Abstract
Background
Soybean oil is a major source of edible oil, and the domestication of wild soybean has resulted in significant changes in oil content and composition. Extensive efforts have been made to identify genetic loci that are related to soybean oil traits. The objective of this study was to identify quantitative trait loci (QTLs) related to soybean seed oil and compare the fatty acid composition between wild and cultivated soybean.
Results
Using the specific-locus amplified fragment sequencing (SLAF-seq) method, a total of 181 recombinant inbred lines (RILs) derived from a cross between wild soybean ZYD00463 (Glycine soja) and cultivated soybean WDD01514 (Glycine max) were genotyped. Finally, a high-density genetic linkage map comprising 11,398 single-nucleotide polymorphism (SNP) markers on 20 linkage groups (LGs) was constructed. Twenty-four stable QTLs for seed oil content and composition were identified by model-based composite interval mapping (CIM) across multiple environments. Among these QTLs, 23 overlapped with or were adjacent to previously reported QTLs. One QTL, qPA10_1 (5.94–9.98 Mb) on Chr. Ten is a novel locus for palmitic acid. In the intervals of stable QTLs, some interesting genes involved in lipid metabolism were detected.
Conclusions
We developed 181 RILs from a cross between wild soybean ZYD00463 and cultivated soybean WDD01514 and constructed a high-density genetic map using the SLAF-seq method. We identified 24 stable QTLs for seed oil content and compositions, which includes qPA10_1 on Chr. 10, a novel locus for palmitic acid. Some interesting genes in the QTL regions were also detected. Our study will provide useful information for scientists to learn about genetic variations in lipid metabolism between wild and cultivated soybean.
Similar content being viewed by others
Background
Soybean (Glycine max L. Merr.) is one of the most important protein and oil crops [1], the yields of which accounted for nearly 60% of the world’s oilseed production in 2018 [2]. Soybean oil is mainly composed of five fatty acids, which include palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), and linolenic acid (C18:3), which are present at approximate average concentrations of 10, 4, 18, 55, and 13%, respectively [3, 4]. The quality of soybean oil depends on the composition of the fatty acids, which affects the nutritional value, flavor, and stability of the soybean oil. Unsaturated fatty acids play an important role in immune system regulation, blood clotting, neurotransmission, cholesterol metabolism, and the structure of membrane phospholipids in the brain and retina [5]. However, unsaturated fatty acids are susceptible to oxidation, resulting in an off-flavor and reducing oil shelf life [6, 7]. Soybeans are thus currently bred to have higher amounts of monounsaturated fatty acids (oleic acid) and lower amounts of polyunsaturated fatty acids (linoleic and linolenic acid), which increases the oxidative stability and is also better for human health [4, 8].
Soybean seed oil content and composition are controlled by multiple quantitative trait loci (QTLs)/genes and are also affected by environmental factors [9, 10]. To date, the QTLs associated with seed oil and fatty acids in soybean have been extensively investigated [11,12,13,14,15,16,17,18,19]. Since the first research attempted to discover oil QTLs in soybean [10], more than 322 oil QTLs and 228 fatty acid QTLs have been identified across all 20 chromosomes (Chr.) in the SoyBase database [20]. Among these QTLs, some are stable and have been detected in different bi-parental populations and environments, including the QTL regions of 1.64–2.09 Mb and 33.35–35.95 Mb on Chr. Twenty for seed oil content [14, 21,22,23,24,25] and the QTL region of 44.58–48.58 Mb on Chr. Fourteen for seed linolenic acid [11, 13, 24, 26]. However, most of these identified QTLs have low selection accuracy and have not been effectively used in marker-assisted selection (MAS) in soybean for seed oil due to insufficient linkage disequilibrium with desirable QTL alleles and the genetic complexity of the trait [27, 28].
With the completion of the genome sequencing of soybean cv. Williams 82 [29] and the rapid development of next-generation sequencing (NGS) technology, single-nucleotide polymorphism (SNP) markers have been used to construct high-density linkage maps to identify QTL intervals [25, 30]. Based on a high-density genetic map consisting of 2062 SNP markers, Cao et al. (2017) identified one QTL, qOil-5, for seed oil, which was mapped to Chr. 05, with a physical distance of 2.5 Mb [30]. Using the Illumina Infinium BeadChip sequencing platform, Patil et al. (2018) reported stable QTLs for oil content on Chr. 02 (qOil_02), Chr. 08 (qOil_08), Chr. 15 (qOil_15), and Chr. 20 (qOil_20) using 3343 polymorphic SNPs (3 K-SNP) [25]. Specific-locus amplified fragment sequencing (SLAF-seq) technology has been used to construct high-density genetic maps, constituting an efficient method for large-scale de novo SNP discovery and genotyping in soybean [31, 32]. Li et al. (2017) detected 26 stable QTLs for five fatty acids using 3541 SLAF markers with an average distance of 0.72 cM [31]. Zhang et al. (2018) created a high-density genetic map containing 8597 SNP loci with an average distance of 0.57 cM, from which two QTLs, qOil10–1 and qOil10–2, for oil content were mapped [32]. In addition, some genes associated with soybean seed oil content and composition have been discovered through genome-wide association studies (GWAS) by SNP genotyping [30, 33,34,35,36,37,38]. All these genes provide useful information for the improvement of seed oil in soybean breeding programs.
In soybean, the key functional genes associated with lipid biosynthesis have been investigated, including the fatty acid desaturase genes FAD2-1A (Glyma.10 g278000), FAD2-1B (Glyma.20 g111000), FAD3A (Glyma.14 g194300), FAD3B (Glyma.02 g227200), FAD3C (Glyma.18 g062000), FAD7 (Glyma.18 g202600 and Glyma.07 g151300) [39,40,41], 3-ketoacyl-ACP synthase II genes (KAS II, Glyma.17 g047000 and Glyma.13 g112700) [42, 43], and the diglyceride acyltransferase gene DGAT (Glyma.17 g053300) [44]. However, overexpression of a single gene could not significantly increase fatty acid biosynthesis flux [45]. It seems likely that lipid metabolism requires the regulation of multiple related genes. Several important transcription factors have also been found to participate in the regulation of lipid accumulation by directly binding to the promoters of lipid biosynthesis genes. For example, the overexpression of GmNFYA, GmDof4, GmDof11, GmbZIP123, and GmMYB73 significantly increases seed lipid accumulation in transgenic plants [46,47,48,49]. Studying these factors can improve our understanding of the mechanisms of lipid metabolism in soybean.
Cultivated soybean seeds have an oil content of approximately 18–22%, whereas wild soybean seeds contain about 8–10% oil [25]. In an attempt to identify genes controlling seed oil content and composition, QTL analyses for oil content between cultivated and wild soybean need to be conducted. In the present study, large-scale SNP markers were developed using SLAF-seq to construct the linkage group and map the QTLs controlling seed oil and composition using a population of 181 recombinant inbred lines (RILs) developed from an interspecific cross between wild soybean ZYD00463 (Glycine soja) and the cultivated soybean WDD01514 (Glycine max). Our results could inform scientists about the genetic variation in seed oil between wild and cultivated soybean during the domestication process and further improve oil quantity and quality by molecular breeding.
Results
Phenotypic variation
The seed oil content and five predominant fatty acid compositions of the parents and progenies were determined from 2015 to 2016 in Wuhan, Hubei Province and Xuchang, Henan Province. Table 1 shows that the average seed oil content of WDD01514 is 23.76%, which is approximately two-fold higher than ZYD00463 (11.89%). In terms of seed oil composition, WDD01514 has higher oleic acid (20.57%) and lower linolenic acid (7.60%) content than ZYD00463, which are 11.39 and 16.60%, respectively. Large variations in oil content and composition were observed among the 181 RILs, ranging from 11.36 to 13.11% for oil content, 17.07 to 22.68% for oleic acid content, and 11.05 to 14.41% for linolenic acid content.
The normality test using the Shapiro-Wilk (w) statistic indicated that oil content, palmitic acid, stearic acid (except 2015/X), and linolenic acid were normally distributed with P-values > 0.05. However, oleic acid and linoleic acid were not normally distributed (P < 0.05) (Table 1; Additional file 1). Oleic acid was skewed toward ZYD00463, while linoleic acid was skewed toward WDD01514 (Additional file 1). Moreover, transgressive segregation was observed in the progenies for palmitic, stearic, oleic, and linoleic acid (Table 1; Additional file 1), suggesting that comprehensive genetic recombination of alleles had occurred between parents.
The broad-sense heritability (h2) of the oil and fatty acids ranged from 80.2 to 92.0% in the combined environment, which indicated that the genetic variations accounted for a major proportion of the observed phenotypic variations (Table 1). ANOVA showed that the F-value of the G × E interaction was significant for all traits (P < 0.001). However, the F-value was less than the genotype (Table 2).
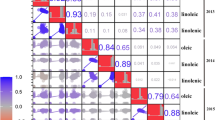
A positive correlation was observed between oil content and oleic acid in all four environments (P < 0.01), whereas a negative correlation was observed between oil content and palmitic acid, oil content and linolenic acid, palmitic acid and oleic acid, oleic acid and linoleic acid, and oleic acid and linolenic acid in all four environments (P < 0.01) (Additional file 2). This suggested that the important genetic factors controlling these traits are tightly linked.
Construction of a genetic map with SNP markers developed using SLAF-seq
The SLAF-seq method was applied to develop SNP markers between the two parents. Ultimately, a total of 11,398 SNP markers distributed over 20 linkage groups (LGs) were used to construct the genetic linkage map (Additional files 3, Additional files 4). These SNP markers encompassed 2913.78 cM of the soybean genome, with a mean distance of 0.26 cM between markers. The genetic distances of 20 LGs spanned from 126.23 cM (Chr. 11) to 226.60 cM (Chr. 03), with mean marker intervals ranging from 0.15 cM to 0.60 cM. The largest LG (Chr. 01) contained 891 SNP markers, while the smallest one (Chr. 13) had 248 SNP markers (Additional file 5).
QTLs for oil content
A total of 22 QTLs for seed oil content were mapped in this study (Additional file 6). Among these, seven stable QTLs with an LOD > 3.6 were identified across multiple environments (Fig. 1; Table 3) and were mapped to Chr. 02 (qOC2_1), 08 (qOC8_1 and qOC8_2), 15 (qOC15_1 and qOC15_2), and 20 (qOC20_1 and qOC20_2) (Additional file 7). The QTL qOC2_1 on Chr. 02 explained an average of 6.7% of the phenotypic variance for oil content; qOC8_1 and qOC8_2 on Chr. 08 explained an average of 8.9 and 6.6% of the phenotypic variance, respectively; qOC15_1 and qOC15_2 on Chr. 15 explained 12.9 and 11.0% of the phenotypic variance on average, respectively; and qOC20_1 and qOC20_2 on Chr. 20 explained 12.2 and 19.3% of the phenotypic variance on average, respectively. All of the QTLs showed negative additive effects, indicating the negative effect on oil content for the allele from the wild soybean parent ZYD00463. In comparison with the reported QTL regions, six QTLs, including qOC8_1, qOC8_2, qOC15_1, qOC15_2, qOC20_1, and qOC20_2, overlapped with previous QTLs (Fig. 2; Additional file 8). The QTL qOC2_1, which was located within the interval of 5.08–6.27 Mb on Chr. 02, was adjacent to the mapped QTL within the interval of 6.86–9.67 Mb (Fig. 2; Additional file 8).
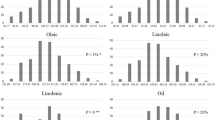
LOD curves on chromosomes for oil content and composition in four different environments. The x-axis indicates genetic position (cM) across the chromosome, the y-axis represents the LOD scores. Horizontal dashed line on the chart represents LOD threshold (3.6). Different environments are represented by different lines: red line (Wuhan in 2015), green line (Xuchang in 2015), yellow line (Wuhan in 2016) and blue line (Xuchang in 2016). LOD curves for oil content (a-d), palmitic acid (e-h), stearic acid (i), oleic acid (j and k), linoleic acid (l and m), and linolenic acid (n-p) in different environments. The arrow represents position of a QTL identified under a single environment, and the number is the genetic position (cM) of a QTL on chromosome
Distribution of stable QTLs in this study and reported QTLs. The QTL regions are marked with bars. The reported QTLs are showed in black bar. The stable QTLs in this study are showed in different color bar. Red bar: oil content; green bar: palmitic acid; blue bar: stearic acid; yellow bar: oleic acid; pale red bar: linoleic acid; light green bar: linolenic acid
Six enzyme genes involved in lipid metabolism were identified in these stable QTL regions (Additional file 9), which included two pyruvate kinase genes (GmPK, Glyma.02 g071000 and Glyma.02 g071100) within the genomic region of qOC2_1, a 3-ketoacyl-ACP reductase gene (GmFabG, Glyma.08 g102100) within qOC8_1, two 3-ketoacyl-CoA synthase genes (GmKCS, Glyma.15 g042500, and Glyma.15 g046300), and a 3-hydroxyacyl-ACP dehydrase gene (GmFabZ, Glyma.15 g052500) within qOC15–1. In addition, two transcription factor genes that might be involved in lipid metabolism were found within the genomic region of qOC8_2 (Additional file 10), including a homeobox-leucine zipper gene (GmZIP, Glyma.08 g124400) and a nuclear factor Y subunit A gene (GmNF-YA, Glyma.08 g124200).
QTLs for oil composition
A total of 66 QTLs related to seed oil composition were mapped (Additional file 6). Among these QTLs, 17 stable QTLs with an LOD > 3.6 were identified across multiple environments (Fig. 1; Table 3). These QTLs were mapped to Chr. 05, 10, 11, 13, 14, 15, and 16 (Additional file 7). For palmitic acid, four stable QTLs, including qPA10_1 on Chr. 10, qPA13_1 on Chr. 13, qPA15_1 on Chr. 15, and qPA16_1 on Chr. 16, contributed an average of 12.9, 8.4, 9.7, and 8.3% of the phenotypic variance, respectively. All of the QTLs for palmitic acid, except for qPA15_1 and qPA16_1, showed negative additive effects, indicating the negative effect on palmitic acid for the allele from the wild soybean parent ZYD00463. For stearic acid, three stable QTLs on Chr. 14, namely, qSA14_1, qSA14_2, and qSA14_3, accounted for an average of 18.6, 16.0, and 17.0% of the phenotypic variance, respectively. All of the three QTLs showed negative additive effects. For oleic acid, two stable QTLs, including qOA11_1 on Chr. 11 and qOA15_1 on Chr. 15, explained 12.3 and 19.1% of the phenotypic variance, respectively. The additive effect of these two QTLs was negative. For linoleic acid, four stable QTLs, including qLA5_1 and qLA5_2 on Chr. 05 and qLA11_1 and qLA11_2 on Chr. 11, were identified and explained an average of 9.9, 9.6, 10.4, and 10.0% of the phenotypic variance, respectively. Except for qLA11_1 and qLA11_2, qLA5_1 and qLA5_2 showed negative additive effects. For linolenic acid, four stable QTLs, namely qLNA5_1 on Chr. 05, qLNA14_1 on Chr. 14, qLNA15_1 on Chr. 15, and qLNA15_2 on Chr. 15, explained an average of 12.5, 10.0, 12.9, and 16.1% of the phenotypic variance, respectively. All of the four QTLs had positive additive effects, which indicated the positive effect on these alleles from the wild soybean parent ZYD00463. Among the 17 stable QTLs for fatty acid composition, qPA10_1 (located within 5.94–9.98 Mb) did not overlap with or was not in the vicinity of the reported QTLs and quantitative trait nucleotides (QTNs) (Fig. 2; Additional files 8 and 11). This suggested that this is a novel locus for palmitic acid.
We identified 21 enzyme-encoding genes that might be involved in lipid metabolism within stable QTL regions (Additional file 9). These genes included two pyruvate kinase genes (GmPK, Glyma.10 g065000 and Glyma.13 g149800), two diacylglycerol acyltransferase genes (GmDGAT, Glyma.13 g106100 and Glyma.13 g118300), three phospholipid diacylglycerol acyltransferase genes (GmPDAT, Glyma.13 g108100, Glyma.16 g005800, and Glyma.11 g190400), two 3-ketoacyl-ACP-synthase I genes (GmFabB, Glyma.13 g128000 and Glyma.05 g218600), a 3-ketoacyl-ACP-synthase II gene (GmFabF, Glyma.13 g112700), two 3-ketoacyl-CoA synthase genes (GmKCS, Glyma.15 g042500 and Glyma.15 g046300), a 3-hydroxyacyl-ACP dehydrase gene (GmFabZ, Glyma.15 g052500), two acyl carrier protein genes (GmACP, Glyma.16 g011300 and Glyma.05 g201300), a malonyl-CoA:ACP malonyltransferase gene (GmFabD, Glyma.11 g164500), an omega-3-fatty acid desaturase 3 gene (GmFAD3, Glyma.11 g174100), an acetyl-CoA carboxylase gene (GmACCase, Glyma.05 g221100), an acyl-CoA synthase gene (GmACS, Glyma.11 g194500), a pyruvate dehydrogenase gene (GmPDH, Glyma.14 g186900), and a lysophosphatidic acid acyltransferase gene (GmLPAAT, Glyma.15 g034100). In addition, nine transcription factor genes involved in lipid metabolism were also identified (Additional file 10), including a helix-loop-helix gene (GmHLH, Glyma.05 g200900), a WRKY protein gene (GmWRKY, Glyma.05 g203900), a C3H protein gene (GmC3H, Glyma.05 g224400), two homeobox-leucine zipper genes (GmZIP, Glyma.10 g071700 and Glyma.11 g145800), a B3 domain protein gene (GmB3, Glyma.10 g076100), two MYB protein genes (GmMYB, Glyma.13 g109100 and Glyma.16 g007200), and a DBB protein gene (GmDBB, Glyma.15 g029500).
Co-localization of the QTLs for different traits was also observed on Chr. Eleven and Fifteen (Additional file 7). The QTLs qOA11_1 and qLA11_1 were co-located on Chr. Eleven within the physical interval of 11.0–25.6 Mb. The QTLs qOC15_1, qPA15_1, qOA15_1, and qLNA15_2 were co-located on Chr. Fifteen within the physical interval of 2.80–5.63 Mb.
Discussion
In the present study, we identified 24 stable QTLs for seed oil and composition. By comparing their mapped regions with previous reports on the soybean reference genome (Fig. 2; Additional files 8 and 11), we discovered that qPA10_1 did not overlap with or was not adjacent to any of the previously reported QTLs. Furthermore, it did not contain QTNs associated with palmitic acid obtained by GWAS. Due to a higher density genetic map constructed with 11,398 SNP markers, the intervals of the QTL regions were significantly reduced in comparison with those previously reported. For example, for oil content, the physical distance of qOC8_1, qOC8_2, qOC15_2, and qOC20_2 in our study was 7.52–9.44 Mb, 9.44–10.8 Mb, 4.15–5.63 Mb, and 32.5–33.8 Mb, respectively. In comparison, the intervals were 5.58–10.28 Mb for qOC8_1 [16], 5.58–10.3 Mb for qOC8_2 [16], 3.23–4.07 Mb for qOC15_2 [17], and 27.0–34.3 Mb for qOC20_2 [54]. For stearic acid, the physical distance of qSA14_1 and qSA14_3 in our study was 32.2–37.5 Mb and 42.2–43.4 Mb, respectively. In comparison, the interval was 16.3–45.9 Mb [56]. For linoleic acid, the physical distance of qLA5_1 and qLA5_2 in our study was 37.5–39.9 Mb and 38.5–40.7 Mb, while the interval was 37.6–42.2 Mb [57].
In comparison with previously reported QTLs (http://www.soybase.org) (Additional files 8 and 11), 23 out of the 24 QTLs were close to or overlapped with previously reported QTLs (Fig. 2). For example, the QTL qOC2_1 for oil content on Chr. 02 was located around 5.08–6.27 Mb, which was adjacent to a reported QTL (6.86–9.67 Mb) [50]. In addition, qOC8_1 on Chr. 08, qOC8_2 on Chr. 08, qOC15_1 on Chr. 15, and qOC15_2 on Chr. 15 were located around 7.52–9.44 Mb, 9.44–10.8 Mb, 2.80–5.63 Mb, and 4.15–5.63 Mb, which overlapped with the reported 5.52–12.64 Mb [16, 51, 52], 5.52–14.21 Mb [51,52,53], 3.23–4.07 Mb [17], and 4.52–5.21 Mb [25], respectively. For palmitic acid, qPA13_1 (21.8–26.8 Mb) on Chr. 13 and qPA16_1 (0.42–1.19 Mb) on Chr. 16 were close to the reported 26.41–29.08 Mb and 2.67–5.06 Mb, respectively [55, 56]. For stearic acid, qSA14_1, qSA14_2, and qSA14_3 on Chr. 14 were located around 32.2–37.5 Mb, 16.3–40.5 Mb, and 42.2–43.4 Mb, respectively, which overlapped with the region of 16.30–45.90 Mb [56]. For linolenic acid, qLNA5_1 (33.8–35.4 Mb) partially overlapped with 31.98–34.65 Mb [18]. Although these QTLs are located in similar regions, whether their responsible genes are identical requires further investigation. In contrast, qPA10_1 on Chr. 10 was mapped to the region of 5.94–9.98 Mb, which was distal from the reported 0.98–1.87 Mb [24] (Fig. 2; Additional file 8) and did not contain any reported QTNs that were associated with palmitic acid by GWAS [58] (Additional file 11). This result indicated that this QTL is a novel locus for palmitic acid.
By annotating all genes in 24 stable QTL intervals with the Gene Annotation Tool of the SoyBase database [20], 12 important enzyme-encoding genes involved in lipid metabolism were identified (Additional file 9). For example, GmACCase (Glyma.05 g221100) encoded an acetyl-CoA carboxylase that catalyzes the formation of malonyl-CoA from acetyl-CoA as the direct substrate of the de novo biosynthesis of fatty acids [59]. GmFabD (Glyma.11 g164500) encoded a malonyl-CoA:ACP malonyltransferase that is responsible for transferring the malonyl group of malonyl-CoA to an acyl carrier protein (ACP). GmACP (Glyma.05 g201300 and Glyma.16 g011300) encoded an acyl carrier protein that transports the growing fatty acid chain between the enzymatic domains of fatty acid synthase [60]. GmFabB (Glyma.05 g218600 and Glyma.13 g128000) and GmFabF (Glyma.13 g112700) encoded a ketoacyl-ACP synthases I and II, respectively, which are mainly used to produce palmitoyl-ACP and stearoyl-ACP as the condensing enzyme of fatty acid chain elongation, respectively. GmFabG (Glyma.08 g102100) and GmFabZ (Glyma.15 g052500) encoded a 3-ketoacyl-ACP reductase and 3-hydroxyacyl-ACP dehydrase, which catalyze the reduction and dehydration reaction of 3-ketoacyl-ACP, respectively [61]. In addition, GmFAD3 (Glyma.11 g174100) encoded an omega-3 fatty acid desaturase 3 that catalyzes a third double bond into linoleic acid to produce linolenic acid [40, 62]. GmDGAT (Glyma.13 g106100 and Glyma.13 g118300) encoded a diacylglycerol acyltransferase that catalyzes the formation of TAGs from fatty acids and glycerol 3-phosphate [63, 64]. The presence of these genes within the stable QTL suggests that these may contribute to soybean seed lipid metabolism. However, whether these enzyme-encoded genes are responsible for the corresponding QTLs requires confirmation using transgenic methods. In addition to the key enzyme genes involved in lipid metabolism, some transcription factor genes also have important roles in regulating fatty acid biosynthesis. We identified 11 transcription factor genes involved in lipid metabolism within the 24 stable QTL intervals (Additional file 10), including GmB3, GmC3H, GmDBB, GmHLH, GmMYB, GmNF-YA, GmWRKY, and GmZIP. Several transcription factors involved in regulating oil and fatty acid biosynthesis have been studied in soybean. For example, the overexpression of GmNFYA in Arabidopsis significantly increases seed oil content [46]. GmMYB73 promotes lipid accumulation in transgenic Arabidopsis, possibly through the suppression of GLABRA2, a transcription factor of HD-ZIP [49]. The roles of transcription factors in our QTL regions in the domestication of the seed oil trait in soybean should be further studied.
In the interval of the novel QTL qPA10_1, three candidate genes potentially involved in lipid metabolism were also identified, including GmPK (Glyma.10 g065000), GmB3 (Glyma.10 g076100), and GmZIP (Glyma.10 g071700). GmPK (Glyma.10 g065000) encodes a pyruvate kinase (PK). During soybean seed development, phosphoenolpyruvate carboxylase and pyruvate kinase activities contribute to a complex interaction that regulates the metabolic flow of glycolytic carbon into precursors for both protein and oil biosynthesis [65]. In Arabidopsis thaliana seeds, reducing the plastidic pyruvate kinase activity by disruption of the gene encoding the β1 subunit of pyruvate kinase resulted in a 60% reduction of seed oil content [66]. GmB3 (Glyma.10 g076100) encodes a B3 domain family of transcription factor. In Brassica napus, the BnFUSCA3 (BnFUS3) mutant, a member of B3 domain transcription factors, repressed seed oil levels and increased levels of linoleic acid, possibly due to the reduced expression of ω-3 FADESATURASE (FAD3) [67]. In soybean, GmLEC2a, a B3 domain transcription factor, complemented the defects of the Arabidopsis atlec2 mutant in seedling development and triacylglycerol accumulation. The overexpression of GmLEC2a in Arabidopsis seeds increased triacylglycerol content by 34% and the composition of long chain fatty acids by 4% relative to the control seeds [68]. GmZIP (Glyma.10 g071700) encodes a homeobox-leucine zipper protein. Song et al. (2013) reported that GmbZIP123 (Glyma.06 g010200) enhances lipid content in transgenic Arabidopsis seeds by promoting expression of sucrose transporter genes (SUC1 and SUC5) and cell wall invertase genes (cwINV1, cwINV3, and cwINV6) [48]. In A. thaliana, bZIP67 binds to G-boxes in the FATTY ACID DESATURASE3 (FAD3) promoter, enhances FAD3 expression, and increases linolenic acid seed content [69].
Conclusions
By means of SLAF-seq technology, we constructed a high-density genetic map comprising 11,398 SNP markers and identified 24 stable QTLs for soybean seed oil content and fatty acid composition using a 181 RIL population derived from a wild soybean and a cultivated soybean. Among these QTLS, one QTL qPA10_1 did not overlap with or was not close to previously reported QTLs and also did not contain any QTNs associated with palmitic acid, indicating that it is a novel locus. Some interesting genes in the QTL regions were also identified and are worthy of further investigation. Our study provides a valuable information that may contribute to the elucidation of oil biosynthesis in soybean.
Methods
Plant materials
The RIL mapping population comprised 181 F7 progenies derived from a cross between the wild soybean ZYD00463 (Glycine soja) and the cultivated soybean WDD01514 (Glycine max). The parents of ZYD00463 (Glycine soja) and WDD01514 (Glycine max) were provided by the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences (Wuhan, China). All 181 RILs and their parents were planted at two experimental stations in Hubei Province, Wuhan (N30°35′, E114°33′) and Henan Province, Xuchang (N34°02′, E113°81′) in 2015 and 2016. The two sites possess different climatic conditions, with Wuhan experiencing higher temperatures and more rain and Xuchang experiencing lower temperatures and less rain. Three replicates of the parents and progenies were planted following a randomized complete block design. Each plot comprised a 2.5-m row with 1.0-m spacing between rows and 0.5-m spacing between adjacent plants. For each genotype, seeds were harvested from five plants from each plot at the R8 growth stage (full-maturity stage) [70]. The soybean seeds were air-dried until a constant weight, then the traits were assessed as described below.
Oil and fatty acid determination
The seed oil content and fatty acid composition were determined according to Wei et al. [71] with minor modifications. Briefly, 30 soybean seeds from each line were ground to a fine powder. Twenty milligrams of each powdered sample were transferred to a 10-mL glass tube. Sulfuric acid-methanol (5%, 2 mL), butylated hydroxytoluene (BHT) (0.2%, 25 μL), methylbenzene (300 μL), and internal standard (IS) (methyl heptadecanoate, Sigma Aldrich, St. Louis, USA, 2.5–5 mg/mL, 100 μL) were added to the samples for fatty acid methyl ester (FAME) preparation, and the sample was esterified in a water bath at 90–95 °C for 1.5 h. After cooling to room temperature, sodium chloride (0.9%, 1 mL) and n-hexane (1 mL) were added to the extracts. The supernatant was obtained for gas chromatography (GC) analysis.
The esters were separated by a GC (Agilent 6890 N, USA) fitted with a capillary column (FFAP, 30 m, 0.25 mm i.d., 0.50-μm film thickness). Nitrogen was used as the carrier gas at an inlet pressure of 25 psi. The temperatures of the injection port and detector (FID) were maintained at 250 °C and 260 °C, respectively, and the temperature program for the column was as follows: 210 °C (1 min), increasing to 230 °C at 10 °C/min (22 min). The computer software used for statistical analysis was STATISTICA version 6.0 (Statsoft Inc., Oklahoma, USA). The peaks were identified based on their retention times using authentic standards of fatty acid methyl esters. The relative peak area was used for quantification of the contents of the fatty acids. The formula for calculating seed oil content was as follows:
where At and As are the total peak area and the internal standard peak area identified based on their retention times, respectively; and ms and mi are the weights of the internal standard and dry seed. The samples of each line were determined thrice.
Statistical analysis
All the phenotypic data were analyzed using PROC MIXED program in SAS 9.3 (SAS Institute Inc., Cary, NC, USA). Pearson’s correlation coefficients among all traits were calculated from lines in single environments using the PROC CORR function in SAS 9.3.
The heritability in a single environment was estimated as follows:
The heritability across environments was calculated as follows:
where \( {\updelta}_{\mathrm{g}}^2 \) is the genotypic variance component for traits per plot among the RILs, \( {\updelta}_{\mathrm{e}}^2 \) is the error variance, r is the number of replications for the trait, and n is the number of environments [72, 73]. ANOVA was conducted across environments to determine the significance of genotype, environment, and their interactions. The error components of variance (\( {\updelta}_{\mathrm{e}}^2 \)), genotype × environment interaction (\( {\updelta}_{\mathrm{gy}}^2 \)), and genotype (\( {\updelta}_{\mathrm{g}}^2 \)) were analyzed using the general linear model procedure (PROC GLM) in SAS 9.3 (SAS Institute Inc.). All parameters were estimated from the expected mean squares in the ANOVA.
Genetic map construction
The SLAF-seq method was applied to develop SNP markers between the two parents [74]. The SNP markers were used to construct the high-density genetic linkage map using the Kosambi mapping function of the JoinMap version 4.0 software [75]. The SNP markers were grouped based on a LOD score of 3.0 and then ordered by the input algorithm to estimate the recombination frequencies. Recombination frequencies between linked loci were transformed into distances (cM) [76]. The collinearity of the LGs with the soybean reference genome was analyzed by aligning the sequence of each SNP marker with the genome sequences of Williams 82 [77].
QTL mapping and candidate gene prediction
Composite interval mapping (CIM) incorporated into WinQTL cartographer version 2.5 was used to detect additive QTLs [78]. For each trait, the threshold for the identification of a significant QTL with a LOD > 3.6 was estimated by permutation tests with 1000 repetitions at P < 0.05. Cofactors were taken into account, and a window size of 10 cM around the test interval was selected for CIM analysis. The distribution of the QTLs on the genetic linkage map was mapped using MapChart version 2.2 [79]. The detected QTLs were denoted by combining a letter or letters representing the abbreviation of traits with a chromosome number [80]. The QTLs that were repeatedly detected in at least two environments, Wuhan and Xuchang across 2 years, were defined as stable QTLs in this study. The QTLs that were previously reported by other groups were defined as reported QTLs. The predicted genes within the stable QTL intervals were obtained from the SoyBase database according to the annotation of the soybean reference genome (Wm82.a2.v1.1). Gene Ontology (GO) enrichment analysis of the predicted genes was performed using the GO website with default settings [81].
Availability of data and materials
All data supporting the conclusions of this article are included within the article and its additional files.
Abbreviations
- ANOVA:
-
Analysis of variance
- BHT:
-
Butylated hydroxytoluene
- CIM:
-
Composite interval mapping
- FAME:
-
Fatty acid methyl ester
- GC:
-
Gas chromatography
- GO:
-
Gene ontology
- GWAS:
-
Genome-wide association study
- IS:
-
Internal standard
- LA:
-
Linoleic acid
- LG:
-
Linkage group
- LNA:
-
Linolenic acid
- LOD:
-
Logarithm of odds
- MAS:
-
Marker-assisted selection
- NGS:
-
Next-generation sequencing
- OA:
-
Oleic acid
- OC:
-
Oil content
- PA:
-
Palmitic acid
- QTL:
-
Quantitative trait locus
- QTN:
-
Quantitative trait nucleotide
- RIL:
-
Recombinant inbred line
- SA:
-
Stearic acid
- SLAF-seq:
-
Specific-locus amplified fragment sequencing
- SNP:
-
Single-nucleotide polymorphism
References
Kim MY, Van K, Kang YJ, Kim KH, Lee SH. Tracing soybean domestication history: from nucleotide to genome. Breed Sci. 2012;61:445–52.
SoyStats. American Soybean Association (ASA), St. Louis, Missouri. 2019. http://soystats.com/international-world-oilseed-production/. Accessed 10 Sep 2019.
Lee JD, Bilyeu KD, Shannon JG. Genetics and breeding for modified fatty acid profile in soybean seed oil. J Crop Sci Biotechnol. 2007;10(4):201–10.
Clemente TE, Cahoon EB. Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol. 2009;151:1030–40.
Abedi E, Sahari MA. Long-chain polyunsaturated fatty acid sources and evaluation of their nutritional and functional properties. Food Sci Nutr. 2014;2(5):443–63.
Smouse TH. A review of soybean oil reversion flavor. J Am Oil Chem Soc. 1979;56(11):747–51.
Mounts TL, Warner K, List GR, Kleiman R, Fehr WR, Hammond EG, Wilcox JR. Effect of altered fatty acid composition on soybean oil stability. J Am Oil Chem Soc. 1988;65(4):624–8.
Lee JD, Bilyeu KD, Pantalone VR, Gillen AM, So YS, Shannon JG. Environmental stability of oleic acid concentration in seed oil for soybean lines with FAD2-1A and FAD2-1B mutant genes. Crop Sci. 2012;52(3):1290–7.
Burton JW. Quantitative genetics: results relevant to soybean breeding. Am Soc Agron. 1987;16:211–47.
Diers BW, Keim P, Shoemaker RC, Fehr WR. RFLP analysis of soybean seed protein and oil content. Theor Appl Genet. 1992;83(5):608–12.
Spencer M, Landau-Ellis D, Meyer E, Pantalone V. Molecular markers associated with linolenic acid in soybean. J Am Oil Chem Soc. 2004;81(6):559–62.
Shibata M, Takayama K, Ujiie A, Yamada T, Abe J, Kitamura K. Genetic relationship between lipid content and linolenic acid concentration in soybean seeds. Breed Sci. 2008;58(4):361–6.
Bachlava E, Dewey R, Burton J, Cardinal A. Mapping and comparison of quantitative trait loci for oleic acid seed content in two segregating soybean populations. Crop Sci. 2009;49(2):433–42.
Qi Z, Wu Q, Han X, Sun Y, Du X, Liu C, Jiang H, Hu G, Chen Q. Soybean oil content QTL mapping and integrating with meta-analysis method for mining genes. Euphytica. 2011;179(3):499–514.
Sun Y, Luan H, Qi Z, Shan D, Liu C, Hu G, Chen Q, Qi Z, Wu Q, Han X, Sun Y, Du X, Liu C, Jiang H, Hu G, Chen Q, Shan D. An integrated quantitative trait locus map of oil content in soybean, Glycine max (L.) Merr., generated using a meta-analysis method for mining genes. Euphytica. 2011;179(3):499–514.
Mao T, Jiang Z, Han Y, Teng W, Zhao X, Li W. Identification of quantitative trait loci underlying seed protein and oil contents of soybean across multi-genetic backgrounds and environments. Plant Breed. 2013;132(6):630–41.
Pathan S, Vuong T, Clark K, Lee J, Shannon J, Roberts C, Ellersieck M, Burton J, Cregan P, Hyten D, Nguyen H, Sleper D. Genetic mapping and confirmation of quantitative trait loci for seed protein and oil contents and seed weight in soybean. Crop Sci. 2013;53(3):765–74.
Ha B, Kim H, Velusamy V, Vuong TD, Nguyen HT, Shannon JG, Lee J. Identification of quantitative trait loci controlling linolenic acid concentration in PI483463 (Glycine soja). Theor Appl Genet. 2014;127(7):1501–12.
Kim H, Kim Y, Kim S, Son B, Choi Y, Kang J, Park Y, Cho Y, Choi I. Analysis of quantitative trait loci (QTLs) for seed size and fatty acid composition using recombinant inbred lines in soybean. J Life Sci. 2010;20(8):1186–92.
Soybase: Integrating Genetics and Genomics to Advance Soybean Research. Iowa State University, Ames. 2017. https://www.soybase.org/. Accessed 10 Oct 2017.
Csanadi G, Vollmann J, Stift G, Lelley T. Seed quality QTLs identified in a molecular map of early maturing soybean. Theor Appl Genet. 2001;103(6–7):912–9.
Wang X, Jiang G, Green M, Scott R, Song Q, Hyten D, Cregan P. Identification and validation of quantitative trait loci for seed yield, oil and protein contents in two recombinant inbred line populations of soybean. Mol Gen Genomics. 2014;289(5):935–49.
Han Y, Teng W, Wang Y, Zhao X, Wu L, Li D, Li W. Unconditional and conditional QTL underlying the genetic interrelationships between soybean seed isoflavone, and protein or oil contents. Plant Breed. 2015;134(3):300–9.
Reinprecht Y, Poysa V, Yu K, Rajcan I, Ablett G, Pauls K. Seed and agronomic QTL in low linolenic acid, lipoxygenase-free soybean (Glycine max (L.) Merrill) germplasm. Genome. 2006;49(12):1510–27.
Patil G, Vuong TD, Kale S, Valliyodan B, Deshmukh R, Zhu C, Wu X, Bai Y, Yungbluth D, Lu F, Kumpatla S, Shannon JG, Varshney RK, Nguyen HT. Dissecting genomic hotspots underlying seed protein, oil, and sucrose content in an interspecific mapping population of soybean using high-density linkage mapping. Plant Biotechnol J. 2018;16(11):1939–53.
Xie D, Han Y, Zeng Y, Chang W, Teng W, Li W. SSR-and SNP-related QTL underlying linolenic acid and other fatty acid contents in soybean seeds across multiple environments. Mol Breed. 2012;30(1):169–79.
Darvasi A, Weinreb A, Minke V, Weller JI, Soller M. Detecting marker-QTL linkage and estimating QTL gene effect and map location using a saturated genetic-map. Genetics. 1993;134(3):943–51.
Collard BCY, Mackill DJ. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B. 2008;363(1491):557–72.
Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83.
Cao Y, Li S, Wang Z, Chang F, Kong J, Gai J, Zhao T. Identification of major quantitative trait loci for seed oil content in soybeans by combining linkage and genome-wide association mapping. Front Plant Sci. 2017;8:1222.
Li B, Fan S, Yu F, Chen Y, Zhang S, Han F, Yan S, Wang L, Sun J. High-resolution mapping of QTL for fatty acid composition in soybean using specific-locus amplified fragment sequencing. Theor Appl Genet. 2017;130(7):1467–79.
Zhang Y, Li W, Lin Y, Zhang L, Wang C, Xu R. Construction of a high-density genetic map and mapping of QTLs for soybean (Glycine max) agronomic and seed quality traits by specific length amplified fragment sequencing. BMC Genomics. 2018;19:641.
Li Y, Reif JC, Ma Y, Hong H, Liu Z, Chang R, Qiu L. Targeted association mapping demonstrating the complex molecular genetics of fatty acid formation in soybean. BMC Genomics. 2015;16:841.
Bandillo N. Arquin D, Song Q, Nelson R, Cregan P, Specht J, Lorenz a. a population structure and genome-wide association analysis on the USDA soybean germplasm collection. Plant Genome. 2015;8(3):1–13.
Sonah H, O’Donoughue L, Cober E, Rajcan I, Belzile F. Indentification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soybean. Plant Biotechnol J. 2015;13(2):211–21.
Leamy LJ, Zhang H, Li C, Chen CY, Song B. A genome-wide association study of seed composition traits in wild soybean (Glycine soja). BMC Genomics. 2017;18:18.
Fang C, Ma Y, Wu S, Liu Z, Wang Z, Yang R, Hu G, Zhou Z, Yu H, Zhang M, Pan Y, Zhou G, Ren H, Du W, Yan H, Wang Y, Han D, Shen Y, Liu S, Liu T, Zhang J, Qin H, Yuan J, Yuan X, Kong F, Liu B, Li J, Zhang Z, Wang G, Zhu B, Tian Z. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 2017;18:161.
Zhang J, Wang X, Lu Y, Bhusal SJ, Song Q, Cregan PB, Yen Y, Brown M, Jiang G. Genome-wide scan for seed composition provides insights into soybean quality improvement and the impacts of domestication and breeding. Mol Plant. 2018;11(3):460–72.
Schlueter J, Vasylenko-Sanders I, Deshpande S, Yi J, Siegfried M, Roe B, Schlueter S, Scheffler B, Shoemaker R. The FAD2 gene family of soybean: insights into the structural and functional divergence of a paleopolyploid genome. Plant Genome. 2007;47(1):S14–26.
Bilyeu KD, Palavalli L, Sleper DA, Beuselinck PR. Three microsomal omega-3 fatty-acid desaturase genes contribute to soybean linolenic acid levels. Crop Sci. 2003;43(5):1833–8.
Andreu V, Lagunas B, Collados R, Picorel R, Alfonso M. The GmFAD7 gene family from soybean: identification of novel genes and tissue-specific conformations of the FAD7 enzyme involved in desaturase activity. J Exp Bot. 2010;61(12):3371–84.
Aghoram K, Wilson RE, Burton JW, Dewey RE. A mutation in a 3-keto-acyl-ACP synthase II gene is associated with elevated palmitic acid levels in soybean seeds. Crop Sci. 2006;46(6):2453–9.
Head K, Galos T, Fang Y, Hudson K. Mutations in the soybean 3-ketoacyl-ACP synthase gene are correlated with high levels of seed palmitic acid. Mol Breed. 2012;30(3):1519–23.
Roesler K, Shen B, Bermudez E, Li C, Hunt J, Damude HG, Ripp KG, Everard JD, Booth JR, Castaneda L, Feng L, Meyer K. An improved variant of soybean type 1 diacylglycerol acyltransferase increases the oil content and decreases the soluble carbohydrate content of soybeans. Plant Physiol. 2016;171:878–93.
Dehesh K, Tai H, Edwards P, Byrne J, Jaworski JG. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001;125:1103–14.
Lu X, Li Q, Xiong Q, Li W, Bi Y, Lai Y, Liu X, Man W, Zhang W, Ma B, Chen S, Zhang J. The transcriptomic signature of developing soybean seeds reveals the genetic basis of seed trait adaptation during domestication. Plant J. 2016;86(6):530–44.
Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 2007;52(4):716–29.
Song QX, Li QT, Liu YF, Zhang FX, Ma B, Zhang WK, Man WQ, Du WG, Wang GD, Chen SY, Zhang JS. Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J Exp Bot. 2013;64(14):4329–41.
Liu YF, Li QT, Lu X, Song QX, Lam SM, Zhang WK, Ma B, Lin Q, Man WQ, Du WG, Shui GH, Chen SY, Zhang JS. Soybean GmMYB73 promotes lipid accumulation in transgenic plants. BMC Plant Biol. 2014;14:73.
Kabelka EA, Diers BW, Fehr WR, LeRoy AR, Baianu IC, You T, Neece DJ, Nelson RL. Putative alleles for increased yield from soybean plant introductions. Crop Sci. 2004;44(3):784–91.
Mansur LM, Lark KG, Kross H, Oliveira A. Interval mapping of quantitative trait loci for reproductive, morphological, and seed traits of soybean (Glycine max L.). Theor Appl Genet. 1993;86(8):907–13.
Liang H, Yu Y, Wang S, Lian Y, Wang T, Wei Y, Gong P, Liu X, Fang X, Zhang M. QTL mapping of isoflavone, oil and protein contents in soybean (Glycine max L. Merr.). Agric Sci China. 2010;9(8):1108–16.
Tajuddin T, Watanabe S, Yamanaka N, Harada K. Analysis of quantitative trait loci for protein and lipid contents in soybean seeds using recombinant inbred lines. Breed Sci. 2003;53(2):133–40.
Chung J, Babka HL, Graef GL, Staswick PE, Lee DJ, Cregan PB, Shoemaker RC, Specht JE. The seed protein, oil, and yield QTL on soybean linkage group I. Crop Sci. 2003;43(3):1053–67.
Wang X, Jiang GL, Green M, Scott RA, Hyten DL, Cregan PB. Quantitative trait locus analysis of saturated fatty acids in a population of recombinant inbred lines of soybean. Mol Breed. 2012;30(2):1163–79.
Li H, Zhao T, Wang Y, Yu D, Chen S, Zhou R, Gai J. Genetic structure composed of additive QTL, epistatic QTL pairs and collective unmapped minor QTL conferring oil content and fatty acid components of soybeans. Euphytica. 2011;182:117–32.
Diers BW, Shoemaker RC. Restriction fragment length polymorphism analysis of soybean fatty acid content. J Am Oil Chem Soc. 1992;69(12):1242–7.
Priolli RHG, Campos JB, Stabellini NS, Pinheiro JB, Vello NA. Association mapping of oil content and fatty acid components in soybean. Euphytica. 2015;203(1):83–96.
Ohlrogge JB, Jaworski JG. Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:109–36.
Chan DI, Vogel HJ. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem J. 2010;430(1):1–19.
Wu GZ, Xue HW. Arabidopsis β-ketoacyl-[acyl carrier protein] synthase I is crucial for fatty acid synthesis and plays a role in chloroplast division and embryo development. Plant Cell. 2010;22:3726–44.
Bilyeu K, Palavalli L, Sleper D, Beuselinck P. Mutations in soybean microsomal omega-3 fatty acid desaturase genes reduce linolenic acid concentration in soybean seeds. Crop Sci. 2005;45(5):1830–6.
Kennedy EP. Biosynthesis of complex lipids. Fed Proc. 1961;20:934–40.
Settlage SB, Kwanyuen P, Wilson RF. Relation between diacylglycerol acyltransferase activity and oil concentration in soybean. J Am Oil Chem Soc. 1998;75(7):775–81.
Smith AJ, Rinne RW, Seif RD. Phosphoenolpyruvate carboxylase and pyruvate kinase involvement in protein and oil biosynthesis during soybean seed development. Crop Sci. 1989;29(2):349–53.
Andre C, Froehlich JE, Moll MR, Benning C. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell. 2007;19:2006–22.
Elahi N, Duncan RW, Stasolla C. Decreased seed oil production in FUSCA3 Brassica napus mutant plants. Plant Physiol Biochem. 2015;96:222–30.
Manan S, Ahmad MZ, Zhang G, Chen B, Haq BU, Yang J, Zhao J. Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front Plant Sci. 2017;8:1604.
Mendes A, Kelly AA, van Erp H, Shaw E. Powers Sj, Kurup S, Eastmond PJ. bZIP67 regulates the Omega-3 fatty acid content of Arabidopsis seed oil by activating FATTY ACID DESATURASE3. Plant Cell. 2013;25(8):3104–16.
Fehr WR, Caviness CE, Burmood DT, Pennington JS. Stage of development descriptions for soybeans, Glycine max (L.). Merrill. Crop Sci. 1971;11(6):929–31.
Wei F, Gao G, Wang X, Dong X, Li P, Hua W, Wang X, Wu X, Chen H. Quantitative determination of oil content in small quantity of oilseed rape by ultrasound-assisted extraction combined with gas chromatography. Ultrason Sonochem. 2008;15(6):938–42.
Xing GN, Zhou B, Wang YF, Zhao TJ, Yu DY, Chen SY, Gai JY. Genetic components and major QTL confer resistance to bean pyralid (Lamprosema indicate Fabricius) under multiple environments in four RIL populations of soybean. Theor Appl Genet. 2012;125(5):859–75.
Hanson HL, Robinson HF, Comstock RE. Biometrical studies of yield in segregating populations of Korean Lespodeza. Agron J. 1956;48:268–72.
Sun XW, Liu DY, Zhang XF, Li WB, Liu H, Hong WG, Jiang CB, Guan N, Ma CX, Zeng HP. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One. 2013;8(3):e58700.
Van Ooijen JW. JoinMap 4: software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma BV, Netherlands. 2006.
Kosambi DD. The estimation of map distances from recombination values. Ann Eugenics. 1944;12(1):172–5.
Liu D, Ma C, Hong W, Huang L, Liu M, Liu H, Zeng H, Deng D, Xin H, Song J, Xu C, Sun X, Hou X, Wang X, Zheng H. Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS One. 2014;9(6):e98855.
Wang S, Basten CJ, Zeng ZB. Windows QTL cartographer version 2.5. Department of Statistics. Raleigh: North Carolina State University; 2012.
Voorrips RE. MapChart: software for the graphical presentation of the linkage maps and QTLs. J Hered. 2002;93(1):77–8.
Asekova S, Kulkarni KP, Kim M, Kim JH, Song JT, Shannon JG, Lee JD. Novel quantitative trait loci for forage quality traits in a cross between PI 483463 and ‘Hutcheson’ in soybean. Crop Sci. 2016;56(5):1–12.
Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(S2):64–70 declare.
Acknowledgments
We greatly thank Dr. Dan Wu for editing the English text of a draft of this manuscript. We also thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by National Key Research and Development Program of China (No. 2016YFD0100504), National Natural Science Foundation of China (Nos. 31522042 and 31371654), National Transgenic Project (No. 2016ZX08004003), Natural Science Foundation of Hubei (No. 2016CFA049), Chinese Academy of Agricultural Sciences (No. Y2017JC14), and Henan Agricultural University. The role of the funding body is to provide financial support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Y.Y and Y.J. designed the research and wrote the article. Q.Y., G.D., J.R., S.C., J.Z., X.L., X.Z. performed the data analysis and experiments. Y.J. coordinated the study. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no competing financial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Frequency distribution of seed oil content and composition in four environments. The arrows indicate traits in two parental lines. Wild soybean ZYD00463 is the male parent (P1), cultivated soybean WDD01514 is female parent (P2). The frequency distribution of seed oil content are shown in the chart (A-D), palmitic acid (E-H), stearic acid (I-L), oleic acid (M-P), linoleic acid (Q-T) and linolenic acid (U-X) in four different environments.
Additional file 2.
Pearson’s correlation coefficients of seed oil content and composition in four environments.
Additional file 3.
High-density genetic map constructed by SNP markers. The x-axis and y-axis indicate linkage group number and genetic distance (centimorgan, cM), respectively.
Additional file 4.
Positions of SNP markers on 20 linkage groups and their corresponding physical positions (bp) in the soybean reference genome.
Additional file 5.
Description of characteristics of the 20 LGs in the high-density genetic map.
Additional file 6.
Additive QTLs associated with oil content and composition in single environment.
Additional file 7.
Location of stable additive QTLs on genetic linkage map across environments. QTLs are marked with bars. The bar length represents the physical interval of the QTL. The stable QTLs are showed in different color bar. Red bar: oil content; green bar: palmitic acid; blue bar: stearic acid; yellow bar: oleic acid; pale red bar: linoleic acid; light green bar: linolenic acid.
Additional file 8.
Information of previously reported QTLs for soybean seed oil content and composition.
Additional file 9.
Candidate genes involved in lipid metabolism within the 24 stable QTL intervals.
Additional file 10.
Candidate genes encoding transcription factor involved in lipid metabolism within the 24 stable QTL intervals.
Additional file 11.
QTNs for soybean seed oil content and composition detected in previous GWAS.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yao, Y., You, Q., Duan, G. et al. Quantitative trait loci analysis of seed oil content and composition of wild and cultivated soybean. BMC Plant Biol 20, 51 (2020). https://doi.org/10.1186/s12870-019-2199-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-019-2199-7