Abstract
Background
The gibberellic acid-stimulated Arabidopsis (GASA) gene encodes a class of cysteine-rich functional proteins and is ubiquitous in plants. Most GASA proteins are influence the signal transmission of plant hormones and regulate plant growth and development, however, their function in Jatropha curcas is still unknown.
Results
In this study, we cloned JcGASA6, a member of the GASA family, from J. curcas. The JcGASA6 protein has a GASA-conserved domain and is located in the tonoplast. The three-dimensional structure of the JcGASA6 protein is highly consistent with the antibacterial protein Snakin-1. Additionally, the results of the yeast one-hybrid (Y1H) assay showed that JcGASA6 was activated by JcERF1, JcPYL9, and JcFLX. The results of the Y2H assay showed that both JcCNR8 and JcSIZ1 could interact with JcGASA6 in the nucleus. The expression of JcGASA6 increased continuously during male flower development, and the overexpression of JcGASA6 was associated with filament elongation of the stamens in tobacco.
Conclusion
JcGASA6, a member of the GASA family in J. curcas, play an important role in growth regulation and floral development (especially in male flower). It is also involved in the signal transduction of hormones, such as ABA, ET, GA, BR, and SA. Also, JcGASA6 is a potential antimicrobial protein determined by its three-dimensional structure.
Similar content being viewed by others
Background
The gibberellic acid-stimulated Arabidopsis (GASA) gene family is a plant-specific group of genes. This family comprises many genes, and most members are regulated by gibberellin (GA). By far, a great number of GASA genes have been isolated and identified from Populus trichocarpa, Glycine max, Arabidopsis, and Solanum tuberosum [1,2,3,4]. The structure and function of this kind of proteins are deeply understood by the analysis of different GASA members identified from a variety of plant species. GASA protein family is a class of cysteine-rich functional proteins. They all have a highly conserved GASA domain (marked by 12 cysteines), which is essential for their normal function [1, 2].
The GASA proteins regulate plant growth and development, including seed germination, lateral root formation, stem elongation, flowering, flower and fruit development, biotic and abiotic stress responses, and signal transduction of hormones [5,6,7,8,9]. The functions of the genes in the GASA family have been mostly studied in Arabidopsis. Most GASA proteins are involved in hormone signal transduction. GAST1, GASA4, GASA6, GASA9 and GASA14 are involved in gibberellin signal transduction and are located downstream of DELLA protein [5, 10]. The GASA proteins are also involved abscisic acid (ABA) signal transduction. The expression of NtGASA1, NtGASA2, NtGASA9 and AtGASA14 can be induced by ABA [5, 11]. However, no study has shown that proteins in the GASA family can perform ethylene (ET) signal transduction, although crosstalk between ET and gibberellin is common in seed germination, stem elongation, and flowering [12, 13]. Several GASA proteins also regulate flower development. For example, the overexpression of AtGASA5 causes delayed flowering, whereas the mutation of AtGASA5 leads to early flowering [14]. Functional deletions in AtGASA6 and AtGASA4 were found to cause late flowering, but early flowering was caused by only the overexpression of AtGASA6 [6].
Jatropha curcas (Euphorbiaceae) is widely distributed in tropical and subtropical areas and is an ideal bioenergy crop for its oil-rich seeds that are especially rich in unsaturated fatty acids [15, 16]. However, due to low seed yield, the economic benefits of further expanding the Jatropha-based biodiesel industry are limited. The low ratio of female to male flowers (1:10–1:30) is a critical factor attributed to the low seed yield of J. curcas [15, 17]. The development of the flower of J. curcas has received much attention. In another study, using transcriptome data, we found that JcGASA6, a member of the GASA family, is differentially expressed in the flower buds during the development of the flowers of J. curcas [18]. However, the JcGASA6 functions have not been further investigated.
In this study, JcGASA6 was cloned from J. curcas and expressed in vitro. Upstream regulators of JcGASA6 and interactive proteins with JcGASA6 protein have been evaluated and screened. Moreover, we observed the expression pattern of JcGASA6 at several key stages during flower development and further analyzed the phenotypic of overexpression JcGASA6 in tobacco. Our results clarified the function of JcGASA6 from the protein and gene levels, and also enriched the signal network of GASA protein in hormone crosstalk.
Results
Bioinformatics analysis and subcellular localization of JcGASA6 protein
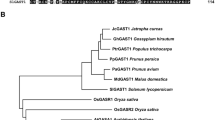
We obtained the coding sequence (CDS) of JcGASA6 from cDNA library of flower bud. The CDS of JcGASA6 contains 327 bp and encodes 108 amino acids. The start codon is ATG and the stop codon is TAG (Fig. S1). Amino acid sequence alignment between JcGASA6 and 14 members of the GASA family from Arabidopsis show that JcGASA6 protein has a special sequence composed of 12 cysteines (C-3X-C-2X-RC-8X-C-3X-C-2X-2C-2X-C-X/2X-CV-2X-G-2X-G-4X-C-X/2X-CY-10X-KCP). This special sequence is a conserved domain of GASA family (Fig. 1A). The phylogenetic tree of protein sequences between JcGASA6 protein and Arabidopsis GASA family indicated that JcGASA6 protein is most closely related to AtGASA4 protein sequences (Fig. 1B). The first 24 amino acids at the N-terminal of JcGASA6 protein were the putative signal peptides according to UniProt (Fig. S2). This suggests that JcGASA6 may transfer after synthesis. Further study of the localization of JcGASA6 protein. Under the excitation of 480 nm, the results of laser confocal microscopy showed that the control protein (pBWA(V)HS-GFP) could emit light normally and located in the cell membrane, while the fusion protein (pBWA(V)HS-GASA6-GFP) could emit light in the tonoplast besides the cell membrane. Co-localization of the marker protein in tonoplast and the fusion protein showed that green and red fluorescence could be detected at tonoplast (Fig. 1C). This indicated that JcGASA6 protein was located in the tonoplast.
Amino acid sequence alignment analysis and subcellular localization of JcGASA6 protein. A Multiple amino acid sequence alignment of JcGASA6 with GASA family. B Phylogenetic analysis of the JcGASA6 protein. C The subcellular localization of JcGASA6 protein. Red arrow indicates the cell membrane, and white arrow indicates the tonoplast
Expression and identification of JcGASA6 in vitro
To further study JcGASA6 from the protein level, JcGASA6 was cloned into pColdII vector, then was overexpressed in BL21 and ESLA to get JcGASA6 protein. JcGASA6 encodes a total of 108 amino acids and the signal peptide consists of the first 24 amino acids. Therefore, the molecular weight of the recombinant protein JcGASA6 was 10.97 kDa after adding the tag sequence of vector and removing the signal peptide. SDS-PAGE analysis showed that the expression products from BL21 (with pColdII-JcGASA6) or ESLA (with pColdII-JcGASA6) contain a 10.97 kDa protein compared with the control (Fig. 2A). This indicates that JcGASA6 was successfully expressed in BL21 and ESLA. Additionally, the 10.97 kDa protein was distributed in the precipitates released from E.coli rupture (Fig. 2B) and was not detected in the supernatants of E.coli rupture (Fig. 2C). This indicates that JcGASA6 was expressed in inclusion bodies way. The 10.97 kDa protein was further identified by ESI-LC-MS/MS. The results showed that a total of 17 peptides from the 10.97 kDa protein matched the amino acid sequence of JcGASA6 protein (Table 1) and the coverage of amino acids match with JcGASA6 protein reached 83% (Fig. 2D). Therefore, the 10.97 kDa protein is JcGASA6 protein.
Prokaryotic expression and identification of JcGASA6. A Whole bacteria SDS-PAGE analysis of JcGASA6 expression. B SDS-PAGE analysis of precipitation from bacteria. C SDS-PAGE analysis of supernatant from bacteria. M, marker; lane 0, control (empty pColdII); lane 1–3, expression of JcGASA6 in BL21(DE3); lane 4–6, expression of JcGASA6 in ESLa; lane 1and 4, induction with 0.10 mM IPTG for 16 hours; lane 2 and 5, induction with 0.25 mM IPTG for 16 hours; lane 3 and 6, induction with 0.25 mM IPTG for 4 hours; red arrow indicates JcGASA6 protein. D Mass spectrometry identification results of expression product from JcGASA6
JcGASA6 protein and Snakin-1 protein (crystal 5e5t.1A) have the highest homology according to the results from the SWISS-MODEL database. Therefore, taking the crystal of 5e5t.1A as the template, we get the speculative three-dimensional structure of JcGASA6 protein after homologous modeling. Both JcGASA6 protein and Snakin-1 protein have two important structures-short helices. The first short helix is composed of α1 and α2, and the second short helices is composed of α3 and α4 (Fig. 3A). These two structures are essential for the function of GASA family protein [19]. Moreover, like Snakin-1 protein, JcGASA6 protein contains 12 cysteines. The 12 cysteine residues form 6 disulfide bonds (Fig. 3B, C), which is of great significance to maintaining the stability of protein three-dimensional structure [20].
Promoter sequence characteristics and upstream regulatory factors of JcGASA6
To further reveal the potential signal transduction pathway involved by JcGASA6, the promoter of JcGASA6 was isolated from J. curcas genome DNA. Except for conserved motifs (AT-TATATA-box, CAAT-box, and TATA-box), JcGASA6 promoter included several motifs with unknown function. Additionally, JcGASA6 promoter also included an ethylene-responsive motif which suggested the expression of JcGASA6 may be regulated by ET (Fig. S3). Then, the upstream regulatory factors of JcGASA6 were screened by yeast one-hybrid (Y1H) system using JcGASA6 promoter as bait. Three genes, which may bind to the promoter of JcGASA6, were screened from cDNA library of J. curcas flower. The three genes included JcFLX-like (ID: XM_012232748.2), JcERF1 (ID: XM_012213416.2), and JcPYL9 (ID: XM_012227842.2). To confirm that the three genes can bind to the promoter of JcGASA6, the three genes were tested by Y1H individually. Self-activation of promoters is common, so 3-amino-1,2,4-triazole (3-AT) with suitable concentration is used to inhibit the self-activation of promoters [21]. In our study, 3-AT with a concentration of 130 mM completely inhibit the self-activation of JcGASA6 promoter (Fig. 4A). Additionally, the yeast cells, which contain pGADT7-JcFLX and JcGASA6-Pro-pHis2, pGADT7-JcPYL9 and JcGASA6-Pro-pHis2, or pGADT7-JcAP2 and JcGASA6-Pro-pHis2, can grow normally on SD/−His-Leu-Trp medium with 130 mM 3-AT (Fig. 4A). These results indicated that all of JcFLX, JcPYL9, and JcERF1 could interact with JcGASA6 promoter to regulate the expression of JcGASA6. Furthermore, the regulation of three genes on JcGASA6 was determined by dual-luciferase assays. The results of dual-luciferase assays showed that all of JcFLX, JcPYL9, and JcERF1 could enhance the activity of Luc driven by JcGASA6 promoter and the activity of LUC was the strongest in JcERF1/JcGASA6 group (Fig. 4B). This suggested that all of the three genes (JcFLX, JcPYL9 and JcERF1) could interact with JcGASA6 promoter to activate the expression of JcGASA6, and JcERF1 has the strongest activation effect on JcGASA6.
All of JcFLX, JcPYL9 and JcERF1 binds to the promoter of JcGASA6 and activate its expression. A Yeast one-hybrid assay shows all of JcFLX, JcPYL9 and JcERF1 bind to JcGASA6 promoter. B Dual-luciferase assays showed all of JcFLX, JcPYL9 and JcERF1 can activate the expression of JcGASA6. Diverse lowercase letters indicate significant differences (P < 0.05). The significance of difference was analyzed by Tukey’s test
Proteins interacting with JcGASA6 protein
In addition to screening the upstream regulators of JcGASA6, we also screened the interacting proteins of JcGASA6 protein by using yeast two-hybrid (Y2H) system. On SD/−Ade/−His/−Leu/−Trp medium, the yeast cells with pGBKT7–53 and pGADT7-T plasmids can grow normally, while the yeast cells with pGBKT7 and pGADT7 plasmids (negative control) and yeast cells with pGBKT7-JcGASA6 and pGADT7 plasmids can not grow normally (Fig. S4). These results indicated that JcGASA6 protein without transcriptional self-activation activity. JcGASA6 was used as the bait to screen its interacting proteins from flower bud cDNA library, and five proteins interacting with JcGASA6 were obtained. The five proteins include JcCNR8 (J. curcas cell number regulator 8, ID: XM_012226926.2), JcAMs (J. curcas transcription factor ABORTED MICROSPORES, ID: XM_020682349.1), JcAPRR2 (J. curcas two-component response regulator-like APRR2, ID: XM_012218861.2), JcFRI (J. curcas FRIGIDA-like protein 4a FRI, ID: XM_012214239.2) and JcSIZ1(J. curcas E3 SUMO-protein ligase SIZ1, ID: XM_012209470.2). To verify the interaction between the five proteins with JcGASA6, we used Y2H assay. Yeast cells, which containing JcCNR8 and JcGASA6, JcAMs and JcGASA6, JcAPRR2 and JcGASA6 or JcSIZ1 and JcGASA6, grew normally on SD/−Trp/−Leu/−Ade/−His and SD/−Trp/−Leu/−Ade/−His (X-a-gal) medium, while the yeast cells with JcFRI and JcGASA6 could not grow (Fig. 5A). These results suggested four proteins JcCNR8, JcAMs, JcAPRR2, and JcSIZ1 may interact with JcGASA6 protein. BiFC assay was carried out to further confirm these results. The BIFC assay show in Fig. 5B, the yellow fluorescence could be detected only when the protein pair of JcCNR8 and JcGASA6 or JcSIZ1 and JcGASA6 were transiently co-expressed in leaf epidermal cells, and the yellow fluorescence was detected in the nucleus (Fig. 5B). These results indicated that both JcCNR8 and JcSIZ1 could interact with JcGASA6 in nucleus.
Identification of proteins interacted with JcGASA6. A Y2H assay to verify the interactions of JcGASA6 with JcCNR8, JcAMs, JcAPRR2, JcFRI and JcSIZ1. B BiFC assay of the interactions of JcGASA6 with JcAMs, JcAPRR2, JcCNR8 and JcSIZ1; Bright: bright field without fluorescence signal; NLS: blue fluorescent signal located in the nucleus; YFP: yellow fluorescent signal of interacting proteins; Merged: Overlap of yellow and blue fluorescence signals in bright field, Bars: 48 μm
Expression pattern of JcGASA6 in the flower development of J. curcas
Our previous studies suggest that JcGASA6 may also be involved in flower development [18], we further analyzed the expression pattern of JcGASA6 during flower development. In order to accurately judge the expression pattern of JcGASA6 during flower development of J. curcas, the flower of J. curcas was observed at the morphological and histological. When the bud length of J. curcas is about 0.15 mm, the primordium of petal has appeared, but the sexual differentiation of flower has not yet begun (Fig. 6St0). During the development of female flowers, the carpel did not fully heal when the ovary length is about 0.80 mm (Fig. 6ST1). Histological analysis showed that the female flower was at the stage of megasporocyte (Fig. 6St1). After that, the megasporocyte undergoes two meiosis to form functional megaspore (Fig. 6ST2), at this time, the carpel has basically healed (Fig. 6St2), and the ovary length was about 1.10 mm. The functional megaspore develops further and enters the mononuclear embryo sac stage (Fig. 6St3), the ovary length was about 1.50 mm (Fig. 6ST3). Finally, functional megaspores formed mature embryo sac with 8-core 7 cells through three mitoses (Fig. 6St4), and the ovary length was about 3.20 mm (Fig. 6ST4). During the development of male flower, the male flower was in the stage of microspore mother cell when the flower bud length is about 0.50 mm (Fig. 6St5, ST5). After two times of meiosis, the microsporocyte entered the tetrad stage (Fig. 6St6), the length of bud was 1.20 mm (Fig. 6ST6). Microsporocyte was released to the anther chambers to form single nucleus pollen (Fig. 6St7), the length of the bud was 2.00 mm (Fig. 6ST7). Finally, single nucleus pollen undergoes nuclear division to form mature pollen with two-cell (Fig. 6St8), the length of bud was about 3.00 mm (Fig. 6ST8). In female flowers, the expression of JcGASA6 gradually increased from the undifferentiated stage (St0) to the megasporocyte meiosis stage (St2) and reached the highest lever at the megasporocyte meiosis stage. After that, the expression of JcGASA6 gradually decreased and reached the lowest level at the mature embryo sac stage (St4). These results suggested that JcGASA6 plays an important role in the early development of female flower. In male flowers, the expression of JcGASA6 increased from undifferentiated stage (St0) to mature pollen stage (St8) and reached the highest level at mature pollen stage. These results indicate that JcGASA6 plays an important role in the whole development of male flower. Moreover, the expression of JcGASA6 was not significantly different in the early development between female (St1-St2) and male flower (St5-St6), while in the late development of female (St3-St4) and male flowers (St7-St8), the expression of JcGASA6 increased significantly in male flowers. This suggested that JcGASA6 was more important for the development of male flower than female flower (Fig. 6A).
Morphological and histological observation on flower of J. curcas. ST1 to ST4 represent the morphology of female flower and the corresponding histological structures are St1 to St4. ST5 to ST8 represent the morphology of male flower, and the corresponding histological structures are St5 to St8. St0, represent the morphology of undifferentiated flower. A Expression profile of JcGASA6 during the development of flower, asterisks indicate a significant difference, *p < 0.05, **p < 0.01, ***p < 0.001. Ca, carpel; Pp, petal primordium; S, stamen; Me, megasporocyte; Fm, functional megaspore; Mes, mononuclear embryo sac; Mi, microsporocyte; Snp, single nucleus pollen; Mp, mature pollen. Bars = 50 μm for histological structures of flower. Bars = 270 μm for morphology of female flower. Bars = 20 μm in St0
Overexpression of JcGASA6 promoted the elongation of stamen filament in tobacco
The expression pattern of JcGASA6 during flower development suggests that JcGASA6 plays an important role in flower development, especially in male flower development. To further reveal the role of JcGASA6 in flower development, overexpression of JcGASA6 in tobacco (wild-type tobacco were controls, WT). There was no significant difference in plant height, ground diameter and style length between wild-type tobacco and transgenic tobacco (Table 2). However, the stamen of transgenic tobacco (TR) was higher than the stigma of ovary, and the filament length of TR tobacco was longer than WT tobacco (Table 2 and Fig. 7C, D). This result suggested that JcGASA6 could promote the elongation of stamen filament. Moreover, the leaves of TR tobacco are longer and wider than those of WT tobacco (Table 2 and Fig. 7A, B). This result indicated that JcGASA6 was also involved in plant growth.
Discussion
Gibberellic Acid-stimulated Arabidopsis (GASA) protein, also known as Snakin protein, is a kind of CRP (cysteine-rich peptides) protein [8]. GASA family proteins have been found in many plants and have many members, characterize by a conserved domain containing 12 cysteines [11]. In present study, JcGASA6 encoded a protein identified as JcGASA6 protein. JcGASA6 is a member of GASA family, suggested by its typical domain containing 12 cysteines (Fig. 1A). It was with a length of 108 amino acids and a molecular weight of 10.97 KD, containing a signal peptide at N-terminal. It showed the closest genetic relationship with AtGASA4 (Fig. 1B).
Antimicrobial peptides (AMPs) are excellent candidate drugs against drug-resistant pathogens. The structure of AMPs, especially an α-helical hairpin structure, plays an important role in killing pathogens [20]. Snakin is the only GASA member with antibacterial effect [3, 22]. Two short helices structures (dbHTH) in Snakin-1 protein are similar to the α-helical hairpin in antimicrobial peptide EcAMP1 [23]. The short helices and six disulfide bonds can form a large positive electrostatic surface (Fig. 3C). The positive electrostatic surface makes Snakin-1 protein play an antibacterial role [20]. Similar to the Snakin-1 protein, the speculative three-dimensional structure of JcGASA6 protein have two important structures-short helices and six disulfide bonds (Fig. 3), which might endow it with an antibacterial role.
Protein localization is often closely related to its function. Most GASA proteins have signal peptides at the N-terminal, and they are located in the plasma membrane or cell wall. AtGASA14 protein is located in the plasma membrane which will make it easier to regulate the balance of reactive oxygen [5]. GhGASA10 (a GASA protein in Cotton) was localized in the plasma membrane, which conducive to synthesize proteins in the cell wall to promotes the cell elongation in fiber [24]. AtGASA6 protein is located in the cell wall, which helps to regulate the elongation of hypocotyl and seed germination [13]. In our study, The N-terminal of JcGASA6 protein also has a signal peptide (Fig. S3). But unlike other members of the GASA protein, JcGASA6 is located at the tonoplast (Fig. 1C). This suggests that the function of JcGASA6 protein may be different from other members of GASA family.
GASA family is firstly thought to play important role in gibberellin signal pathway. AtGASA6 and AtGASA4 are located the downstream of DELLA protein in GA signal pathway [10]. To data, most members of the GASA family are found to be widely involved in hormone signal transduction, such as ABA, ET, GA, BR (brassinolide), SA (salicylic acid). OsGSR1 control BR biosynthesis to regulate salt stress [25]. Snakin-1 is involved in hormonal balance and Snakin-1 silencing enhanced GA and SA levels [26]. AtGASA6 is down-regulated by ABA and can integrate GA and ABA to promote cell elongation and seed germination [13]. Our study also found that ABA receptor JcPYL9 protein could directly activate the expression of JcGASA6 (Fig. 4), supporting that the members of GASA family would be involved in ABA signal transduction. A few evidence showed that members of GASA protein family were involved in ET signal transduction. FaGAST2, a member of GASA family in strawberry, was up-regulated by ethephon [7]. ERF1 is the target gene of the core transcription factor EIN3 of the ET signaling pathway. EIN3 protein can activate the expression of ERF1 through binding to the promoter of ERF1 [27]. Our results showed that the expression of JcGASA6 could be activated by ERF1 protein in J. curcas (Fig. 4), which further confirmed that GASA protein family is involved in ET signal transduction.
AtGASA4 (closeted genetic relationship with JcGASA6 protein) and AtGASA6 (the homologous protein of JcGASA6) are involved in flower development. The gasa4 mutant showed more leaf buds before generating flowers, but overexpression of AtGASA4 could not promote flowering transformation. This indicates that AtGASA4 is not enough to induce flowering transformation [28]. AtGASA4 and AtGASA6 also affect flowering time, but AtGASA6 plays a predominant role in causing early flowering [6]. FLX is reported to be involved in the regulation of early flowering [29]. In present study, FLX could directly activate the expression of JcGASA6. Although we did not observe abnormal flowering in transgenic tobacco, overexpression of JcGASA6 caused elongated filaments in tobacco (Figs. 6 and 7C). Furthermore, the expression of JcGASA6 increased continuously during male flower development. All these results suggested that JcGASA6 would contribute to the development of male flowers, especially the filaments of stamens. This affection of JcGASA6 on flower is different from other GASA family (such as AtGASA4 and AtGASA6). On the other hand, JcGASA6 protein could interact with JcCNR8 protein. CNR can control organ size by regulating cell number [30]. These would explain that the leaf size of transgenic tobacco was larger than wild-type tobacco (Fig. 7A, B) (Table 3). Therefore, JcGASA6 is also involved in growth regulation. Also, this is the first time to find that the GASA family can interact with CNR protein, which may support AtGASA10 facilitate wall growth [4].
Additionally, the JcGASA6 protein also could interact with SUMO E3 ligase (JcSIZ1). SUMO (Small Ubiquitin-like Modifier) E3 ligase is an important member involved in protein SUMO modification. SUMO E3 ligase recruits target proteins and promote the binding of SUMO to the target protein, then the activity of the target protein changes reversibly [31, 32]. Therefore, the activity of JcGASA6 protein may be modified by SUMO.
Conclusions
JcGASA6, a member of the GASA family, has an antibacterial structure similar to the antibacterial peptide Snakin-1, and is located in the tonoplast. In addition, JcGASA6 can be activated by the ABA receptor JcPYL9 and the core transcription factor JcERF1, which supports that JcGASA6 can participate in ABA and ET signal transduction. Meanwhile, JcGASA6 is activated by the early flowering gene FLX, and its expression increases continuously during male flower development. Abnormal flowering was not observed after overexpressing JcGASA6, but the elongation of stamen filaments was caused by the overexpression of JcGASA6. These results confirmed that JcGASA6 is involved in flower development. Additionally, overexpression of JcGASA6 increased the leaf size in transgenic plants, and JcGASA6 can interact with CNR (a protein with the regulation cell number). These results indicate that JcGASA6 is also involved in growth regulation. Moreover, JcGASA6 interacts with ubiquitin ligase SIZ1 also implies that its activity may be modified by SUMO. All in all, our results have revealed the functions of JcGASA6 from multiple perspectives.
Methods
Plant materials and flower collection
The flower buds of J. curcas L. were collected from Zhenfeng, Guizhou Province, China (36°14,050.2′N, 87°51,047.8′E). Flower buds for morphological and microscopic observation were temporarily stored in the mixture of acetaldehyde acetic acid 50% alcohol (4: 6: 90, v/v), and for RNA extraction were temporarily stored in RNAlocker (Tiandz, Inc., Beijing China). All samples were placed on ice.
JcGASA6 isolation and sequence analysis
The full-length cDNA of JcGASA6 was cloned from flower bud by RACE-pcr, then the obtained cDNA sequences were aligned in NCBI database (Accession number: KU500008).
Sequence of the GASA family of Arabidopsis thaliana was obtained from NCBI database (Table S1). Multiple sequence alignment used DNAMAN, phylogenetic tree constructed by MEGA 6.0, and signal peptide predicted by UniProt. (https://www.uniprot.org/peptidesearch/).
Subcellular localization, expression and identification of JcGASA6 protein
The full-length cDNA of JcGASA6 was fused with the pBWA(V)HS-osGFP then recombinant plasmid pBWA(V)HS-JcGASA6-osGFP was transfected transformed into rice protoplasts by PEG (polyethylene glycol). The protoplasts were observed by confocal laser microscope under the excitation of 480 nm wavelength after dark culture at 28 °C for 48 hours (FV10-ASWOLYMPUS, Japan) [33]. Gamatip protein located in the tonoplast was used as a marker [34]. All primers used for subcellular localization are listed in Table S2.
The coding sequence of JcGASA6 was amplified by specific primers (JcGASA6ex-F and JcGASA6ex-R), then the coding sequence was connected to the expression vector pCold II after digesting with Nde I and Hind III. The sequence of recombinant plasmid JcGASA6-pCold II was examined using vector primers (pCold II-F and pCold II-R) (Table S2). The recombinant plasmid was transformed into E. coli competent cells (BL21 or ESLA) to overexpress JcGASA6, and the empty vector pCold II was transformed into BL21 or ESLA as the control. Then the BL21 or ESLA were cultured at 16 °C, and isopropyl β-D-thiogalactoside (IPTG) was used as an expression inducer. The details of method same as previously reported [35].
The expression products of JcGASA6 in E. coli were analyzed by SDS-PAGE electrophoresis. Cut off the SDS-PAGE glue containing JcGASA6 protein, and wash the SDS-PAGE glue with ultrapure water and decolorization with acetonitrile mixture. The decolorized SDS-PAGE glue was digested by trypsin overnight at 37 °C to form enzymolysis solution, then the enzymolysis solution was identified by LC-MS/MS method. The method was consistent with the previously reported [36]. The three-dimensional structure of protein was analyzed by the SWISS-MODEL database (https://www.swissmodel.expasy.org/interactive).
Promoter isolation and analysis, construction of cDNA library for yeast-hybrid system
The promoter sequence of JcGASA6 was obtained from the published genomic database (ID: 105640538), and PlantCARE was used for promoter sequence analysis (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The cDNA library of Yeast-Hybrid System was constructed by mixed RNA from flowers at different developmental stages. The primary library was constructed using attB2 as linker and ATTB-A1, ATTB1-B and ATTB1-C as primers. The clone number of primary librarywas8.04 × 106 cfu. The plasmid of primary library was extracted and transferred into DH10B by the electrotransfer method, and then the secondary library was obtained. The clone number of secondary library was 1.31 × 107 cfu. The method is consistent with that previously reported [37] (Table S3).
Yeast-one hybrid (Y1H) and dual-luciferase assay
The promoter of JcGASA6 was amplified using specific primer pro-JcGASA6-F/R, then fusion to the pHIS2 (Table S4). Co-transferred fusion plasmid and secondary library to Y187 yeast system, then screen the upstream regulator of JcGASA6. The screening process refers to the previous method [38]. Three upstream regulators (JcFLX, JcERF1 and JcPYL9) were screened from the secondary library. The interaction between JcGASA6 promoter and these three regulators was verified one by one with Y187 yeast system [38]. pGADT7 and p53-pHis2 were co-transformed into Y187 as negative controls. pGADT7–53 and p53-pHis2were co-transformed into Y187 as positive controls. The promoter self-activation of JcGASA6 was detected using Y187 with plasmids pGADT7 and JcGASA6-Pro-pHis2.
The primer JcCASA6-luc-F/R was used to construct pGreenII 0800-JcGASA6-luc, and three primers (AP2-F/R, FLX-F/R and PYL9-F/R) was used to construct regulators (pGreenII 62-JcFLX-SK, pGreenII 62-JcERF1-SK and pGreenII 62-PYL9-SK). Co-transferred pGreenII 0800-proJcGASA6-luc and regulator into tobacco leaves, then detected the fluorescence value (Dual-Luciferase Assay System, Promega) (Table S4) [38]. pGreenII 62-SK and pGreenII0800-Luc were co-transformed into tobacco leaves as negative controls.
Yeast-two hybrid(Y2H) and bimolecular fluorescence complementation (BiFC) assay
Full-cDNA JcGASA6 fusion with pGBKT7 by using GASA6-GBK-F/R primer, then both of pGBKT7-JcGASA6and pGADT7-AD were transferred into AH109 by LiAc method, then the AH109was cultured for the detection of self-activation activity. Co-transferredpGBKT7-JcGASA6 and secondary library to AH109 for screening the interaction proteins of JcGASA6. Five proteins were screened, including JcCNR8, JcAMs, JcAPRR2, JcFRI and JcSIZ1. The interaction between the five proteins andJcGASA6 were verified by one-to-one in AH109 [39] (Table S5). pGADT7 and pGBKT7 were co-transformed into AH109 as negative controls. pGADT7–53 andpGBKT7-T were co-transformed into AH109 as positive controls.
Full-cDNA JcCNR8, JcSIZ1, JcAMs, and JcAPRR2were fused with PSPYCE-35Srespectively by using specific primer, and Full-cDNA JcGASA6was fused with PSPYNE-35S. Co-transferred PSPYNE-35S-JcGASA6 and PSPYCE-35S-JcCNR8/PSPYCE-35S-JcSIZ1/PSPYCE-35S-JcAMs/PSPYCE-35S- JcAPRR2 into EHA105. The five types of EHA105 infected tobacco leaves respectively, then the fluorescence signal was detected 72 hours after infection [39] (Table S5). PSPYCE-35S and PSPYNE-35S were co-transformed into tobacco leaves as negative controls. PSPYCE-35S-bZIP63 and PSPYNE-35S-bZIP63 were co-transformed into tobacco leaves as positive control.
Morphology and microscopic observation of flower
Flower buds were classified according to their length, dissected under stereoscope, observed and photographed. Since the flower bud of undifferentiated stage is too small to be observed clearly under stereoscope, it was observed under the scanning electron microscope. The undifferentiated flower were fixed in the mixture of formaldehyde-acetic acid-50% ethanol, and then dissected and observed by electron microscope [18]. According to the classification, paraffin sections of these flower buds were made [40] (Table 3).
Expression of JcGASA6 during flower development determined by qRT-PCR
The total RNA of flower was extracted by RNA isolation kit (Omega Bio-Tek, Beijing, China), qualified RNA was used to synthesize the first strand cDNA (Takara, Beijing, China). PCR amplification was used Bio-Rad CFX system (Bio-Rad, USA). Beta-tubulin and actin as internal control. The 2 - ΔΔ CT method was used to calculate the relative expression of JcGASA6 [41]. All primers used for PCR amplification are listed in Table S6 and each sample reaction was repeated three times.
Overexpression of JcGASA6 in Nicotiana tabacum L.
Fusion full-length cDNA of JcGASA6 with the pBWA(V)KS-GUS. The recombinant plasmid (pBWA(V)KS-JcGASA6-GUS) was transformed into agrobacterium tumefaciens (GV3101), then positive agrobacterium tumefaciens were screened. Tobacco (K326) leaves infected by GV3101 were cultured in dark at 25 °C for 2 days. After tobacco leaves were differentiated into seedlings, positive tobacco was detected (Fig. S5), and wild-type tobacco was used as control [42] (Table S6).
Availability of data and materials
All data generated or analyzed in our study are available in this article and its supplementary information files. Gene sequences can be down-loaded at NCBI database (https:// www. Ncbi. nlm. Nih. gov/). The GenBank accession number of JcGASA6 is KU500008, AtGASA1 is P46689.2, AtGASA2 is P46688.1, AtGASA3 is P46687.1, AtGASA4 is P46690.2, AtGASA5 is Q84J95.1, AtGASA6 is Q6NMQ7.1, AtGASA7 is O82328.1, AtGASA8 is O80641.1, AtGASA9 is Q8GWK5.1, AtGASA10 is Q8LFM2.1, AtGASA11 is F4IQJ4.1, AtGASA12 is Q6GKX7.1, AtGASA13 is A8MR46.1, and AtGASA14 is Q9LFR3.1.
Abbreviations
- GASA:
-
Gibberellic acid-stimulated Arabidopsis
- GA:
-
Gibberellin
- ABA:
-
Abscisic Acid
- ET:
-
Ethylene
- SA:
-
Salicylic Acid
- BR:
-
Brassinolide
- CDS:
-
Coding Sequence
- Y1H:
-
Yeast One-Hybrid
- 3-AT:
-
3-Amino-1,2,4-Triazole
- Y2H:
-
Yeast Two-Hybrid
- WT:
-
Wild-Type
- TR:
-
Transgenic
- AMPs:
-
Antimicrobial peptides
- SUMO:
-
Small Ubiquitin-like Modifier
References
Wu K, Qu Y, Rong H, Han X, Tian Y, Xu L. Identification and Expression Analysis of the Populus trichocarpa GASA-Gene Family. Int J Mol Sci. 2022;23(3):1507.
Ahmad MZ, Sana A, Jamil A, Nasir JA, Ahmed S, Hameed MU. Abdullah: a genome-wide approach to the comprehensive analysis of GASA gene family in Glycine max. Plant Mol Biol. 2019;100(6):607–20.
Almasia NI, Molinari MP, Maroniche GA, Nahirnak V, Barrios Baron MP, Taboga OA, et al. Successful production of the potato antimicrobial peptide Snakin-1 in baculovirus-infected insect cells and development of specific antibodies. BMC Biotechnol. 2017;17(1):75.
Trapalis M, Li SF, Parish RW. The Arabidopsis GASA10 gene encodes a cell wall protein strongly expressed in developing anthers and seeds. Plant Sci. 2017;260:71–9.
Sun SL, Wang HX, Yu HM, Zhong CM, Zhang XX, Peng JZ, et al. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J Exp Bot. 2013;64(6):1637–47.
Qu J, Kang SG, Hah C, Jang JC. Molecular and cellular characterization of GA-stimulated transcripts GASA4 and GASA6 in Arabidopsis thaliana. Plant Sci. 2016;246:1–10.
Moyano-Canete E, Bellido ML, Garcia-Caparros N, Medina-Puche L, Amil-Ruiz F, Gonzalez-Reyes JA, et al. FaGAST2, a strawberry ripening-related gene, acts together with FaGAST1 to determine cell size of the fruit receptacle. Plant Cell Physiol. 2013;54(2):218–36.
Zhang S, Wang X. One new kind of phytohormonal signaling integrator: up-and-coming GASA family genes. Plant Signal Behav. 2017;12(2):e1226453.
Fan S, Zhang D, Zhang L, Gao C, Xin M, Tahir MM, et al. Comprehensive analysis of GASA family members in the Malus domestica genome: identification, characterization, and their expressions in response to apple flower induction. BMC Genomics. 2017;18(1):827.
Zhang SC, Wang XJ. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chin Sci Bull. 2008;53(24):3839–46.
Li Z, Gao J, Wang G, Wang S, Chen K, Pu W, et al. Genome-wide identification and characterization of GASA gene family in Nicotiana tabacum. Front Genet. 2021;12:768942.
Fu X, Gao X, Liu X. Integration of ethylene and gibberellin signaling. Springer Netherlands. 2015;1:153–73.
Zhong CM, Xu H, Ye ST, Wang SY, Li LF, Zhang SC, et al. Gibberellic acid-stimulated Arabidopsis6 serves as an integrator of gibberellin, Abscisic acid, and glucose signaling during seed germination in Arabidopsis(1[OPEN]). Plant Physiol. 2015;169(3):2288–303.
Zhang S, Yang C, Peng J, Sun S, Wang X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol Biol. 2009;69(6):745–59.
Alburquerque N, Garcia-Almodovar RC, Valverde JM, Burgos L, Martinez-Romero D. Characterization of Jatropha curcas accessions based in plant growth traits and oil quality. Ind Crop Prod. 2017;109:693–8.
Qian JF, Shi HX, Yun Z. Preparation of biodiesel from Jatropha curcas L. oil produced by two-phase solvent extraction. Bioresour Technol. 2010;101(18):7025–31.
Gangwar M, Shankar J. Molecular mechanisms of the floral biology of Jatropha curcas: opportunities and challenges as an energy crop. Front Plant Sci. 2020;11:609.
Xu G, Huang J, Yang Y, Yao YA. Transcriptome Analysis of Flower Sex Differentiation in Jatropha curcas L. Using RNA Sequencing. PLoS One. 2016;11(2):e0145613.
Zhang M, Cheng W, Wang J, Cheng T, Zhang Q. Genome-Wide Identification, Evolution, and Expression Analysis of GASA Gene Family in Prunus mume. Int J Mol Sci. 2022;23(18):10923.
Yeung H, Squire CJ, Yosaatmadja Y, Panjikar S, Lopez G, Molina A, et al. Radiation damage and racemic protein crystallography reveal the unique structure of the GASA/Snakin protein superfamily. Angew Chem Int Ed Engl. 2016;55(28):7930–3.
Deng CY, Wang JY, Lu CF, Li YF, Kong DY, Hong Y, et al. CcMYB6-1 and CcbHLH1, two novel transcription factors synergistically involved in regulating anthocyanin biosynthesis in cornflower. Plant Physiol Bioch. 2020;151:271–83.
Oliveira-Lima M, Benko-Iseppon AM, Neto J, Rodriguez-Decuadro S, Kido EA, Crovella S, et al. Snakin: structure, roles and applications of a plant antimicrobial peptide. Curr Protein Pept Sci. 2017;18(4):368–74.
Barashkova AS, Ryazantsev DY, Rogozhin EA. Rational Design of Plant Hairpin-like Peptide EcAMP1: Structural-Functional Correlations to Reveal Antibacterial and Antifungal Activity. Mol. 2022;27(11):3554.
Chen B, Sun Y, Tian Z, Fu G, Pei X, Pan Z, et al. GhGASA10-1 promotes the cell elongation in fiber development through the phytohormones IAA-induced. BMC Plant Biol. 2021;21(1):448.
Li Q, Xu F, Chen Z, Teng Z, Sun K, Li X, et al. Synergistic interplay of ABA and BR signal in regulating plant growth and adaptation. Nat Plants. 2021;7(8):1108–18.
Nahirnak V, Rivarola M, Almasia NI, Barrios Baron MP, Hopp HE, Vile D, et al. Snakin-1 affects reactive oxygen species and ascorbic acid levels and hormone balance in potato. PLoS One. 2019;14(3):e0214165.
Binder BM. Ethylene signaling in plants. J Biol Chem. 2020;295(22):7710–25.
Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahl-Sorteberg HG. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007;48(3):471–83.
Shen L, Zhang Y, Sawettalake N. A molecular switch for FLOWERING LOCUS C activation determines flowering time in Arabidopsis. Plant Cell. 2022;34(2):818–33.
Wang GL, Zhang CL, Huo HQ, Sun XS, Zhang YL, Hao YJ, et al. The SUMO E3 ligase MdSIZ1 Sumoylates a cell number regulator MdCNR8 to control organ size. Front Plant Sci. 2022;13:836935.
Streich FC, Lima CD. Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature. 2016;536(7616):304.
Benlloch R, Lois LM. Sumoylation in plants: mechanistic insights and its role in drought stress. J Exp Bot. 2018;69(19):4539–54.
Yarra R, Xue Y. Ectopic expression of nucleolar DEAD-box RNA helicase OsTOGR1 confers improved heat stress tolerance in transgenic Chinese cabbage. Plant Cell Rep. 2020;39(12):1803–14.
Besse M, Knipfer T, Miller AJ, Verdeil JL, Jahn TP, Fricke W. Developmental pattern of aquaporin expression in barley (Hordeum vulgare L.) leaves. J Exp Bot. 2011;62(12):4127–42.
Li Q, Zhu TT, Zhang R, Bu QT, Yin J, Zhang L, et al. Molecular cloning and functional analysis of hyoscyamine 6 betahydroxylase (H6H) in the poisonous and medicinal plant Datura innoxia mill. Plant Physiol Bioch. 2020;153:11–9.
Liu G, Tian H, Huang YQ, Hu J, Ji YX, Li SQ, et al. Alterations of Mitochondrial Protein Assembly and Jasmonic Acid Biosynthesis Pathway in Honglian (HL)-type Cytoplasmic Male Sterility Rice. J Biol Chem. 2012;287(47):40051–60.
Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, et al. Efficient yeast one−/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 2010;51(12):2145–51.
Shi SL, Liu Y, He YJ, Li LZ, Li DL, Chen HY. R2R3-MYB transcription factor SmMYB75 promotes anthocyanin biosynthesis in eggplant (Solanum melongena L.). Sci Hortic-Amsterdam. 2021;255(1):12.
Liu YD, Wen L, Shi Y, Su DD, Lu W, Cheng YL, et al. Stress-responsive tomato gene SlGRAS4 function in drought stress and abscisic acid signaling. Plant Sci. 2021;304:110804.
Chen DH, Molitor AM, Xu L, Shen WH. Arabidopsis PRC1 core component AtRING1 regulates stem cell-determining carpel development mainly through repression of class I KNOX genes. BMC Biol. 2016;14(1):112.
Shen XP, Xu LA, Liu YH, Dong H, Zhou D, Zhang YZ, et al. Comparative transcriptome analysis and ChIP-sequencing reveals stage-specific gene expression and regulation profiles associated with pollen wall formation in Brassica rapa. BMC Genomics. 2019;20(1):264.
Gomez LM, Teixeira-Silva NS, Caserta R, Takita MA, Marques MOM, de Souza AA. Overexpression of Citrus reticulata SAMT in Nicotiana tabacum increases MeSA volatilization and decreases Xylella fastidiosa symptoms. Planta. 2020;252(6):103.
Acknowledgements
We are grateful to XIAN CHUNFENG BIOTECHNOLOGY CO., LTD for their excellent technical assistance.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31760198), and the Scientific Research Project of Ordinary Undergraduate Colleges and Universities of Guizhou Provincial Department of Education (Qian Jiao Ji[2022]No. 145).
Author information
Authors and Affiliations
Contributions
Xue Li, Ming-sheng Zhang, Xu Gang conceived and designed the research, Xue Li, Liang-qing Zhao, Qian-qian Ling-hu performed the experiments and analyzed the data. Xue Li wrote the paper. Xu Gang, supervision, funding acquisition. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Before conducting the research, the permission from research institute of forest resources and environment of Guizhou University to collect and analyze the J. curcas L samples documented in this work was obtained. All the methods complied with Chinese regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, X., Zhang, Ms., Zhao, Lq. et al. The study on interacting factors and functions of GASA6 in Jatropha curcas L.. BMC Plant Biol 23, 99 (2023). https://doi.org/10.1186/s12870-023-04067-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04067-4