Abstract
Background
WRKY proteins are one of the largest gene families and are well-known for their regulatory roles in many aspects of plant development, including plant response to both biotic and abiotic stresses. Although the roles of WRKY proteins in leaf blast resistance have been well-documented in rice, their functions in panicle blast, the most destructive type of blast disease, are still largely unknown.
Results
Here, we identified that the transcription of OsWRKY67 was strongly activated by leaf and panicle blast infection. OsWRKY67 is ubiquitously expressed and sub-localized in the nucleus. Rice plants overexpressing OsWRKY67 showed quantitatively enhanced resistance to leaf blast, panicle blast and bacterial blight. In contrast, silencing of OsWRKY67 increased the susceptibility to blast and bacterial blight diseases. RNA-seq analysis indicated that OsWRKY67 induces the transcription of a set of defense-related genes including the ones involved in the salicylic acid (SA)-dependent pathway. Consistent with this, the OsWRKY67-overexpressing plants accumulated higher amounts of endogenous SA, whereas lower endogenous SA levels were observed in OsWRKY67-silenced plants relative to wild-type Nipponbare plants before and after pathogen attack. Moreover, we also observed that OsWRKY67 directly binds to the promoters of PR1a and PR10 to activate their expression.
Conclusions
These results together suggest the positive role of OsWRKY67 in regulating rice responses to leaf blast, panicle blast and bacterial blight disease. Furthermore, conferring resistance to two major diseases makes it a good target of molecular breeding for crop improvement in rice.
Similar content being viewed by others
Background
Rice blast and bacterial blight diseases are two of the leading causes of rice yield loss all over the world [1]. Nowadays, it is a common issue that the rice cultivars always exhibit a short-life span of disease resistance and how to extend the life span of disease resistance is the priority for rice improvement. Employment of host resistance is deemed as the most effective way to address this issue [2]. Host resistance of plants consists of both qualitative (complete) resistance and quantitative (partial) resistance [3, 4]. Quantitative resistance conferred by multiple genes is considered to be more broad-spectrum and durable than qualitative resistance mediated by R genes, based on its non-race specificity [5]. Therefore, quantitative resistance is the favored strategy for sustainable control of plant disease. The quantitative resistance of rice is the result of the comprehensive effect of various kinds of genes, including transcription factors (TF) such as WRKYs.
WRKY TFs are one of the largest gene families and are well-known for their regulatory roles in almost every aspect of plant development, including plant response to both biotic and abiotic stresses [6]. To date, many WRKY proteins have been demonstrated to associate with various defense responses in Arabidopsis, tobacco and barley [7,8,9,10,11,12,13]. In rice, more than 100 WRKY genes have been identified [14], and several of these genes (e.g. OsWRKY13, OsWRKY31, OsWRKY45, OsWRKY53, OsWRKY71, OsWRKY89 and OsWRKY76) have been confirmed to positively regulate rice resistance to blast or bacterial blight via transgenic technology [15,16,17,18,19,20,21,22]. Moreover, OsWRKY62 negatively regulates the basal and Xa21-mediated resistance to bacterial blight [23], and OsWRKY28 is linked to a negative role in blast disease resistance [24].
Despite these results, the regulatory roles of most of the WRKY genes in defense response are still largely unknown. Rice blast disease can be differentiated as leaf blast and panicle blast based on the location of infection in a plant. Panicle blast is more destructive than leaf blast because of its more direct relationship to yield loss. Inconsistent findings of plant response in leaf blast and panicle blast resistance suggest that the mechanisms of rice response to leaf blast and panicle blast likely differ to some extent [25,26,27]. However, majority of studies on WRKY genes in rice blast resistance reported so far have focused on leaf blast except for OsWRKY45, which confers durable resistance to panicle blast [28]. Thus, we still have very limited knowledge about the roles of other WRKY genes in panicle blast resistance in rice.
To investigate the functions of WRKY genes on both leaf and panicle blast resistance, we performed a genome-wide differential gene expression analysis via microarray of a blast resistant advanced backcross line that was infected with leaf and panicle blast. Our results showed that most of the WRKY genes differentially expressed with rice blast infection, implying their crucial roles in regulating rice blast resistance. Among the differentially expressed WRKY genes, we identified that OsWRKY67 (Os05g0183100) was strongly activated by leaf blast and panicle blast infection. The gene overexpression and RNAi silencing experiments suggest that OsWRKY67 positively regulates leaf blast, panicle blast and bacterial blight resistance in rice. Expression profiling in OsWRKY67-overexpressing plants indicates that subsets of defense-related genes were divergently regulated by OsWRKY67. Further analysis also suggests that OsWRKY67 confers disease resistance through the activation of the salicylic acid (SA) signaling pathway and directly targets pathogenesis-related (PR) proteins 1a (PR1a) and 10 (PR10).
Results
Transcription of OsWRKY67 is activated by blast inoculation
Our previous microarray analysis for excavating genes that were regulated by blast infection showed that the expression of OsWRKY67 was remarkably activated by both leaf blast and panicle blast inoculation (Additional file 1: Table S1). Here, to confirm this result, the expression patterns of OsWRKY67 were also analyzed in Nipponbare plants after infection with leaf blast and panicle blast, and real-time PCR was conducted at 6, 12, 24 and 48 h after inoculation. Our results indicated that the transcription levels of OsWRKY67 were remarkably activated at 6, 12 and 24 h in both leaves and panicles after pathogen inoculation, then slightly declined in leaves but remained highly expressed in panicles at 48 h (Fig. 1). These results together suggest the important role of OsWRKY67 in regulating blast disease resistance in rice.
The expression pattern of OsWRKY67 after blast fungus infection. The values are means ± standard deviations (SDs) of three biological replicates, and asterisks represent significant differences relative to the water treatment (0 h) (t-test, **P < 0.01 and *P < 0.05). a The transcription of OsWRKY67 was activated by leaf blast infection. LB = leaf blast. b The transcription of OsWRKY67 was activated by panicle blast infection. PB = panicle blast
OsWRKY67 is ubiquitously expressed and is sub-localized in the nucleus
To determine the spatiotemporal expression of OsWRKY67 in rice, real-time PCR analysis was conducted in different Nipponbare tissues and the result revealed that OsWRKY67 was expressed in all rice tissues inspected, with the highest level in leaf tissues (Fig. 2a). We also fused the promoter of OsWRKY67 to the β-glucuronidase (GUS) gene to generate OsWRKY67pro::GUS transgenic plants in Nipponbare, and the GUS staining analysis was also performed in different tissues of these transgenic plants. The GUS signal was detected in the leaf, bud, node, spikelet hull, one-week old seedling, root, panicles at the booting stage and panicles at the heading stage (Fig. 2b-j). These observations supported the results from the qRT-PCR, suggesting that OsWRKY67 is expressed in different tissues during the entire rice life cycle.
The spatiotemporal expression of OsWRKY67 in Nipponbare plants. a Relative expression levels of OsWRKY67 in different Nipponbare tissues. Young leaf, leaf at the three- to four-leaf stage; old leaf, leaf at the booting stage. The values are means ±SDs of three biological replicates. b-j GUS staining analysis of OsWRKY67 in different Nipponbare tissues. b leaf at the booting stage; c sprouting stage; d hull; e one-week old seedling; f root at the booting stage; g the second node at the booting stage; h the first node at the booting stage; i panicle at the booting stage; j panicle at the heading stage
To investigate the subcellular localization of OsWRKY67, the CDS sequence of OsWRKY67 was fused with the green fluorescence protein (GFP) which is controlled by the CaMV 35S promoter. We transiently expressed the GFP-OsWRKY67 fusion protein and the control protein (empty GFP protein) in onion epidermal cells. The results showed that the GFP signal was localized in the nucleus of the cells transfected with the fusion protein, whereas the GFP signal in the control protein was universally distributed in the nucleus and cytoplasm (Additional file 2: Figure S1a). To further validate this result, the fused GFP-OsWRKY67 protein was also expressed transiently in rice protoplasts and these results also illustrated the nuclear localization of OsWRKY67 protein (Additional file 2: Figure S1b).
Overexpression of OsWRKY67 enhances resistance to blast and bacterial blight in rice
To confirm the function of OsWRKY67 in disease resistance in rice, we produced the OsWRKY67 overexpressing (OX-WRKY67) plants in Nipponbare which are susceptible to blast and Xoo diseases. Twelve independent transgenic lines were generated and the OX-WRKY67 plants showed a dwarfed phenotype relative to wild-type Nipponbare plants. The tiller numbers of OX-WRKY67 plants were also significantly lower than that of wild-type plants (Additional file 3: Figure S2). Two independent homozygous lines (OX 5–2 and OX 6–5) with high transcription levels of OsWRKY67 were selected for disease evaluation (Fig. 3a; Additional file 4: Figure S3). In the spray inoculation experiment, the OX-WRKY67 plants exhibited fewer susceptible-type lesions compared with wild-type Nipponbare plants (Additional file 5: Figure S4). To further validate the role of OsWRKY67 in leaf resistance, we evaluated leaf blast resistance of the OX-WRKY67 plants by inoculation with the same blast isolate using punch method. As expected, the lesions on the leaves of OX-WRKY67 plants were smaller than that on the leaves of Nipponbare plants (Fig. 3b, c). Moreover, the spores in the infected OX-WRKY67 leaves were also fewer than that in the wild-type Nipponbare leaves (Fig. 3d). The two over-expressed lines, OX 5–2 and OX 6–2, also showed enhanced resistance to GD08-T13 at the heading stage, with the infected main axis length ranging from 55 to 56.8%, whereas the infected main axis length was 79.5% for Nipponbare plants (Fig. 3e, f).
Phenotypes of the OsWRKY67 overexpressing (OX-WRKY67) plants infected with blast fungus. The asterisks represent significant differences relative to Nipponbare plants (t-test, **P < 0.01). a Relative transcription levels of OsWRKY67 in wild-type Nipponbare (NPB) and OX-WRKY67 plants. The values are mean ± SD of three biological replicates. b The OX-WRKY67 plants exhibited enhanced leaf blast resistance after inoculation with GD08-T13 using the punch method. c Relative lesion size in Nipponbare and OX-WRKY67 plants after infection by leaf blast. The values are mean ± SD of twelve biological replicates. d Numbers of spores produced on the Nipponbare and OX-WRKY67 plants after punch inoculation. The values are means ± SDs of six biological replicates. e The OX-WRKY67 plants exhibited enhanced panicle blast resistance after inoculation with GD08-T13. f The infected main axis length in Nipponbare and OX-WRKY67 plants after infection by panicle blast. The values are means ± SDs of twenty biological replicates
The OX-WRKY67 plants also showed significantly enhanced resistance to Xoo race 4 (P < 0.01), with lesion lengths ranging from 4.73 to 4.21 cm versus 13.78 cm for the wild type Nipponbare plants (Additional file 6: Figure S5a, b). The number of bacteria on OX-WRKY67 plants was much lower than that on susceptible control Nipponbare at 12 and 16 days after infection (Additional file 6: Figure S5c). These results together suggest that overexpression of OsWRKY67 enhances resistance to blast and bacterial blight in rice.
Silencing of OsWRKY67 increases susceptibility to blast and bacterial blight in rice
To further confirm the functions of OsWRKY67 in rice disease resistance, we produced OsWRKY67-silenced (WRKY67-RNAi) plants of Nipponbare using an RNAi vector. No remarkable differences were observed between the transgenic and wild-type Nipponbare plants. Three independent transgenic lines (RNAi-9, RNAi-11, RNAi-15) in which OsWRKY67 was most severely suppressed were used for disease resistance evaluation (Fig. 4a). Punch inoculation of plants revealed that the lesions on WRKY67-RNAi plants were approximately 50% larger than on Nipponbare plants (Fig. 4b, c) and the WRKY67-RNAi plants had more spores than Nipponbare plants, suggesting that the rate of fungal growth was faster in the WRKY67-RNAi plants relative to Nipponbare plants (Fig. 5d). Moreover, the WRKY67-RNAi plants also showed increased susceptibility to M. oryzae at the heading stage, with the infected main axis length ranging from 63.2 to 90% (82.38%), versus 47.5% for wild-type Nipponbare (Fig. 4e, f). To evaluate the resistance of the WRKY67-RNAi plants to bacterial blight, the leaves of the WRKY67-RNAi and Nipponbare plants were inoculated with isolate Chinese Xoo race 4 at the booting stage. The lesion length in WRKY67-RNAi plants was longer than that in the control plants, and the number of bacteria on RNAi plants was also much more than that on Nipponbare plants at 12 and 16 days after infection (Additional file 7: Figure S6). These results together indicate the positive role of OsWRKY67 in regulating disease resistance in rice.
Phenotypes of the OsWRKY67-silenced (WRKY67-RNAi) plants infected with blast fungus. The asterisks represent significant differences relative to wild-type Nipponbare (NPB) plants (t-test, **P < 0.01 and *P < 0.05). a Relative transcription levels of OsWRKY67 in Nipponbare and WRKY67-RNAi plants. The values are means ± SDs of three biological replicates. b Silencing of OsWRKY67 increased susceptibility to leaf blast infection using punch method. c Relative lesion size in Nipponbare and WRKY67-RNAi plants after leaf blast infection. The values are means ± SDs of twelve biological replicates. d Numbers of spores produced on Nipponbare and WRKY67-RNAi plants after punch inoculation. The values are means ± SDs of six biological replicates. e Silencing of OsWRKY67 increased susceptibility to panicle blast in rice. f The infected main axis length in Nipponbare and WRKY67-RNAi plants after panicle blast inoculation. The values are means ± SDs of twelve biological replicates
OsWRKY67 regulates the transcription of genes involved in the salicylic acid (SA) signaling pathway. The wild-type Nipponbare and transgenic plants were inoculated with GD08-T13 at the three- to four-leaf stage. The values are means ± SDs of three biological replicates and the asterisks represent significant differences relative to Nipponbare plants at **P < 0.01 or *P < 0.05 by t-tests. The transcript level of Nipponbare plants was set to “1” at each time point
Differential expression of defense-related genes in OsWRKY67 overexpressing plants
To learn about the regulatory mechanisms of OsWRKY67 in disease resistance in rice, global gene expression profiling was performed in OX-WRKY67 and wild-type Nipponbare plants by high through-put sequencing. In total, 345 genes were differentially expressed between OX-WRKY67 and Nipponbare plants, including 200 up-regulated genes and 145 down-regulated genes [|log2 Ratio| ≥ 1 and the False Discovery Rate (FDR) ≤ 0.01; Additional file 8: Table S2]. The results of the RNA-seq analysis were verified by real-time PCR for nine up-regulated genes and three down-regulated genes. As shown in Additional file 9: Figure S7, the transcription levels s of two PR4 genes, four chitinases and three peroxidases were markedly induced whereas the three WRKY genes were all suppressed in OX-WRKY plants relative to Nipponbare plants. These results are consistent with the sequencing data, indicating the reliability of results from the RNA-seq analysis (Additional file 10: Figure S8). In addition, the transcript levels of these twelve genes were also analyzed in OX-WRKY and Nipponbare plants after pathogen infection. They all showed a similar differential expression pattern after blast inoculation (Additional file 9: Figure S7), further confirming that these genes were under the control of OsWRKY67.
Among the differentially expressed genes, 59 have been previously demonstrated to be involved or implicated in plant defense response (Additional file 8: Table S2; genes in red). The up-regulated genes in OX-WRKY67 plants included ten PR genes, five chitinase genes, three peroxidase genes and two phenylalanine ammonia-lyases (PAL). Overexpression of OsWRKY67 also activated the transcription of other defense-related genes, such as laccases, cellulose synthases, dirigents and protease inhibitors (Additional file 8: Table S2). In contrast, the transcription of three WRKY genes (WRKY60, WRKY7 and WRKY74) was significantly suppressed in OX-WRKY67 plants compared with those in wild-type plants. Moreover, several genes which are speculated to be associated with defense-related signal transduction, including protein kinases and other transcription factors (MYB, NAC and zinc finger), were also down-regulated in OX-WRKY67 plants (Additional file 8: Table S2).
Gene ontology (GO) analysis of these differentially expressed genes (DEGs) revealed 15 categories of enriched genes. Particularly, “cell wall organization”, “cellulose biosynthesis”, “chitin metabolism” and “response to biotic stimulus” were remarkably enriched for the up-regulated genes, while different “transport” processes were enriched for the down-regulated genes (Additional file 11: Table S3). Furthermore, pathway analysis indicated that these DEGs were mainly involved in ribosomes, biosynthesis of secondary metabolites, metabolic pathway and plant-pathogen interactions (Additional file 12: Table S4).
OsWRKY67 acts in the salicylic acid dependent pathway
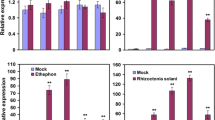
From our observation of the highly induced transcription of PR genes and other defense-related genes, we deduced that OsWRKY67’s functional role in disease resistance may, at least, partially dependent on the SA signaling pathway. To validate this inference, we first analyzed the expression levels of several well-known defense-related genes in the OX-WRKY67, WRKY67-RNAi and wild-type Nipponbare plants. Isochorismate synthase 1 (ICS1) and phenylalanine ammonia-lyase 1 (PAL1) are two SA synthesis genes, while phytoalexin-deficient 4 (PAD4), Arabidopsis NPR1 homolog 1 (NHI), PR1a and PR10 are four genes that function in the SA signaling pathway [17, 27]. Our results showed that ICS1, PAD4, PAL1, NH1, PR1a and PR10 were significantly induced in OX-WRKY67 plants compared with those in Nipponbare plants after inoculation with M. oryzae (Fig. 5a). Among these six genes, the expression of PAL1, PR1a and PR10 were remarkably higher in transgenic plants than in Nipponbare plants both before and after blast inoculation, while the higher transcript levels of ICS1, PAD4 and NH1 were only identified after pathogen inoculation (Fig. 5a). In contrast, the transcription levels of PAD4, PAL1, NH1, PR1a and PR10 were remarkably lower in WRKY67-RNAi plants compared to Nipponbare plants after pathogen inoculation (Fig. 5b). No obvious difference was detected for ICS1 between WRKY67-RNAi and Nipponbare plants (Fig. 5b). Moreover, the transcript levels of these six genes were also analyzed in the transgenic plants and Nipponbare plants at the booting stage after Xoo infection. Consistent with the results from the blast inoculation experiments, the transcription of these genes was induced in OX-WRKY67 plants and down-regulated in WRKY67-RNAi plants relative to the Nipponbare plants (Additional file 13: Figure S9).
Next, we analyzed the transcription of OsWRKY67 after treating the wild-type Nipponbare plants with exogenous SA. The SA treatment significantly induced expression of OsWRKY67 at all measured time points after treatment (Fig. 6a). We also measured the contents of endogenous SA in the seedlings of OX-WRKY67, WRKY67-RNAi and Nipponbare after inoculation with blast isolate GD08-T13 at the three- to four-leaf stage (Fig. 6b). The concentration of SA was elevated due to blast infection in both OX-WRKY67 and Nipponbare plants, but the SA level was remarkably higher in OX-WRKY67 plants compared with wild-type Nipponbare plants before and after blast infection (Fig. 6b). On the contrary, the endogenous SA content was significantly lower (P < 0.05) in the WRKY67-RNAi plants compared to Nipponbare plants after pathogen attack (Fig. 6b). These observations together suggest that there is a close relationship between the OsWRKY67-mediated defense response and the SA signaling pathway.
OsWRKY67 confers disease resistance in a SA-dependent manner. The values are means ± SDs of three biological replicates and the asterisks represent significant differences relative to the water treatment (0 h) or Nipponbare plants at **P < 0.01 and *P < 0.05 by t-test. a The expression of OsWRKY67 was activated by exogenous SA. CK = water treatment. b Transcriptionally modulating OsWRKY67 influenced the accumulation of SA
OsWRKY67 directly regulates the expression of PR1a and PR10
The WRKY transcription factors regulate the expression of downstream genes by binding to the promoters of these genes via the W-box elements. To determine the DNA-binding activity of OsWRKY67, we performed a yeast one-hybrid assay using DNA fragments (BS65 and mBS65) according to previous reports [24, 29]. The assay showed that OsWRKY67 strongly bonded to the probe of BS65, whereas OsWRKY67 did not recognize the mutant probe mBS65, implying the specificity in DNA-binding for OsWRKY67 (Fig. 7a). Interestingly, we also identified multiple W-box sequences in the promoters of OsWRKY67, PR1a, PR10, ICS1 and NH1, and the yeast one-hybrid assays indicated that OsWRKY67 could bind to its own promoter as well as the promoters of PR1a and PR10 through the interaction with the W-boxes (Fig. 7a, b). The direct interactions between OsWRKY67 and the promoters of ICS1 and NH1 were not identified.
OsWRKY67 binds specifically to the W-box elements. a Yeast one-hybrid assays indicated that OsWRKY67 can bind directly to the sequences of W-box (BS65) and its own promoter (67p). P53 is the positive control. NC is the negative control which did not contain the W-box and mBS65 is the mutated probe of BS65 with TGAC sequence changed to CCTA. The detailed sequences of BS65 and mBS65 are shown in section c. b Yeast one-hybrid assays indicated that OsWRKY67 can bind directly to the promoters of PR1a and PR10 through the W-box motif. NC is the negative control from the PR10 promoter which did not contain the W-box motif. c The EMSA experiment showed the binding of OsWRKY67 protein to the sequences of the W-box. An excess of unlabeled-BS65 probe was added as competitors. The negative control was conducted by inoculating the Biotin-labeled BS65 probe without the OsWRKY67-His protein. d The EMSA experiment showed the binding of OsWRKY67 protein to its own promoter through the W-box element. The oligonucleotides 67p and m67p were used as the probes. An excess of unlabeled-67p probe was added as competitors. e The EMSA experiment showed the binding of OsWRKY67 protein to the promoter of PR10. The oligonucleotides P10 and mP10 were used as the probes. An excess of unlabeled-P10 probe was added as competitors. f The EMSA experiment showed the binding of OsWRKY67 protein to the promoter of PR1a. The oligonucleotides P1 and mP1 were used as the probes. An excess of unlabeled-P1 probe was added as competitors
To further validate the binding specificity of OsWRKY67 to these W-box elements, we performed electrophoretic mobility-shift assays (EMSA) with an OsWRKY67-histone (His) fusion protein. As shown in Fig. 7c, d-f, no binding signal was detected for the control (non-fusion protein, lane 1). The fusion protein could bind to the BS65, 67p, P1 and P10 probes, but could not bind to the mBS65, m67p, mP1 and mP10 probes, in which the TGAC motifs were mutated. Moreover, the specific binding of the fusion protein to BS65, 67p, P1 and P10 was eliminated by addition of unlabeled competitors (Fig. 7c, d-f). Taken together, these results indicated that OsWRKY67 directly binds to the W-box elements in the promoters of PR1a and PR10 to activate its expression. Moreover, our results also demonstrated that OsWRKY67 could bind to its own promoter, implying that the expression of OsWRKY67 could be self-regulated.
Discussion
As one of the largest families of transcriptional regulators specifically expressed in plants, the pivotal roles of WRKY proteins in regulating disease resistance and abiotic stress response have been well-documented in many plant species [7,8,9,10,11,12,13]. In rice, more than 100 WRKY proteins have been identified and 11 of them have been demonstrated to be associated with defense responses [15,16,17,18,19,20,21,22]. However, only OsWRKY45 has been reported on regarding its regulatory role in panicle blast [28]. Because panicle blast is more destructive to grain yield than leaf blast, the gaps in knowledge of the roles of other members of this family in panicle blast resistance need to be filled. Here, the functions of OsWRKY67 in leaf blast, panicle blast and bacterial blight have been demonstrated. OsWRKY67 was significantly induced after infection with leaf blast and panicle blast. The plants constitutively expressing OsWRKY67 were more resistant to leaf blast and panicle blast as manifested by smaller lesion size and lower fungal growth when compared with wild-type Nipponbare plants. OsWRKY67-silenced plants, on the other hand, exhibited increased susceptibility to blast disease as manifested by larger lesion size and greater fungal growth. Furthermore, OsWRKY67-overexpressing plants also exhibited enhanced resistance to Xoo as indicated by their smaller lesions and lower bacterial growth rates. Altogether, these results together suggest a positive role of OsWRKY67 in rice-M. oryzae and rice-Xoo interactions.
Furthermore, we have demonstrated that OsWRKY67-overexpressing plants exhibited a dwarfed phenotype and produced lower numbers of tillers relative to the wild-type Nipponbare plants, indicating that OsWRKY67 also functions in regulating plant growth and development. However, there were no significant phenotypic differences between WRKY67-RNAi and Nipponbare plants. Interestingly, we also identified that the transcription of OsWRKY67 was modulated by cold, high salinity, dehydration, oxidative and abscisic acid stresses, and OsWRKY67 overexpressing plants showed increased tolerance to dehydration stress whereas overexpression of OsWRKY67 led to increased sensitivity to dehydration stress (unpublished data). Therefore, OsWRKY67 may have multiple roles in regulating rice development and various stress responses.
WRKY proteins regulate plant defense response by distinct signaling pathways where many WRKY proteins positively regulate plant defense responses while others negatively regulate disease resistance [15,16,17,18,19,20,21,22]. Most of the defense pathways mediated by WRKY proteins were either SA-dependent or jasmonic acid (JA)-dependent. For example, rice WRKY45 functions in benzothiadiazole-inducible blast resistance by modulating the SA signaling pathway [18], while WRKY11 and WRKY17 negatively regulate the basal resistance of Arabidopsis in a JA-dependent pathway [8]. In the present study, our data have demonstrated the positive role of OsWRKY67 in plant disease resistance and the resistance is dependent on the SA signaling pathway. First, many genes that are involved in SA synthesis or SA-conferred defense response were significantly up-regulated in OsWRKY67 overexpressing plants relative to wild-type Nipponbare plants. Second, the transcription of OsWRKY67 was markedly induced by exogenous SA. Third, the endogenous level of SA was remarkably higher in OsWRKY67-overexpressing plants whereas the SA content was lower in OsWRKY67-silenced plants compared with Nipponbare plants. Fourth, OsWRKY67 directly induced the expression of PR1a and PR10 by binding to their promoters via W-box elements. These data strongly suggest that OsWRKY67 positively regulates rice disease resistance in a SA-dependent pathway. In Arabidopsis, NPR1 has been validated to play a major role in transducing the SA signal by activation of downstream defense-related genes [30]. In this study, we identified that NH1, a rice homolog of NPR1, was significantly induced in OsWRKY67 overexpressing plants relative to Nipponbare plants both before and after pathogen infection. This suggests that OsWRKY67-mediated disease resistance may be associated with a NH1-dependent SA signaling pathway. Further research will be needed to verify this inference.
In addition to its function as a positive regulator, OsWRKY67 is also a negative regulator of gene expression. Among the genes down-regulated in OsWRKY67-overexpressing plants, three WRKY (WRKY60, WRKY7 and WRKY74) genes were identified, suggesting that OsWRKY67 may act upstream of these genes. In Arabidopsis, WRKY7 has been demonstrated to negatively regulate plant disease resistance [31]. In this study, the down-regulated expression of WRKY7 in OsWRKY67-overexpressing plants further indicated the positive role of OsWRKY67 in plant defense response.
Many studies have shown that WRKY proteins can directly regulate the transcription of their target genes by binding to the W-boxes in the promoter of these genes. Since the promoters of WRKY60, WRKY7 and WRKY74 contains many W-boxes (data not shown), their expression may be regulated directly by OsWRKY67. Alternatively, these WRKY genes can also be regulated by other WRKY genes that function downstream of OsWRKY67. Notably, in this study, we also observed several W-boxes in the promoter of OsWRKY67, and our results indicated that OsWRKY67 could bind to its own promoter and therefore, could auto-regulate itself. Moreover, except for the possibility of self-regulation, the expression of OsWRKY67 is probably modulated directly by other upstream WRKY proteins. These results exhibit an intricate network of autoregulation and cross-regulation in the OsWRKY67-mediated disease resistance pathways in rice. However, the cross-regulation between distinct WRKY proteins during plant defense response seems to be a general characteristic of the WRKY family. For instance, AtWRKY18 contains many W-box elements in its promoter and these elements function as negative regulators [32], whereas AtWRKY6 negatively regulates not only its own expression but also the transcription of WRKY42 [33]. Together, these observations indicate that WRKY proteins form a highly interconnected network in plants, and this network modulates distinct WRKY gene expression during the defense response and thereby, fine-tune the plant defense response.
Based on the findings of the potential cross-regulation network of WRKY proteins in plant defense responses as well as the important role of OsWRKY45 in regulating rice panicle blast resistance [28, 34], we deduced that if the OsWRKY67-mediated disease resistance is involved in modulating the expression of OsWRKY45 in rice. To verify this inference, the transcription levels of OsWRKY45 were analyzed in the wild-type Nipponbare and OsWRKY67-overexpressing and silenced plants at normal conditions. Simultaneously, the expression levels of OsWRKY13, OsWRKY30 and OsWRKY31, which are involved in leaf blast resistance [17, 20, 35], were also analyzed in these plants. Unfortunately, no significant differences were observed in the expression of these WRKY genes between wild-type Nipponbare and the transgenic plants (Additional file 14: Figure S10), indicating that OsWRKY67-mediated disease resistance is not involved in regulating the expressions of OsWRKY45, OsWRKY13, OsWRKY30 and OsWRKY31 in rice.
Conclusion
In the present study, we have demonstrated that OsWRKY67 positively regulates rice resistance to leaf blast, panicle blast and bacterial blight diseases. OsWRKY67 confers disease resistance by the induction of a series of defense–related genes and the activation of the SA signaling pathway with PR1a and PR10 as the direct targets of OsWRKY67. Moreover, OsWRKY67 can also bind to its own promoter via the W-box element, suggesting self-regulation in OsWRKY67-mediated biological processes. However, we still do not know whether there are other TFs, including WRKYs, acting upstream of OsWRKY67 and how OsWRKY67 may coordinate with the potential WRKY genes to contribute to disease resistance in rice. Further study will be required to address these issues. Resistance to the two major diseases attributable to OsWRKY67 makes it a promising target of molecular breeding for crop improvement in rice.
Methods
Plant growth and hormone treatments
The japonica rice (Oryza sativa L.) Nipponbare and a blast-resistant line BC10 were used in this study. Collection of the two rice varieties was complied with the institutional and national guidelines in China. Surface-sterilized rice seeds were planted onto 1/2 MS medium for germination and growth. Two-weeks later, the seedlings were transplanted into bucket and kept in the net-house of Rice Research Institute, Guangdong Academy of Agricultural Sciences, in Guangzhou (23°06′N, 113°15′E). Hormone treatments were performed using the same method according to our previous report [27]. The leaves of seedlings at the three- to four-leaf stage were gently watered with a SA solution at a concentration of 100 μM. Control treatment seedlings were sprayed with water.
Plasmid construction and transformation
To build the overexpressing plasmid, the coding sequence of OsWRKY67 amplified from BC10 using OsWRKY67-OE-F/R (Additional file 15) was sub-cloned into the pOx overexpressing vector [36]. A 210-bp partial sequence of OsWRKY67 was amplified by RT-PCR using primers OsWRKY67-RNAi-F/R (Additional file 15), and was then inserted into the pRNAi-Ubi vector [27] to generate the OsWRKY67 RNAi plasmid. For promoter analysis, ~ 2.4-kb fragments upstream of OsWRKY67 amplified using primers OsWRKY67-GUS-F/R (Additional file 15) was sub-cloned into the pCAMBIA1381Z vector. All the plasmid constructs were verified by sequencing and then were transferred into Agrobacterium tumefaciens EHA105 for transformation of Nipponbare plants via an Agrobacterium-mediated genetic transformation approach.
Evaluation of disease resistance
Isolate GD08-T13 of Magnaporthe oryzae was used for rice blast inoculation and the Chinese Xanthomonas oryzae pv. Oryzae (Xoo) race 4 isolate was used for Xoo inoculation. Evaluation of blast and Xoo resistance was conducted using the same method described in our previous reports [27, 37]. For spray inoculation, seedlings at the three- to four- leaf stage were sprayed with a spore suspension (1 × 106 spores/mL) containing 0.05% Tween-20. The disease was assessed 7 days after inoculation by counting the number of lesions per leaf. Six-week-old plants were punch inoculated as described previously [38, 39]. Briefly, 5 μL of a spore suspension (5 × 105 spores/mL) containing 0.05% Tween-20 was added to the press-injured spots on fully expanded rice leaves. The inoculated spots were wrapped with transparent scotch tape, and leaves were photographed 12 days after inoculation to measure lesion size by the ImageJ program (http://imagej.nih.gov/ij/). For determination of in planta sporulation after punch inoculation, leaf strips containing a lesion spot were excised and submerged in 100 μL of distilled water in a 1.5 mL microcentrifuge tube. After the suspension was vigorously mixed, spores were counted with a microscope [39].
For panicle blast inoculation, the cotton-wrapping inoculation method was used as described previously [27]. The upper-middle part of a panicle was wrapped with sterile cotton within 2 to 3 days after heading. Two milliliters of a suspension of 1 × 106 spores/ml of M. oryzae GD08-T13 was injected into the cotton after which the cotton was wrapped with foil. The inoculated plants were sprayed with water for 3 min every 3 h to maintain the humidity. Evaluation of panicle blast resistance was conducted at 3 weeks after inoculation by measuring the percent of infected main axis length.
To better distinguish the resistance phenotypes between silenced plants and control plants (Nipponbare), lower concentrations of spore suspension [about 0.5 × 106 spores / ml for panicle blast and 2.5 × 105 spores / ml for leaf blast (punch method)] were used.
GUS staining analysis
Analysis of GUS activity in different rice tissues was conducted as described previously [27].
Subcellular localization of OsWRKY67 protein
The CDS sequence of OsWRKY67 amplified using primers OsWRKY67-GFP-F/R (Additional file 15) was inserted into the 35S-GFP vector to produce the OsWRKY67-GFP fusion protein. Next, the fusion plasmid and the empty 35S-GFP plasmid were transferred into onion epidermal cells or rice stem protoplasts as described previously [37, 40]. Laser confocal microscopy (Zeiss LSM710, Germany) was used to detect the GFP fluorescence after 24 h of incubation at 25 °C.
Yeast one-hybrid assay and EMSA
The yeast one-hybrid assays and EMSA experiments were performed using the same method according to our previous report [27]. For yeast one-hybrid assays, about 350 bp fragments containing at least three W-boxes were amplified from the promoters of OsWRKY67, PR1a and PR10. Then, the cloned sequences were inserted into the pAbAi vector to generate the bait protein. A negative control (NC) was included by using a 100 bp sequence without a W-box from the promoter of OsWRKY67. Primers used for the EMSA experiments were labeled by the EMSA Probe Biotin Labeling Kit (Beyotime, China). The un-labeled primers were employed as the competitors.
Real-time PCR analysis
Total RNA was extracted from different rice tissues using Eastep® Super Total RNA Extraction Kit (Promega Biotech Co., Ltd., USA). Inverse transcription and real-time PCR were conducted using the same method as described previously [27]. Gene-specific primers used in this study are shown in Additional file 15.
Quantification of endogenous SA
The SA was extracted and measured using the same protocol in our previous report [37]. The optimized MS/MS conditions used for quantification of endogenous SA are listed in Additional file 16: Table S6.
RNA-seq analysis
Two-week old leaves of wild-type Nipponbare and OsWRKY67 overexpressing plants were collected for RNA-seq analysis. Solexa sequencing was conducted by the ANNOROAD Gene Technology Co. LTD (Beijing, China). The raw data was submitted to the Sequence Read Archive of the National Center of Biotechnology Information (NCBI) (accession number: SRP096216). For data analysis, clean reads were obtained by removing low quality tags, adaptor sequences and other contaminants. Next, the clean reads were all mapped to the Oryza sativa genome (MSU7) using the bowtie2 program(v2.2.3) [41] and tophat2 program (v2.0.12) [42] with default parameter values. Cufflinks program (v2.2.1) [43] was used for estimation of the FPKM (Fragments Per Kilobase per Million mapped reads) values to obtain the gene expression level. The program edgeR (v3.8.6) [44] was applied to detect differentially expressed genes with an FDR ≤ 0.05 and a relative change threshold of two-fold. An online server named PlantGSEA [45] was used for enrichment analysis of biological processes with the Plant Ontology, Gene Ontology and KEGG pathway databases.
Abbreviations
- DEGs:
-
Differentially expressed genes
- FDR:
-
False discovery rate
- GFP:
-
Green fluorescence protein
- GO:
-
Gene ontology
- GUS:
-
β-glucuronidase
- ICS1:
-
Isochorismate synthase 1
- NC:
-
Negative control
- NCBI:
-
National Center of Biotechnology Information
- NHI:
-
Arabidopsis NPR1 homolog 1
- OX-WRKY67:
-
OsWRKY67 overexpressing
- PAD4:
-
Phytoalexin-deficient 4
- PAL1:
-
Phenylalanine ammonia-lyase 1
- PR10:
-
Pathogenesis-related [PR] protein 10
- SA:
-
Salicylic acid
- TF:
-
Transcription factor
- WRKY67-RNAi:
-
OsWRKY67 silencing
- Xoo :
-
Xanthomonas oryzae pv. Oryzae
References
Liu W, Liu J, Triplett L, Leach J, Wang G. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol. 2014;52:213–41.
Hu K, Qiu D, Shen X, Li X, Wang S. Isolation and manipulation of quantitative trait loci for disease resistance in rice using a candidate gene approach. Mol Plant. 2008;1:786–93.
Ou SH, Nuque FL, Bandong JM. Relation between qualitative and quantitative resistance to rice blast. Phytopathology. 1975;65:1315–6.
Parlevliet JE. Components of resistance that reduce the rate of epidemic development. Annu Rev Phytopathol. 1979;1:203–22.
Kou Y, Wang S. Broad-spectrum and durability: understanding of quantitative disease resistance. Curr Opin Plant Biol. 2010;13:181–5.
Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends in Plant Sci. 2010;15:247–58.
Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–31.
Journot-Catalino N, Somssich IE, Roby D, Kroj T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell. 2006;18:3289–302.
Xu X, Chen C, Fan B, Chen Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 2006;18:1310–26.
Zheng Z, Qamar SA, Chen Z, Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006;48:592–605.
Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–103.
Lai Z, Vinod KM, Zheng Z, Fan B, Chen Z. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 2008;8:68.
Naoumkina M, He X, Dixon R. Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol. 2008;8:132.
Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol. 2007;49:827–42.
Chujo T, Takai R, Akimoto-Tomiyama C, Ando S, Minami E, Nagamura Y, Kaku H, Shibuya N, Yasuda M, Nakashita H, Umemura K, Okada A, Okada K, Nojiri H, Yamane H. Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim Biophys Acta. 2007;1769:497–505.
Liu X, Bai X, Wang X, Chu C. OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol. 2007;164:969–79.
Qiu DY, Xiao J, Ding XH, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant-Microbe Interact. 2007;20:492–9.
Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 2007;19:2064–76.
Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007;65:799–815.
Zhang J, Peng YL, Guo ZJ. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008;18:508–21.
Tao Z, Liu HB, Qiu DY, Zhou Y, Li X, Xu C, Wang S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009;151:936–48.
Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H, Kaku H, Minami E, Nishizawa Y. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot. 2013;64:5085–97.
Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant. 2008;1:446–58.
Chujo T, Miyamoto K, Shimogawa T, Shimizu T, Otake Y, Yokotani N, , Nishizawa Y, Shibuya N, Nojiri H, Yamane H, Minami E, Okada K. OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol Biol 2013;82:23–37.
Zhuang JY, Ma WB, Wu JL, Chai RY, Lu J, Fan YY, Jin MZ, Leung H, Zheng KL. Mapping of leaf and neck blast resistance genes with resistance gene analog, RAPD and RFLP in rice. Euphytica. 2002;128:363–70.
Puri KD, Shrestha SM, Chhetri GBK, Joshi KD. Leaf and neck blast resistance reaction in tropical rice lines under green house condition. Euphitica. 2009;165(3):523–32.
Liu Q, Yang J, Zhang S, Zhao J, Feng A, Yang T, Wang X, Mao X, Dong J, Zhu X, Leung H, Leach JE, Liu B. OsGF14b positively regulates panicle blast resistance, but negatively regulates leaf blast resistance in Rice. Mol Plant-Microbe Interact. 2016;29:46–56.
Shimono M, Koga H, Akagi A, Hayashi N, Goto S, Sawada M, Kurihara T, Matsushita A, Sugano S, Jiang CJ, Kaku H, Inoue H, Takatsuji H. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol Plant Pathol. 2012;13:83–94.
Mare’ C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L. Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol Biol. 2004;55:399–416.
Cao H, Glazebrook J, Clarke JD, Volko S, Dong X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63.
Kim KC, Fan B, Chen Z. Pathogen-induced Arabidopsis WRKY7 is a trans- criptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 2006;142:1180–92.
Chen C, Chen Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 2002;129:706–16.
Robatzek S, Somssich IE. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002;16:1139–49.
Inoue H, Hayashi N, Matsushita A, Xinqiong L, Nakayama A, Sugano S, Jiang CJ, Takatsuji H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-proteininteraction. Proc Natl Acad Sci U S A. 2013;110:9577–82.
Peng X, Hu Y, Tang X, Zhou P, Deng X, Wang H, Guo Z. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acidaccumulation, PR gene expression and resistance to fungal pathogens in rice. Planta. 2012;236:1485–98.
Liu Q, Yang T, Yu T, Zhang S, Mao X, Zhao J, Wang X, Dong J, Liu B. Integrating Small RNA Sequencing with QTL Mapping for Identification of miRNAs and Their Target Genes Associated with Heat Tolerance at the Flowering Stage in Rice. Front Plant Sci. 2017;8:43.
Liu Q, Yang J, Yan S, Zhang S, Zhao J, Wang W, Yang T, Wang X, Mao X., ., Dong, J, Zhu X, Liu B. The germin-like protein OsGLP2-1 enhances resistance to fungal blast and bacterial blight in rice. Plant Mol Biol 2016;92: 411–423.
Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci U S A. 2001;98:759–64.
Ding B, Bellizzi Mdel R, Ning Y, Meyers BC, Wang GL. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histoneH4acetylation of defense-related genes in rice. Plant Cell. 2012;24:3783–94.
Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7:30.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Yi X, Du Z, Su Z. PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 2013;41:W98–103.
Acknowledgements
We thank Jia Jian for technical support of sub-localization and Zhang Weina (Center for Agrobiological Gene Research, Guangdong Academy of Agricultural Sciences) for help with laser confocal microscopy.
Funding
This research was supported partially by the NSFC-IRRI project (31461143019), Science and Technology Projects of Guangdong Province (2015B020231002, 2016A050502030 and 2014B070706013), the Natural Science Foundation of Guangdong Province (2014A030310489), and the Common Technical Innovation Team of Guangdong Province on Agricultural Seed Industry (2017LM2148).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files. The raw data of RNA-seq was available in NCBI (accession number: SRP096216).
Author information
Authors and Affiliations
Contributions
XZ and BL conceived of the study, drafted proposal and corrected manuscript. QL and XL conducted the quantitative qRT-PCR assay, transgenic functional confirmation and chemical treatment experiments, drafting the manuscript and proposal writing. SY conducted the quantitative analysis of the endogenous SA. TY analyzed the data of RNA-seq. JY evaluated the blast and Xoo resistance of rice plants. JD, SZ, JZ, TY and XM participated in RNA extraction and quantitative qRT-PCR assays. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. The microarray data of OsWRKY67 after leaf blast and panicle blast infection. (XLSX 9 kb)
Additional file 2:
Figure S1. Subcellular localization of OsWRKY67. Bar = 2 μm. a Laser confocal microscopy images deriving from GFP of onion epidermal cells transiently expressing GFP or GFP-OsWRKY67 fusion protein. Arrows indicate the nucleus. b Laser confocal microscopy images deriving from GFP of rice protoplasts transiently expressing GFP or GFP-OsWRKY67 fusion protein. (JPG 1354 kb)
Additional file 3:
Figure S2. Phenotypes of wild-type Nipponbare (NPB) and OsWRKY67-overexpressing (OX-WRKY67) plants at normal conditions. The values are means ± SDs of twenty biological replicates and the asterisks represent significant differences relative to Nipponbare plants at **P < 0.01 by t-test. a Phenotypes of Nipponbare and OX-WRKY67 transgenic lines OX 5–2 and OX 6–5 at the heading stage. b Plant heights of Nipponbare and OX-WRKY67 transgenic lines OX 5–2 and OX 6–5. c Tiller numbers of Nipponbare and OX-WRKY67 transgenic lines OX 5–2 and OX 6–5. (JPG 1090 kb)
Additional file 4:
Figure S3. Southern blot analysis of the copy numbers of transgene. (JPG 439 kb)
Additional file 5:
Figure S4. Phenotypes of OX-WRKY67 and Nipponbare plants infected with leaf blast using spraying method. a The OX-WRKY67 plants exhibited enhanced leaf blast resistance after inoculation with GD08-T13 using spraying method. b The lesion numbers per leaf of Nipponbare and OX-WRKY67 plants after inoculation with GD08-T13. The values are means ± SDs of twelve biological replicates. The asterisks represent significant differences relative to Nipponbare plants (t-test, **P < 0.01). (JPG 348 kb)
Additional file 6:
Figure S5. Phenotypes of the OX-WRKY67 and Nipponbare plants infected with bacterial blight. The values are means ± SDs of twenty biological replicates and the asterisks represent significant differences relative to Nipponbare plants at **P < 0.01 by t-test. a Disease phenotypes of OX-WRKY67 and Nipponbare plants after Xoo inoculation. b Lesion lengths in OX-WRKY67 and Nipponbare plants after Xoo inoculation. c Growth rates of Xoo race 4 in the leaves of OX-WRKY67 and Nipponbare plants. Bacterial populations were determined from three leaves 12 or 16 days after inoculation by counting colony-forming units (cfu). Similar results were obtained in two independent experiments. (JPG 665 kb)
Additional file 7:
Figure S6. Phenotypes of Nipponbare and OsWRKY67-silenced (WRKY67-RNAi) plants infected with bacterial blight. The values are means ± SDs of twenty biological replicates and the asterisks represent significant differences relative to Nipponbare plants at **P < 0.01 and *P < 0.05 by t-tests. a Disease phenotypes of WRKY67-RNAi and Nipponbare plants after Xoo inoculation. b Lesion lengths in WRKY67-RNAi and Nipponbare plants after Xoo inoculation. c Growth rates of Xoo race 4 in the leaves of WRKY67-RNAi and Nipponbare plants. Bacterial populations were determined from three leaves 12 or 16 days after inoculation by counting colony-forming units (cfu). Similar results were obtained in two independent experiments. (JPG 711 kb)
Additional file 8:
Table S2. Differentially expressed genes in OsWRKY67 overexpressing plants by RNA-seq analysis. (XLSX 26 kb)
Additional file 9:
Figure S7. Overexpression of OsWRKY67 influences the expression of defense-related genes. The values are means ± SDs of three biological replicates and asterisks represent significant differences relative to Nipponbare plants before blast inoculation (0 h) or at 12, 24 and 48 h after inoculation with M. oryzae (t-test, **P < 0.01 and *P < 0.05). a Nine defense-related genes were over-presented in OX-WRKY67 plants. b Three WRKY genes were suppressed in OX-WRKY67 plants. (JPG 918 kb)
Additional file 10:
Figure S8. The results of qRT-PCR showed an excellent concordance with the sequencing data. (JPG 214 kb)
Additional file 11:
Table S3. Gene ontology analysis of up- and down-regulated genes in OsWRKY67-overexpressing plants detected by RNA-seq analysis. (XLSX 12 kb)
Additional file 12:
Table S4. A statistic pathway enrichment analysis of differentially expressed genes between OsWRKY67-overexpressing plants and Nipponbare plants. (XLSX 11 kb)
Additional file 13:
Figure S9. OsWRKY67 regulates the expression of a set of defense-related genes at 12 h and 36 h after bacterial blight infection. The wild-type Nipponbare and transgenic plants were inoculated with Xoo race 4 at the booting stage. The values are means ± SDs of three biological replicates and the asterisks represent significant differences relative to Nipponbare plants at **P < 0.01 or *P < 0.05 by t-tests. The expression of Nipponbare plants was set to “1” at each time point. a Relative expression levels of defense-related genes in Nipponbare and OX-WRKY67 plants. b Relative expression levels of defense-related genes in Nipponbare and WRKY67-RNAi plants. (JPG 1135 kb)
Additional file 14:
Figure S10. Relative expression levels of four WRKY genes in wild-type Nipponbare and OsWRKY67 transgenic plants. The values are means ± SDs of three biological replicates. (JPG 381 kb)
Additional file 15:
Table S5. Primers used for quantitative RT-PCR and vector construction. (XLSX 10 kb)
Additional file 16:
Table S6. Characteristic fragment ions of the SA standard and its optimized MS/MS condition. (XLSX 10 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, Q., Li, X., Yan, S. et al. OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice. BMC Plant Biol 18, 257 (2018). https://doi.org/10.1186/s12870-018-1479-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-018-1479-y