Abstract
Background
The aim of this study was to identify the common H. pylori virulence genes among dyspeptic Southwestern Saudi patients and their association with clinical outcomes and histopathological findings to help practitioners and researchers in the region for better management of infections caused by such bacteria.
Methods
Four hundred two gastric biopsy specimens were analyzed using histopathological examination and real time-PCR. The positive 187 specimens by RT-PCR were genotyped using PCR targeting cagA, vacA and iceA genes.
Results
One hundred twenty-eight gastric biopsy specimens were positive in genotyping PCRs. The cagA, vacA, iceA1 and iceA2 genes were detected in rates of 49.2% (63/128), 100%(128/128), 42.2% (54/128), 32.8% (42/128), respectively. The vacA s1as1bm2 subtype was the highest 23.4% (30/128), followed by m2 and s1a1b subtypes which were equally detected [16.4% (21/128) for each]. The iceA genes were significantly associated with gastritis and gastric ulcer. Overall, vacA genotypes were significantly associated with gastritis, gastric and duodenal ulcers. The vacA subtypes: s1as1bm2, s1a1b and s2 m2 showed chronic active gastritis in percentages of 90.0, 81, and 84.2%, respectively. All vacA mixed genotypes showed chronic active gastritis.
Conclusions
H. pylori virulence genes are highly prevalent and diverse among patients with dyspepsia in Southwestern region of Saudi Arabia. The iceA genes and the different vacA subtypes are significantly associated with the clinical outcomes and histopathological changes especially chronic active gastritis.
Similar content being viewed by others
Background
The World Health Organization (WHO) considers H. pylori as a bacterial carcinogen [1]. It has been reported that infection with H. pylori accounts for 75% of non-cardia gastric malignancy worldwide [2]. The H. pylori-induced gastritis can lead to atrophic gastritis, which in turn may progress to intestinal metaplasia, dysplasia and neoplasia, gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (MALT) [3, 4]. The mechanism of pathogenic effect of H. pylori is unclear, but it may be related to complex host bacterial interactions triggered by virulence genes. These effects may also be enhanced by the invasiveness of H. pylori [5]. The variation in the clinical outcomes may be attributed to the considerable genetic variation exists between strains of H. pylori [6, 7]. The major H. pylori virulence genes are vacuolating cytotoxin A (vacA) and cytotoxin associated antigen A (cagA), and genes induced by contact with gastric epithelium (ice genes), which are of two types, iceA1 and iceA2 [8]. The vacA gene has many alleles such as vacA s1, s2, m1 and m2 [9]. The cagA gene is not present in all H. pylori strains, but is associated with clinical outcomes such as gastritis and peptic ulcer disease (PUD) as well as higher risk of occurrence of gastric carcinoma. [10, 11]. The vacA gene is found in all H. pylori strains, and some of its subtypes are associated with chronic inflammation of gastric mucosa and development of PUD [12]. This pathogenicity island is usually absent in H. pylori strains isolated from persons who are carriers of H. pylori, but are asymptomatic [13]. An important study on Saudi patients, which was conducted to detect the presence of cagA, iceA1, and iceA2 virulence genes in H. pylori from gastric biopsies, has reported a correlation between these genotypes and the development of PUD and gastritis [14]. A recent study in the Kingdom of Saudi Arabia has reported a high prevalence rate of H. pylori among dyspeptic patients and a strong correlation with duodenal ulcer [15]. Some studies have shown that the occurrence of gastric cancer is independent of cagA status and of other virulence factors (cagE, cagT, vacA, babA and hrgA) [16,17,18]. However, a large number of other studies have shown an increased risk of gastric cancer in people with cagA positive H. pylori [19,20,21]. Recently, significant association between vacA s1 am1 and babA2 genotypes with cases of gastric ulcer and cancer has been reported in Saudi Arabia [22].
Furthermore, many studies across the world reported that the patients who are infected with vacA s1 or m1 H. pylori strains have an increased risk of PUD and gastric carcinoma, in comparison to individuals infected with s2, m2 H. pylori strains [23, 24]. Moreover, another study has shown the role of vacA s1 in the development of gastric carcinoma in the absence of cagA genes [25].
The main objectives of this study were to identify the distribution of H. pylori cagA, vacA and iceA genotypes among Southwestern Saudi dyspeptic patients, and to correlate them with the clinical outcomes and histopathological changes.
Methods
Patients and clinical specimens
Gastric biopsies were collected from 404 Saudi patients who attended gastroenterology clinics at general hospitals in Jazan (Southwestern region of Saudi Arabia). Gastric biopsies were obtained from each patient through the upper gastrointestinal endoscopy. The endoscopic examination findings were: gastritis, gastric ulcer, duodenal ulcer and gastric cancer.
DNA extraction
There were non-sufficient biopsy materials in two cases, and from all the remaining 402 cases, DNA was extracted. First, the biopsy specimens were minced into small pieces by sterile scalpels, then the DNA was extracted by using DNeasy blood & tissue kit (Qiagen) according to manufacturer’s instructions.
Real time (RT) PCR detection of H. pylori
All DNA samples extracted from the gastric biopsies (402 specimens) were submitted for quantitative real time PCR amplification using “genesig Quantification of Helicobacter pylori” kit (PrimerDesign Ltd. Southampton, United Kingdom) which is primer-probe based and targets the RNA polymerase beta-subunit (rpoB) gene of H. pylori. The reactions were carried in 20 μl volumes containing 10 μl of “oasig™ 2× qPCR Mastermix” (PrimerDesign Ltd.), 1 μl H. pylori specific primer/probe mix, 1 μl internal control primer/probe mix, 2 μl of internal control DNA, 3 μl of the sample extracted DNA, made up to a total volume of 20 μl by adding RNase/DNase free water supplied with the kit. The reactions were carried out using the Smart Cycler (Cepheid, Italy). Positive control (H. pylori DNA supplied with the kit) and negative control (contains RNase/ DNase free water) reactions were included in each PCR run. The PCR cycling conditions were 50 cycles of denaturation at 95 °C for 10 s and data collection at 60 °C for 60 s.
Genotyping by detection of virulence genes of H. pylori
Overall, RT-PCR was positive in 187 specimens. The DNAs from these all 187 specimens were submitted for detection for cagA, vacA and iceA virulence genes. These genes were used for genotyping of the studied H. pylori strains.
Detection of these virulence genes was carried out by PCR using wide array of primers listed in Table 1. Each virulence gene was detected by a single PCR using its specified primers pair. All amplification reactions were carried out in total volumes of 50 μl containing reaction buffer 5 μl of 10× PCR buffer supplemented with MgCl2 (50 mM KCl, 10 mM Tris-HCl [pH 9.0], 15 mM MgCl2], deoxynucleoside triphosphate mix [0.2 mM each of dATP, dCTP, dGTP, and dTTP] (Roche, Germany), 0.4 μM of each primer, 2.5 U of FastStart Taq DNA polymerase (Roche, Germany), 5 μl of template DNA, made up to a total volume of 50 μl with sterile RNase and DNase free water. The PCR cycling conditions were 30 cycles of 94 °C for 10 min, 94 °C for 2 min, 55 °C for 2 min, and 72 °C for 2 min, followed by an extension step of 72 °C for 10 min. PCR was carried out using a thermal cycler (Eppendorf Mastercycler gradient, Germany). The resulting products were separated in 1.5% agarose gels and Tris-acetate-EDTA buffer (Promega, Madison, USA) and stained with ethidium bromide, and then images were captured using the Uvitec imaging system (Cambridge, UK).
Histopathological microscopic examination
All biopsies were submitted for histopathological examination to investigate the tissue inflammatory changes associated with infection. The biopsies were fixed in 10% formalin overnight, processed and embedded in paraffin wax. Four micron–thick tissue sections were taken and stained with the routine hematoxylin and eosin (H&E) stain, and modified Giemsa stain (Sheedhan’s modified method) [26] then examined according to the classification and grading system of Sydney [27].
Statistical analysis
The SPSS version 20.0 (IBM Corp., NY, USA) was used for data analysis. Statistical analysis involved descriptive statistics as well as inferential statistics. Descriptive statistics included simple tabulation, frequencies and proportion for categorical variables including cross-tabulations. Differences in proportions were evaluated for significance using Chi Square/Fisher Exact test. P-values less than 0.05 were used to indicate statistical significance.
Results
A total of 187 gastric biopsies were positive for presence of H. pylori by RT-PCR. Only 128 of them gave positive results in the genotyping PCR reactions.
Genotyping PCR results of the three tested genes
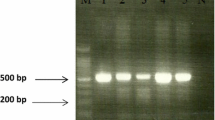
We used two primer pairs to study the cagA gene status; one pair targeted cagA gene itself with production of amplified product of 349 bp size (Fig. 1a) and the other called cag empty site (550 bp in size) which is positive in cases negative for cagA gene (Fig. 1b). The PCR targeted iceA1 produced 247 bp-sized products (Fig. 1c); while the PCR amplified products of iceA2 gene were either 229 or 334 bp in size (Fig. 1d). Regarding vacA status (genotyping), the tested DNA samples were surveyed for s (signal) and m (middle) regions of vacA gene by multiple sets of primers targeted (s1a, s1b, s1c and s2 alleles of s region, and m1, m2 alleles of m region). The amplified products of these different genes are shown in Fig. 2. The specimens carried s1 and s2 or m1 and m2 were considered as mixed infection. The s1a1b was considered as a subtype of the s1 genotype.
Agarose gel electrophoresis of the PCR-based genotyping amplified products of (a) cagA gene (349 bp). Lanes; M; 100 bp DNA ladder (Solis Biodyne). Lanes 1, 2, 3, 5, 7, 9, 11, and 15 are cagA positive while remaining lanes are negative for this gene. b cag empty site (550 bp). Lanes; M; 100 bp molecular DNA marker (Cleaver Scientific Ltd.). Lanes 1, 2, 4, 5, 7, 8 are cagA empty site positive while the remaining lanes are negative for this site. c iceA1 gene (247 bp). Lanes; M; 100 bp molecular DNA marker (Cleaver Scientific Ltd.). Lanes 1, 2, 3, 4, 5, 7, 8, 9, 11, 13, 14 and 15 are iceA1 positive while remaining lanes are negative for this gene. Note that PCR bands in lanes 4 and 11 are faint. d iceA2 gene (229 or 334 bp). Lanes; M; 100 bp molecular DNA marker (Cleaver Scientific Ltd.). Lanes 1, 4, 6, 10, 11, and 12 are iceA2 positive while lanes 2, 3, 5, 7–9, 13–15 are negative for this gene. Note that PCR products in lanes 1, 6 and 12 are of 334 bp size, while lanes 4, 10 and 11 have 229 bp PCR products
Agarose gel electrophoresis of the PCR-based genotyping amplified products of vacA gene. a PCR products of vacA s1a gene (212 bp). Lanes; M; 100 bp molecular DNA marker (Cleaver Scientific Ltd.). Lanes 4, 5, 8, 9, and 10 are positive while lanes 1, 2, 3, 6, 7 and 9 are negative. b PCR products of vacA s1b gene (187 bp). Lanes; M; 100 bp molecular DNA marker (Cleaver Scientific Ltd.). Lanes 2, 5, 7, 8 and 9, while lanes 10 are positive while lanes 1, 3, 4 and 10 are negative. c products of vacA s2 gene (199 bp). a) Positive photo. b) Negative photo. Lanes; M; 100 bp molecular DNA marker (Cleaver Scientific Ltd.). Lanes 7 and 9 are positive while lanes 1, 2, 3, 4, 5, 6 and 8 are negative for this gene. d amplified products of vacA m1 gene (290 bp). a Positive photo. b Negative photo. Lanes; M; 100 bp molecular DNA marker (Cleaver Scientific Ltd.). Lanes 4 and 8 are positive while lanes 1, 2, 3, 5, 6, 7 and 9 are negative for this gene. e products of vacA m2 gene (352 bp). Lanes; M; 100 bp molecular DNA marker (Cleaver Scientific Ltd.). Lanes 1, 2, 3, 5, 6, 7 and 10 are positive while lanes 4, 8 and 9 are negative for this gene
Prevalence and distribution of H. pylori cagA, iceA and vacA genes
The results revealed that 49.2% (63/128) of tested H. pylori were cagA positive, 48.4% (62/128) were cagA negative and 2.3% (3/128) were mixed (positive for both cagA and cagA empty site genes). Regarding iceA gene: 42.2% (54/128) were iceA1 positive, 32.8% (42/128) were iceA2 positive, 3.9% (5/128) were iceA1 and iceA2 positive (mixed). Numerous subtypes of vacA gene were obtained as shown in Table 2. s1as1bm2 subtype was the highest 23.4% (30/128), followed by m2 and s1a1b subtypes which were equally detected 16.4% (21/128) for each. As regards the distribution of vacA genotypes according cagA status, s1a1bm2 and s1a1b alleles were detected in higher levels in cagA positive subtypes, 31.7 and 27%, respectively. The distribution of vacA alleles according to iceA status revealed that s1a1b and s1as1bm2 were highly detected in iceA1 subtypes (in rates of 20.4 and 18.5%, respectively), most of the iceA2 subtypes had s1as1bm2, m2 and s2 m2 alleles (28.6, 23.8, and 16.7%, respectively), while three of the iceA mixed five subtypes were having s1as1bm2 alleles, and s1as1b allele was the mostly found allele in iceA negative subtypes (Table 2).
Distribution of genotypes according to the patients’ gender and age
Table 3 shows distribution of H. pylori genes (vacA, cagA and iceA) according to gender and age groups. Regarding the gender, overall, there was no significant difference in distribution of vacA, cagA and iceA genes between males and females. However, vacA subtype m2 was higher among females (66.7% in females versus 33.3% in males). In contrast to that, vacA s1a1b was higher in males (61.9%), whereas in females it was 38.1%. As regards to patients’ age groups, vacA m2 was higher (61.9%) in the young adults (age group between 13 and 29 years), while with an advance in age (age groups between 30 and 49 years and age groups above 50 years), the prevalence of this gene was decreasing (23.8 and 14.3%, respectively). However, no significant differences were found in distribution of other vacA subtypes, cagA and iceA genes among the different age groups.
Association of cagA, vacA and iceA with clinical outcomes and histopathological changes
The clinical outcomes were assessed endoscopically as normal, gastritis, gastric ulcer and duodenal ulcers, while the histopathological changes were determined by histopathological examination and were classified into: mild chronic gastritis, moderate chronic gastritis, severe gastritis, chromic active gastritis. The relationship between H. pylori genes (cagA, iceA and vacA) and the clinical outcomes (endoscopic findings) and histopathological findings are presented in Table 4 and Fig. 3.
Although no significant differences of cagA status according to clinical outcome or histopathological changes, more than 80% of duodenal ulcers were cagA positive and all 3 mixed cagA genotypes were positive for chronic active gastritis. The iceA1and iceA2 genes were significantly associated with gastritis and gastric ulcer (p-value of 0.026) and there was an apparent association between iceA1 and iceA2 genes and chronic active gastritis with percentages of 38.5 and 34.4%, respectively, in comparison to iceA negative (24.0%). Moreover, all mixed iceA cases showed gastritis either moderate chronic or chronic active.
The prevalence of H pylori genes vacA according endoscopic findings and histopathological changes is shown in Fig. 3. Overall, vacA genotype was significantly associated with clinical outcomes (p-value of 0.000), as vacA subtypes: s1a1b, s1as1bm2, s2 m2 and m2, s1bm2 were significantly correlated to gastritis, whereas, subtypes s1 am1, s1 am2, m1were significantly associated with gastric and duodenal ulcers. The subtype vacA s1bm2 was associated with gastritis in 5 cases (45.5%) and with duodenal ulcer in 4 cases (36.4%). Eighty percent of mixed vacA genotypes showed gastritis. Regarding the association between vacA genotype and the histopathological changes, although there was no statistical significance (p- value of 0.232), vacA subtypes: s1as1bm2, s1a1b and s2 m2 showed chronic active gastritis in percentages of 90.0, 81, 84.2%, respectively. The subtype vacA s1bm2 was associated mainly with chronic active gastritis (9 cases, 81.8%), while the subtype vacA m2 was presented with chronic active gastritis in 10 (47.6%) cases and with moderate active gastritis in 9 (42.9%) cases. All vacA mixed genotypes showed chronic active gastritis.
Discussion
Previous studies on the association of H. pylori genotypes with the clinical outcomes in Saudi Arabia are so scarce [22]. Our data revealed that the overall prevalence of cagA gene was 49.2% (63/128). Regarding the distributions of cagA gene in relation to endoscopic and histopathological findings, more than 80% of duodenal ulcer cases were cagA positive and all the 3 cagA mixed types were positive in cases of chronic active gastritis. However, no statistical significance was found. The distribution of cagA genes and its association with the clinical outcomes in our study is consistent with the previous studies on H. pylori genes in Saudi Arabia by Momenah et al, Marie et al.and Kadi et al., in which, they found that the overall prevalence of cagA was 52.4, 62 and 81.8%, respectively [10, 14, 28]. In agreement with our findings, Momenah et al. and Marie et al. found an association between cagA gene and peptic ulcers (100 and 71%, respectively). However, Kadi et al. revealed no significant association. In contrast to our findings, Seriki et al and colleagues reported a high prevalence of cagA gene among the studied patients (100%). As similar to our findings, they found no association of stastistical significance between cagA genes and clinical outcomes [29]. On the other hand, a recent study from Brazil by Sallas et al. found cagA gene in 50% of H.pylori isolates [30]. This percentage is consistent with ours. Also, similar to our data another study from Ecuador reported cagA prevalence rate of 45.9% [30, 31]. In contrary to our results Sallas et al. [30] found no correlation between cagA gene alone and clinical outcomes. However, they found significant asscoiation between cagA/vacA genes and chronic gastritis and gastric cancer. Our results are different from a recent Mexican study by Cantu et al. and colleagues, who investigated for H.pylori genotypes in oral cavities of 100 asymptomatic children [32]. They reproted cagA prevalence rate of 80.8% among the studied subjects. This difference may be due to variations in study population, ages and the site of sample collection. However, in agreement with our data, they found correlation between cagA status and clinical outcomes. More recently, an Egyptian study by Abu-Taleb et al. reported cagA prevalence of 57.4% in the studied subjects [33]. In other parts of the world, the rates of H. pylori cagA genes were 90% in East Asia (Japan and Korea) and 60% in North America, Europe and Cuba [34]. The wide variation of cagA prevalence rates a cross the globe could be attributed to the differences in: study sizes, socioeconomic, geographical as well as genetic factors.
In the present study, vacA gene was detected in all the isolates (100%). Similar to our results, El Khadir et al. found vacA detection rate of 99% in Morocco, whereas low rates of vacA gene (90, 93%) were reported in Ethiopia and Netherland, respectively [35]. In the current study, the most predominant vacA subtype were s1as1bm2 (23.4%), m2 (16.4%) and s1a1b (16.4%). Whereas, s2 m2 subtype was found in 14.8% (19/128). On the other hand, our data showed that: s1 am1, s1bm1 and m1 were the least vacA subtypes (representing 0.8%). In agreement with our findings, Pajavand et al. [9] reported that the predominant vacA subtypes were s1 m2 and s2 m2 with frequency of 39.5 and 50%, respectively, whereas the least subtype was s1 m1 with frequency of only 7%. However, vacAs1 m1 and vacAs1 m2 were the predominat according to Cantu et al. report [32]. Furthermore, Marie et al. [10] investigated vacA genes in Saudi population. He reported frequencies as follows: vacA s1 m1 (28%), s1 m2 (40%) and s2 m2 (26%) in subjects with peptic ulcer and gastritis. Our results were consistent with his findings regarding distribution of vacA s1 m2 and s2 m2 subtypes. In contrast to Marie et al. [10], Sallas et al. [30] and Cantu et al. [32] results, we found very low s1 am1 rate in the H. pylori + ve strains. The low vacA s1 m1 in the present study could be attributed to the differences in: study sizes, regions and population.
It has been well established that vacA s1/m1 is associated with severe damage to the gastric epithelium [36]. Worldwide, numerous studies had reported vacAs1 m1 frequency of 24–84% [9].
With respect to the association of these vacA subtypes with the clinical outcomes, our results revealed a significant association (with p-value of 0.000 for all). In addition, vacA s1 am1,s1 am2, s1bm2 were the predominant in gastric ulcer and duodenal ulcer. These findings were in agreement with Sallas et al. [35] and Pajavand et al. [9] results. A recent study from Saudi Arabia, showed high vacA s1 in subjects with gastric ulcer and gastric cancer (80 and 100%, respectively) [22]. Several studies from China, Middle East, Africa and Western countries have shown that individuals infected with vacA s1 or m1 H. pylori strains have an increased risk of peptic ulcer or gastric cancer compared with individuals infected with s2 or m2 strains, so s2 and m2 strains are considered as less virulent [24, 37, 38]. Surprisingly, Sedaghat et al. [8] found no significant correlation between vacA genes and clinical outcomes. In the present study we detected high rate of vacAs1a1b (16%).This finding is inconsistent with the previous studies. Interestingly, vacAs1a1b was found to be significantly associated with chronic active gastritis. It is well-known that chronic active gastritis can evolve into atrophic gastritis, intestinal metaplasia, dysplasia, which may eventually lead to the development of gastric cancer [39].
In the current study, mixed vacA, cagA and iceA genotypes were seen in 3.9%, 2.3, 3.9% of cases. The rates of mixed genotypes vary from 0 to 85% in different populations across the world [40]. In the present study, we found correlation between cagA positive strains and the most virulent vacA subtypes such as s1a1bm2 and s1a1b, whereas in cagA negative strains, the less virulent subtypes such s2 m2 and m2 were the most predominant. Boukharis et al. found a significant association between vacA s1 m1 and intestinal metaplasia [41]. Also, Matsunari et al. found that the association of cagA strains with vacA s1 increase the risk of atrophic gastritis and gastric cancer [42]. However, Hussein et al revealed no correlation between H. pylori virulence genes (vacA, cagA and dupA) and histopathological changes in Iraqi patients [36].
With respect to iceA genotype/subtypes, our results revealed that 42.2% of H. pylori were harboring iceA1, 32.8% were iceA2, whereas 3.9% were mixed (iceA1/iceA2). Similar to our findings Abu-Taleb et al. [33] and Sedaghat et al. [8] reported iceA gene prevalence rates of 46.29 and 48.6%, respectively. Our results showed a significant association of iceA1 and iceA2 genes with gastritis and gastric ulcer (p-value of 0.026). Furthermore, there was a clear association between iceA1, iceA2 and chronic active gastritis (38.5, 34.4%, respectively), although it was not statistically significant. Kadi et al. [14], Momenah et al. [28], and Abu-Taleb et al. [33] were in agreement with our data regarding the correlation of iceA1 with gastritis and peptic ulcer. Also, numerous studies have shown a similar association between iceA genes and the clinical outcomes [43]. In contrast to our results, some studies from Brazil and Iran showed no association between iceA1 gene and gastritis [8, 44].
As similar to other studies, our results revealed no gender differences in terms of distribution of H. pylori cagA and iceA genotypes [14]. However, some vacA subtypes revealed gender variations in the present study. We found high rates of the less virulent vacA m2 among females (65%), compared to 35% in males. Whereas, vacA s1a1b was predominant in males (61.9%), compared to 38.1% in females. In concordance with our results, El Khadir et al. found that vacA subtypes (s1, i1 and m1) were more prevalent in males than in females [35]. In contrary to our results, he found no differences in relation to age. However, we found that vacA m2 gene was higher (61.9%) in young adults (age group between 13 and 29 years). While, in older age groups, the prevalence of this gene was decreasing (23.8 and 14.3% in the age group between 30 and 49 years and above 50 years, respectively). Similar observations were reported by Feliciano et al. [34].
Conclusions
This study showed a high prevalence and diversity of H. pylori virulence genes among patients with dyspepsia in Southwestern region of Saudi Arabia. There was a significant association between different vacA subtypes and iceA genes with the clinical outcomes. Moreover, there were some variations in the distribution of virulence genes with respect to age and gender. Furthermore, there was a significant association between some vacA subtypes, iceA genes and histopathological changes.
Abbreviations
- Bp:
-
Base pair
- cagA :
-
cytotoxin-associated gene A
- dATP:
-
Deoxyadenosine triphosphate
- dCTP:
-
Deoxycytidine triphosphate
- dGTP:
-
Deoxyguanosine triphosphate[
- DNA:
-
Deoxyribonucleic acid
- DNase:
-
Deoxyribonuclease enzyme
- dTTP:
-
Deoxythymidine triphosphate
- EDTA:
-
Ethylenediaminetetraacetic acid
- H&E:
-
Hematoxylin and eosin
- H. pylori :
-
Helicobacter pylori bacteria
- PCR:
-
Polymerase chain reaction
- PUD:
-
peptic ulcer disease
- RNase:
-
Ribonuclease enzyme
- RT-PCR:
-
Real time polymerase chain reaction
- vacA :
-
Vacuolating cytotoxin gene A
- WHO:
-
World Health Organsation
References
De Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–15.
Dixon MF. Pathology of gastritis and peptic ulceration. In: Mobley HLT, Mendz GL, Hazell SL, editors. Helicobacter pylori: physiology and genetics. Washington, DC: ASM Press; 2001. p. 459–69.
Wang HC, Cheng FC, Wu MS, Shu HY, Sun HS, Wang YC, et al. Genome sequences of three Helicobacter pylori strains from patients with gastric mucosa-associated lymphoid tissue lymphoma. Genome Announc. 2015;3:e00229–15.
Souod N, Sarshar M, Dabiri H, Momtaz H, Kargar M, Mohammadzadeh A, et al. The study of the oipA and dupA genes in Helicobacter pylori strains and their relationship with different gastroduodenal diseases. Gastroenterol Hepatol Bed Bench. 2015;8(Suppl1):S47–54.
Amieva MR. El–Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306–23.
Semino-Mora C, Doi SQ, Marty A, Simko V, Carlstedt I, Dubois A. Intracellular and interstitial expression of Helicobacter pylori virulence genes in gastric precancerous intestinal metaplasia and adenocarcinoma. J Infect Dis. 2003;187:1165–77.
Camorlinga-Ponce M, Romo C, Gonzalez-Valencia G, Munoz O, Torres J. Topographical localisation of cagA positive and cagA negative Helicobacter pylori strains in the gastric mucosa; an in situ hybridisation study. J Clin Pathol. 2004;57:822–8.
Sedaghat H, Moniri R, Jamali R, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, babA2, and oipA genotypes in patients with upper gastrointestinal diseases. Iran J Microbiol. 2014;6(1):14–21.
Pajavand H, Alvandi A, Mohajeri P, et al. High frequency of vacA s1m2 genotypes among Helicobacter pylori isolates from patients with gastroduodenal disorders in Kermanshah, Iran. Jundishapur J Microbiol. 2015;8(11):e25425.
Marie MA. Relationship between Helicobacter pylori virulence genes and clinical outcomes in Saudi patients. J Korean Med Sci. 2012;27:190–3.
Kim SS, Ruiz VE, Carroll JD, Moss SF. Helicobacter pylori in the pathogenesis of gastric cancer and gastric lymphoma. Cancer Lett. 2012;305:228–38.
Siddique I, Al-Qabandi A, Al-Ali J, Alazmi W, Memon A, , Mustafa AS, et al. Association between Helicobacter pylori genotypes and severity of chronic gastritis, peptic ulcer disease and gastric mucosal interleukin-8 levels: evidence from a study in the Middle East. Gut Pathog 2014;6: 41.
Baldwin DN, Shepherd B, Kraemer P, Hall MK, Sycuro LK, Pinto-Santini DM, et al. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun. 2007;75:1005–16.
Kadi RH, Halawani EM, Abdelkader HS. Prevalence of H. pylori strains harbouring cagA and iceA virulence genes in Saudi patients with gastritis and peptic ulcer disease. Microbiol Discov. 2014;2:2.
Alhussaini M. Prevalence of H.pylori among patients with different gastrointestinal disorders in Saudi Arabia. Med J Indones. 2016;25:214–420.
Fischbach W, Chan AO, Wong BC. Helicobacter pylori and gastric malignancy. Helicobacter. 2005;10(s1):34–9.
Tiwari SK, Khan AA, Manoj G, Ahmed S, Abid Z, Habeeb A, et al. A simple multiplex PCR assay for diagnosing virulent Helicobacter pylori infection in human gastric biopsy specimens from subjects with gastric carcinoma and other gastro-duodenal diseases. J App Microbiol. 2007;103:2353–60.
Koehler CI, Mues MB, Dienes HP, Kriegsmann J, Schirmacher P, Odenthal M. Helicobacter pylori genotyping in gastric adenocarcinoma and MALT lymphoma by multiplex PCR analyses of paraffin wax embedded tissues. Mol Pathol. 2003;56:36–42.
Amjad N, Osman HA, Razak NA, Kassian J, Din J, Abdullah NB. Clinical significance of Helicobacter pylori cagA and iceA genotype status. World J Gastroenterol. 2010;16:4443–7.
Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–5.
Kim JY, Kim N, Nam RH, Suh JH, Chang H, Lee JW, et al. Association of polymorphisms in virulence factor of Helicobacter pylori and gastroduodenal diseases in South Korea. J Gastroenterol Hepatol. 2014;29:984–91.
Bibi F, Alvi SA, Sawan SA, Yasir M, Sawan A, Jiman-Fatani AA, et al. Detection and genotyping of Helicobacter pylori among gastric ulcer and cancer patients from Saudi Arabia. Pak J Med Sci. 2017;33:320–4.
Momtaz H, Souod N, Dabiri H, Sarshar M. Study of Helicobacter pylori genotype status in saliva, dental plaques, stool and gastric biopsy samples. World J Gastroenterol. 2012;18:2105–11.
da Costa DM, dos Santos PE, Rabenhorst SH. What exists beyond cagA and vacA? Helicobacter pylori genes in gastric diseases. World J Gastroenterol. 2015;21:10563–72.
Carmo AP, Rabenhorst SH. Importance of vacAs1 gene in gastric cancer patients infected with cagA-negative Helicobacter pylori. APMIS. 2011;119:485–6.
Gray SF, Wyatt JI, Rathbone BJ. Simplified techniques for identifying Campylobacter pyloridis. J Clin Pathol. 1986;39:1279.
Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney system. International workshop on the histopathology of gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–81.
Momenah AM, Tayeb MT. Helicobacter pylori cagA and iceA genotypes status and risk of peptic ulcer in Saudi patients. Saudi Med J. 2007;28(3):382–5.
Seriki AT, Smith SI, Adeleye AI, Fowora MA, Lesi O, et al. Helicobacter pylori cytotoxin-associated gene a protein among adult dyspeptic patients in South-Western Nigeria. Afr J Microbiol Res. 2017;11:681–6.
Sallas M, Melchiades J, Zabaglia L, Moreno J, et al. Prevalence of Helicobacter pylori vacA,cagA,,dupA and oipA genotypes in patients with Gastric Disease. Adv Mircobiol. 2017;7(1):1–9.
Sasaki T, Hirai I, Yamamoto Y. Analysis of Helicobacter pylori infection in a healthy Panamanian population. Kansenshogaku Zasshi. 2009;83:127–32.
Mendoza-Cantú A, Urrutia-Baca V, Urbina-Ríos C, De la Garza-Ramos M. et al. Prevalence of Helicobacter pylori vacA Genotypes and cagA Gene in Dental Plaque of Asymptomatic Mexican Children. BioMed Res Int. 2017. Article ID 4923640, 10 pages.
Abu-Taleb AMF, Abdelattef RS, Abdel-Hady AA, et al. Prevalence of Helicobacter pylori cagA and iceA genes and their association with gastrointestinal diseases. Int J Microbiol. 2018;2018:4809093.
Feliciano O, Gutierrez O, Valdés L, Fragoso T, Calderin AM, Valdes AE, et al. Prevalence of Helicobacter pylori vacA, cagA, and iceA genotypes in Cuban patients with upper gastrointestinal diseases. Biomed Res Int. 2015;2015:753710.
El Khadir M, Alaoui Boukhris S, Benajah D-A, El Rhazi K, Ibrahimi SA, El Abkari M, et al. VacA and CagA status as biomarker of two opposite end outcomes of Helicobacter pylori infection (gastric Cancer and duodenal ulcer) in a Moroccan population. PLoS One. 2017;12:e0170616.
Hussein NR, Napaki SM, Atherton JC. A study of Helicobacter pylori-associated gastritis patterns in Iraq and their association with strain virulence. Saudi J Gastroenterol. 2009;15:125–7.
Aziz F, Chen X, Yang X, Yan Q. Prevalence and correlation with clinical diseases of Helicobacter pylori cagA and vacA genotype among gastric patients from Northeast China. Biomed Res Int. 2014;2014:142980.
Uchida T, Nguyen LT, Takayama A, Okimoto T, Kodama M, Murakami K, et al. Analysis of virulence factors of Helicobacter pylori isolated from a Vietnamese population. BMC Microbiol. 2009;9:175.
Pinto-Ribeiro I, Ferreira RM, Batalha S, Hlaing T, Wong SI, Carneiro F, et al. Helicobacter pylori vacA Genotypes in Chronic Gastritis and Gastric Carcinoma Patients from Macau, China. Toxins (Basel). 2016;8:142.
Lai CH, Huang JC, Chiang-Ni C, Li JP, Wu LT, Wu HS, et al. Mixed infections of Helicobacter pylori isolated from patients with gastrointestinal diseases in Taiwan. Gastroenterol Res Pract. 2016;2016:7521913.
Boukhris S, Amarti A, El Rhazi K, El Khadir M, Benajah DA, Ibrahimi SA, et al. Helicobacter pylori genotypes associated with gastric Histo-pathological damages in a Moroccan population. PLoS One. 2013;8:e82646.
Matsunari O, Miftahussurur M, Shiota S, Suzuki R, Vilaichone RK, Uchida T, et al. Rare Helicobacter pylori virulence genotypes in Bhutan. Sci Rep. 2016;6:22584.
Shiota S, Suzuki R, Yamaoka Y. The significance of virulence factors in Helicobacter pylori. J Dig Dis. 2013;14:341–9.
Gatti LL, Módena JL, Payão SL, Smith MD, Fukuhara Y, Módena JL, et al. Prevalence of Helicobacter pylori cagA, iceA and babA2 alleles in Brazilian patients with upper gastrointestinal diseases. Acta Trop. 2006;100:232–40.
Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37:2274–9.
Atherton JC, Cao P, Peek RM Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–7.
Tummuru MK, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799–809.
Peek RM Jr, Thompson SA, Donahue JP, Tham KT, Atherton JC, Blaser MJ, et al. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians. 1998;110:531–44.
Mukhopadhyay AK, Kersulyte D, Jeong JY, Datta S, Ito Y, Chowdhury A, et al. Distinctiveness of genotypes of Helicobacter pylori in Calcutta. India J Bacteriol. 2000;182:3219–27.
Acknowledgements
We would like to show our deep gratitude to KASCT for the financial and technical support. Our gratitude is extended to our colleagues in gastrointestinal units at general Hospitals in Jazan region for their support.
Funding
This research received financial support from King Abdulaziz City for Science and Technology (KACST), grant number ARP-47-32 for 20 months under the annual grant program. In addition to acting as funding body, KACST also has a role of monitoring the progress of the project by regular technical and financial reports. The funding body has no role in the design of the study or collection, analysis, or interpretation of data or in writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
MA, AS, AE, EE, TA and HA conceptualized and designed the research project, and carried out the practical work. MM performed the statistical analysis and finalized the manuscript. All authors provided significant input in writing of the manuscript, read, revised and approved the final version of it.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Informed written consent was obtained from all enrolled patients, per the ethics guidelines in Saudi Arabia. Ethical approval for the current study was obtained from the Ethics Committee of the Faculty of Medicine, Jazan University (FMRERC).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Akeel, M., Shehata, A., Elhafey, A. et al. Helicobacter pylori vacA, cagA and iceA genotypes in dyspeptic patients from southwestern region, Saudi Arabia: distribution and association with clinical outcomes and histopathological changes. BMC Gastroenterol 19, 16 (2019). https://doi.org/10.1186/s12876-019-0934-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-019-0934-z