycolatum, which other reports have shown to be common non-diphtheriae corynebacteria [35], were not available for this study; this might have been due to differences in the geographical distribution of non-diphtheriae corynebacteria.
Despite these limitations, our study has some strengths. First, the HRM method can identify and differentiate between C. striatum, C. propinquum, and C. simulans directly from clinical sputum samples without isolating and culturing the pathogens. This meets the need for rapid diagnosis and, more importantly, yields high diagnostic accuracy in culture-negative samples. Second, this assay could potentially be applied to recognize novel or unusual Corynebacterium spp. in clinical specimens by targeting single loci, with no need to design new assays; this is supported by previous research [45]. Third, this technique produced results in 2 h with no need for gel electrophoresis of the PCR products, avoiding sample cross-contamination. Finally, HRM analysis is a cost-effective method using common and widely available reagents and equipment, making it especially suitable for resource-limited settings.
Conclusions
In conclusion, the HRM assay described in this study is the first developed for the detection and identification of C. striatum, C. propinquum, and C. simulans from pure cultures and clinical specimens. It outperformed the culture method, proving to be capable of detecting many samples the culture method missed. The present study demonstrated this HRM assay to be a simple, rapid, accurate, sensitive, specific, and cost-effective diagnostic approach. The HRM assay is a promising alternative for identifying Corynebacterium spp., effectively complementing current methods used in clinical microbiology laboratories, and has the potential to contribute to early diagnosis and epidemiological surveillance.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Abbreviations
- HRM:
-
high-resolution melting
- MCDA:
-
multiple cross-displacement amplification
- TP:
-
true positive
- TN:
-
true negative
- FP:
-
false positive
- FN:
-
false negative
References
Neemuchwala A, Soares D, Ravirajan V, Marchand-Austin A, Kus JV, Patel SN. In vitro antibiotic susceptibility pattern of non-diphtheriae Corynebacterium isolates in Ontario, Canada, from 2011 to 2016. Antimicrob Agents Chemother. 2018;62:e01776.
Verroken A, Bauraing C, Deplano A, Bogaerts P, Huang D, Wauters G, et al. Epidemiological investigation of a nosocomial outbreak of multidrug-resistant Corynebacterium striatum at one Belgian university hospital. Clin Microbiol Infect. 2014;20:44.
Wang J, Wang Y, Du X, Cui J, Wang K, Zhang L, et al. Rapid transmission of multidrug-resistant Corynebacterium striatum among susceptible patients in a tertiary hospital in China. J Infect Dev Ctries. 2016;10:1299.
Renom F, Garau M, Rubi M, Ramis F, Galmes A, Soriano JB. Nosocomial outbreak of Corynebacterium striatum infection in patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2017;45:2064.
Nhan TX, Parienti JJ, Badiou G, Leclercq R, Cattoir V. Microbiological investigation and clinical significance of Corynebacterium spp in respiratory specimens. Diagn Microbiol Infect Dis. 2012;74:236.
Díez-Aguilar M, Ruiz-Garbajosa P, Fernández-Olmos A, Guisado P, Campo R, Quereda C, et al. Non-diphtheriae Corynebacterium species: an emerging respiratory pathogen. Eur J Clin Microbiol Infect Dis. 2013;32:769.
Hahn WO, Werth BJ, Butler-Wu SM, Rakita RM. Multidrug-resistant Corynebacterium striatum associated with increased use of parenteral antimicrobial drugs. Emerg Infect Dis. 2016;22(11):1908–14.
McMullen AR, Anderson N, Wallace MA, Shupe A, Burnham C-AD. When good bugs go bad: epidemiology and antimicrobial resistance profiles of Corynebacterium striatum, an emerging multidrug-resistant, Opportunistic Pathogen. Antimicrob Agents Chemother. 2017;61:e01111.
Alibi S, Ferjani A, Boukadida J, Cano ME, Fernandez-Martinez M, Martinez-Martinez L, et al. Occurrence of Corynebacterium striatum as an emerging antibiotic-resistant nosocomial pathogen in a Tunisian hospital. Sci Rep. 2017;7:9704.
Campanile F, Carretto E, Barbarini D, Grigis A, Falcone M, Goglio A, et al. Clonal multidrug-resistant Corynebacterium striatum strains, Italy. Emerg Infect Dis. 2009;15:75.
Hinic V, Lang C, Weisser M, Straub C, Frei R, Goldenberger D. Corynebacterium tuberculostearicum: a potentially misidentified and multiresistant Corynebacterium species isolated from clinical specimens. J Clin Microbiol. 2012;50:2561.
Santos AS, Ramos RT, Silva A, Hirata R, Mattos-Guaraldi AL, Meyer R, et al. Searching whole genome sequences for biochemical identification features of emerging and reemerging pathogenic Corynebacterium species. Funct Integr Genomics. 2018;18:593.
Alibi S, Ferjani A, Gaillot O, Marzouk M, Courcol R, Boukadida J. Identification of clinically relevant Corynebacterium strains by Api Coryne, MALDI-TOF-mass spectrometry and molecular approaches. Pathol Biol. 2015;63:153.
Bernard K. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol. 2012;50:3152.
Khamis A, Raoult D, La Scola B. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J Clin Microbiol. 2005;43:1934.
Khamis A, Raoult D, La Scola B. rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol. 2004;42:3925.
Santos CS, Ramos JN, Vieira VV, Pinheiro CS, Meyer R, Alcantara-Neves NM, et al. Efficient differentiation of Corynebacterium striatum, Corynebacterium amycolatum and Corynebacterium xerosis clinical isolates by multiplex PCR using novel species-specific primers. J Microbiol Methods. 2017;142:33.
Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597.
Talmi-Frank D, Nasereddin A, Schnur LF, Schonian G, Toz SO, Jaffe CL, et al. Detection and identification of old world Leishmania by high resolution melt analysis. Plos Negl Trop Dis. 2010;4:e581.
Papavasileiou A, Madesis PB, Karaoglanidis GS. Identification and differentiation of Monilinia species causing Brown rot of pome and stone fruit using high-resolution melting (HRM) analysis. Phytopathology. 2016;106:1055.
Winchell JM, Wolff BJ, Tiller R, Bowen MD, Hoffmaster AR. Rapid identification and discrimination of Brucella isolates by use of real-time PCR and high-resolution melt analysis. J Clin Microbiol. 2010;48:697.
Khosravi AD, Hashemzadeh M, Hashemi Shahraki A, Teimoori A. Differential identification of Mycobacterial species using high-resolution melting analysis. Front Microbiol. 2017;8:2045.
Sun Y, Cheng Y, Lin P, Zhang H, Yi L, Tong M, et al. Simultaneous detection and differentiation of Canine parvovirus and Feline parvovirus by high resolution melting analysis. BMC Vet Res. 2019;15:141.
Schonhuber W, Le Bourhis G, Tremblay J, Amann R, Kulakauskas S. Utilization of tmRNA sequences for bacterial identification. BMC Microbiol. 2001;1:20.
Qiu X, Chen D, Wang X, Zhou H, Hou X, Zhang J, et al. A novel isothermal amplification-based method for detection of Corynebacterium striatum. J Microbiol Methods. 2019;164:105675.
Zhao F, Niu L, Yan L, Nong J, Wang C, Jing W, et al. Establishment and application of multiple cross displacement amplification coupled with lateral flow biosensor (MCDA-LFB) for visual and rapid detection of Candida albicans in clinical samples. Front Cell Infect Microbiol. 2019;9:102.
Watson PF, Petrie A. Method agreement analysis: a review of correct methodology. Theriogenology. 2010;73:1167.
Shin JH, Pride DT. Comparison of three nucleic acid amplification tests and culture for detection of group B Streptococcus from enrichment broth. J Clin Microbiol. 2019;57:e01958.
Goodrich JS, Miller MB. Comparison of culture and 2 real-time polymerase chain reaction assays to detect group B Streptococcus during antepartum screening. Diagn Microbiol Infect Dis. 2007;59:17.
Miller SA, Deak E, Humphries R. Comparison of the AmpliVue, BD max system, and illumigene molecular assays for detection of group B Streptococcus in antenatal screening specimens. J Clin Microbiol. 2015;53:1938.
Baio PVP, Mota HF, Freitas AD, Gomes DLR, Ramos JN, Sant’Anna LO, et al. Clonal multidrug-resistant Corynebacterium striatum within a nosocomial environment, Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2013;108:23.
Boltin D, Katzir M, Bugoslavsky V, Yalashvili I, Brosh-Nissimov T, Fried M, et al. Corynebacterium striatum—a classic pathogen eluding diagnosis. Eur J Intern Med. 2009;20:e49.
Alatoom AA, Cazanave CJ, Cunningham SA, Ihde SM, Patel R. Identification of non-diphtheriae Corynebacterium by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50:160.
Gomila M, Renom F, Gallegos M del C, Garau M, Guerrero D, Soriano JB, et al. Identification and diversity of multiresistant Corynebacterium striatum clinical isolates by MALDI-TOF mass spectrometry and by a multigene sequencing approach. BMC Microbiol 2012;12:52.
Los-Arcos I, Len O, Martin-Gomez MT, Baroja A, Berastegui C, Deu M, et al. Clinical characteristics and outcome of lung transplant recipients with respiratory isolation of Corynebacterium spp. J Clin Microbiol. 2018;56.
Wattiau P, Janssens M, Wauters G. Corynebacterium simulans sp. nov., a non-lipophilic, fermentative Corynebacterium. Int J Syst Evol Microbiol. 2000;50:347–53.
Wang Y, Li H, Wang Y, Li H, Luo L, Xu J, et al. Development of multiple cross displacement amplification label-based gold nanoparticles lateral flow biosensor for detection of Listeria monocytogenes. Int J Nanomedicine. 2017;12:473–86.
Prats-Sánchez I, Soler-Sempere MJ, Sánchez-Hellín V. Chronic obstructive pulmonary disease exacerbation by Corynebacterium propinquum. Arch Bronconeumol. 2015;51:154–5.
Malkoçoğlu G, Gencer H, Kaya A, Dalgıç N, Bulut ME, Aktaş E. Corynebacterium propinquum bronchopneumonia in a child with ataxia telangiectasia. TurkJPediatr. 2016;58:558.
Shariff M, Aditi A, Beri K. Corynebacterium striatum: an emerging respiratory pathogen. J Infect Dev Ctries. 2018;12:581–6.
Iaria C, Stassi G, Costa GB, Biondo C, Gerace E, Noto A, et al. Outbreak of multi-resistant Corynebacterium striatum infection in an Italian general intensive care unit. J Hosp Infect. 2007;67:102.
Nagassar RP, Nicholson AM, Williams W, Bridgelal-Nagassar RJ. Diphtheroids as a cause of endocarditis in a haemodialysis patient. BMJ Case Rep. 2012;8:bcr1020114894.
Cazanave C, Greenwood-Quaintance KE, Hanssen AD, Patel R. Corynebacterium prosthetic joint infection. J Clin Microbiol. 2012;50:1518–23.
Ogasawara M, Matsuhisa T, Kondo T, Oshima R, Sugiura F, Niwa T. Pyogenic spondylitis with acute course caused by Corynebacterium simulans. J Infect Chemother. 2020;26:294–7.
Mandviwala T, Shinde R, Kalra A, Sobel JD, Akins RA. High-throughput identification and quantification of Candida species using high resolution derivative melt analysis of panfungal amplicons. J Mol Diagn. 2010;12:91.
Acknowledgements
Not applicable.
Funding
This study was supported by National Key R&D Program of China, grant numbers 2019YFC1200705 and 2017YFC1200303, and the China Special Grant for the Prevention and Control of Infectious Diseases, grant number 2017ZX10303401. The funding agencies had the role of pay the consumption materials used in this study, and had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
SX and ZL conceived and designed the experiments. SX performed the experiments, and was a major contributor in writing the manuscript. XQ, XH, HZ, DC, XW, LH, XJ, ML, JZ, and ML contributed reagents/materials/analysis tools. DL and LS supervised the research. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written informed consent was obtained from all patients included in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, and was in accordance with the medical research regulations of the Ministry of Health, China [Approval No.ICDC-2018005].
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing financial interests in relation to this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Bacterial strains used in this study.
Additional file 2.
Representative results of clinical isolates tested by high-resolution melting graphs
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xu, S., Qiu, X., Hou, X. et al. Direct detection of Corynebacterium striatum, Corynebacterium propinquum, and Corynebacterium simulans in sputum samples by high-resolution melt curve analysis. BMC Infect Dis 21, 21 (2021). https://doi.org/10.1186/s12879-020-05633-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-020-05633-z