Abstract
Background
The emergence of Streptococcus agalactiae infections in patients with bacteremia is increasing. It is crucial to investigate the epidemiology, molecular characteristics, biofilm status, and virulence analysis of Streptococcus agalactiae in these patients.
Methods
In this cross-sectional study, 61 S. agalactiae isolated from blood infection were subjected to characterization through antimicrobial susceptibility tests, biofilm formation, multilocus sequence typing (MLST), and PCR analysis for detecting resistance (tet and erm family) and virulence genes (alp2/3, alp4, bca, bac, eps, rib, lmb, cylE, and pilus island genes).
Results
Overall, 32.7% of the isolates demonstrated an inducible clindamycin resistance phenotype. The results showed that 49.2, 24.6, and 8.2% of confirmed Streptococcus agalactiae strains were classified as strong, intermediate, and weak biofilm-forming strains, respectively. tet(M) (57.1%) was recovered most, followed by tet(M) + tet(L) (14.3%), tet(S) + tet(K) (10.7%), tet(M) + tet(K) (8.9%), tet(M) + tet(K) + tet(O) (5.4%), and tet(S) + tet(L) + tet(O) (3.6%). Three virulence gene profiles of cylE, lmb, bca, rib (24.6%; 15/61), cylE, lmb, rib, alp3 (19.7%; 12/61), and cylE, lmb, bac, rib (16.4%; 10/61) were detected in approximately two-thirds of the isolates. MLST revealed that the 61 isolates belonged to six clonal complexes, including CC1 (49.2%), followed by CC12 (18%), CC19 (13.1%), CC22 (9.8%), CC17 (6.6%), and CC283 (3.3%), and 11 different sequence types (STs), including ST1 (24.6%), ST7 (14.8%), ST918 (13.1%), ST2118 (9.8%), ST19 (9.8%), ST48 (6.6%), ST1372 (4.9%), ST22 (4.9%), ST40 (4.9%), ST734 (3.3%), and ST283 (3.3%). Remarkably, all CC1 and CC12 isolates, three-fourths of CC19, and half of CC22 were confirmed as biofilm producers. Conversely, CC17 and CC28 isolates were found to be nonproducers. The occurrence of strong biofilm formation was limited to specific CCs, namely CC1 (34.4%), CC12 (8.2%), CC19 (3.3%), and CC22 (3.3%).
Conclusion
The high prevalence of CC1 and CC12 clones among S. agalactiae strains reflects the emergence of these lineages as successful clones in Iran, which is a serious concern and poses a potential threat to patients.
Similar content being viewed by others
Background
Streptococcus agalactiae strains are common residents of the gastrointestinal and genitourinary tracts of humans and cause various infections, such as skin and soft tissue infections, urinary tract infections, endocarditis, pneumonia, meningitis, and bacteremia [1]. Evidence has recently designated S. agalactiae isolated from blood infections as a high-priority pathogen with the potential to increase fatality rates globally [2]. Bloodstream infections (BSIs) caused by S. agalactiae strains are relatively rare, accounting for only 1–2% of all such infections. Previous studies in European countries have reported an incidence rate of 1.2–4.1 per 100,000 for S. agalactiae BSIs, depending on the year and population studied. However, according to a 2016 analysis, the United States seems to have a higher incidence rate, with an estimated 11 cases per 100,000 [3]. In the United States, nonpregnant adults now account for over 75% of invasive S. agalactiae cases and approximately 90% of associated deaths. A population-based surveillance conducted in Canada, from 2011 to 2018 revealed an incidence rate of 4.7 per 100,000 for S. agalactiae BSIs with an increasing annual incidence of β-hemolytic streptococcal bloodstream infections from 2011 to a peak in 2016, followed by a decrease [4,5,6,7].
Reports have also indicated that numerous S. agalactiae virulence factors, including surface proteins, adhesion factors, capsules, immune evasion factors, and toxins, are related to the pathogenesis of this bacterium [2, 8,9,10,11]. Among these virulence factors, it seems that surface antigens belonging to the alpha-like protein (Alp) family and the pilus islands (PIs) significantly contribute to the pathogenesis of this bacterium [12,13,14,15,16,17]. Alp proteins play a crucial role in the invasion of human epithelial cells by S. agalactiae through their interaction with glycosaminoglycans present on the surface of host cells, allowing them to traverse through the cell layer. Pili in S. agalactiae have been extensively studied, indicating their significant involvement in adhesion and invasion mechanisms. They have been suggested as promising candidates for vaccines due to their crucial roles in these processes. Evidence commonly regards pili as significant factors in colonization. The pili of S. agalactiae are appendages anchored to the bacterial cell wall, extending outward from the surface. Within each gene, there are three structural subunit proteins encoded, which could be considered promising candidates for vaccines [2, 13, 16].
Information on molecular characteristics of S. agalactiae related to bacteremia is very important. The epidemiological studies are imperative to monitor the possible dissemination of these strains and to adopt rational infection control strategies for the prevention of sequential outbreaks [10, 12, 14,15,16]. Various S. agalactiae population epidemiology-based studies have identified five main genetically unique lineages, namely CC1, CC10, CC17, CC19, and CC23. These lineages are commonly associated with both invasive disease and asymptomatic colonization in individuals of different age groups. Earlier evidence indicated that ST17 and ST19 strains were more frequently associated with invasive disease. However, in adult individuals with invasive infections caused by S. agalactiae, CC17 strains are less commonly identified compared to CC19 [7, 18,19,20]. Molecular epidemiology studies have indicated various clonal distributions of invasive S. agalactiae strains. For example, CC1 was the most frequently identified invasive type in Taiwan [18], CC23 in East African [19], CC19 in China [8], CC17 in Japan [7], CC10 in Ethiopia [20], and CC17 in Poland [21]. According to the earlier data, the major invasive S. agalactiae clone found in Iranian hospitals was the CC/ST19 clone [22]. Although several studies have recently focused on understanding the serotypes and antibiotic resistance pattern of S. agalactiae isolated from clinical samples in Iran, there is very little information about genetic diversity and biofilm formability of S. agalactiae isolated from BSIs. Therefore, it is crucial to gain insight into the molecular characteristics of this bacterium. This study attempted to identify the biofilm production, virulence determinants, antibiotic resistance genes, and genetic variability of S. agalactiae strains isolated from blood infections.
Results
Identification and antibacterial susceptibility pattern
In the current study, 61 clinical isolates of S. agalactiae obtained from BSIs were used. The samples were collected from patients with an age range of 18–65 years. The mean age of the patients was 39.2 years. The results revealed that all isolates displayed sensitivity toward vancomycin and penicillin. Overall, resistance to all of the antibacterial agents was not detected among the tested isolates. The majority of isolates (91.8%) were resistant to tetracycline. This was followed by erythromycin (86.9%), chloramphenicol (67.2%), clindamycin (42.6%), levofloxacin (19.7%), and linezolid (13.1%). In total, 77% (47/61) of isolates were found to be multidrug resistance (MDR). We found eleven resistance patterns in the examined isolates, wherein erythromycin, tetracycline, clindamycin, chloramphenicol (19.7%; 12/61), erythromycin, tetracycline, chloramphenicol (19.7%; 12/61), erythromycin, tetracycline, clindamycin (16.4%; 10/61), tetracycline, chloramphenicol (9.9%; 6/61), and erythromycin, tetracycline, levofloxacin (8.2%; 5/61) were the top five frequently detected profiles (Table 1). In the current research, 20 (32.7%) of the isolates were confirmed as clindamycin resistance (ICR). Constitutive clindamycin-resistant (CCR) isolates were detected at a prevalence rate of 42.6%.
Biofilm formation
By the microtiter plate (MtP) assay, 50 isolates (82%) were found to be able to produce biofilms to different degrees. Eleven isolates (18%) did not have the ability to form biofilms and were classified as non-biofilm producers. Table 2 gives information about different degrees of biofilm production in the S. agalactiae isolates tested.
In Table 3, the highest resistance rate among strong biofilm producers belonged to erythromycin (96.7%; 29/30) and tetracycline (93.3%; 28/30), while the lowest resistance rate was recorded for levofloxacin (30%; 9/30) and linezolid (23.3%; 7/30). One isolate, which showed susceptibility to all tested antibiotics, did not produce biofilm. However, all of the weak producer isolates were resistant to tetracycline and chloramphenicol.
Resistance gene detection
Table 4 depicts the prevalence of resistance genes in clinical isolates of S. agalactiae. In this investigation, the findings indicated that all erythromycin-resistant S. agalactiae carried at least one macrolide resistance gene. Resistance to macrolides was mainly due to erm(B), which was contained in 39 isolates (63.9%), followed by erm(A) in 20 isolates (32.8%) and mef(A/E) in 14 isolates (22.9%). Among the 20 ICR S. agalactiae strains, 11 isolates (55%) carried both erm(B) + erm(A), 5 isolates (25%) carried erm(A) alone, and 4 isolates (20%) carried erm(B) alone, while in 26 S. agalactiae isolates with simultaneous resistance to clindamycin and erythromycin, 15 isolates (57.7%) carried erm(B) alone, one isolate (3.8%) carried mef(A/E) alone, one isolate (3.8%) carried erm(A) alone, 7 isolates (27%) carried erm(B) + mef(A/E), and two isolates (7.7%) carried erm(A) + erm(B). Regarding 56 tetracycline-resistant isolates, the results indicated that tet(M) (57.1%) was recovered the most, followed by tet(M) + tet(L) (14.3%), tet(S) + tet(K) (10.7%), tet(M) + tet(K) (8.9%), tet(M) + tet(K) + tet(O) (5.4%), and tet(S) + tet(L) + tet(O) (3.6%). At least one tet gene was identified in each tetracycline-resistant isolate.
Pilus island and virulence gene detection
Our analysis indicated that among the 61 S. agalactiae strains, 40 isolates (65.6%) carried the rib gene, 29 isolates (47.5%) carried bca, 24 isolates (39.3%) carried apl2, 22 isolates (36.1%) carried alp3, 14 isolates (23%) carried bac, and 3 isolates (4.9%) carried alp4. Simultaneous carriage of the cylE and lmb genes was confirmed in all tested isolates. We identified seven different patterns of virulence gene combinations, wherein cylE, lmb, bca, rib (24.6%; 15/61), cylE, lmb, rib, alp3 (19.7%; 12/61), cylE, lmb, bac, rib (16.4%; 10/61), cylE, lmb, alp2/3 (16.4%; 10/61), and cylE, lmb, bca, alp2 (16.4%; 10/61) were the top five identified patterns. Screening of PI-encoding genes showed that the combination pattern PI-1 + PI-2a (59%; 36/61) was detected most frequently, followed by PI-1 + PI-2a (24.6%; 15/61), whereas the carriage rate of PI-2b or PI-2a alone was found in 6 (9.8%) and 4 (6.6%) isolates, respectively.
MLST
In the current research, all S. agalactiae strains were successfully typed using the MLST technique. A total of eleven particular sequence types (STs), including ST1 (24.6%; 15/61), ST7 (14.8%; 9/61), ST918 (13.1%; 8/61), ST2118 (9.8%; 6/61), ST19 (9.8%; 6/61), ST48 (6.6%; 4/61), ST1372 (4.9%; 3/61), ST22 (4.9%; 3/61), ST40 (4.9%; 3/61), ST734 (3.3%; 2/61), and ST283 (3.3%; 2/61), were identified and categorized into six clonal complexes (CCs). As presented in Table 4, the most prevalent CC was CC1 (49.2%; 30/61), followed by CC12 (18%; 11/61), CC19 (13.1%; 8/61), CC22 (9.8%; 6/61), CC17 (6.6%; 4/61), and CC283 (3.3%; 2/61). CC1, CC12, CC19, and CC22 represented different STs. Table 4 shows that ICR S. agalactiae strains were distributed to three particular STs, including ST1 (55; 11/20), ST7 (30; 6/20), and ST2118 (15; 3/20), all of which belonged to CC1. Overall, the most common STs in CCR S. agalactiae isolates included ST2118 (11.5%; 3/26), ST918 (30.8%; 8/26), ST1372 (11.5%; 3/26), ST19 (23.1%; 6/26), ST734 (7.7%; 2/26), ST22 (11.5%; 3/26), and ST40 (3.9%; 1/26).
Isolates simultaneously harboring tet(M), tet(K), and tet(O) belonged to ST1, ST7, and ST48 (one isolate each). The findings also showed that the isolates carrying tet(S) + tet(L) + tet(O) belonged to ST2118 and ST40 (one isolate each). Isolates harboring tet(M) + tet(L) genes were mainly found in ST1 (5 isolates), ST7 (2 isolates), and ST2118 (one isolate). Isolates harboring tet(M) + tet(K) simultaneously belonged to ST734 (n = 2), ST918 (n = 1), ST1372 (n = 1), and ST22 (n = 1). Overall, macrolide resistance genes were strongly linked with specific STs. erm(B) + erm(A) were present in ST1 (n = 5), ST2118 (n = 4), ST7 (n = 3), and ST22 (n = 1). All 7 isolates carrying erm(B) + mef(A/E) belonged to ST918. The results are presented in Table 5.
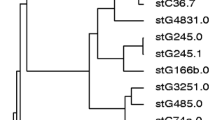
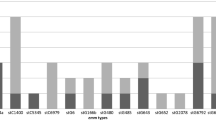
Variability in PI types was observed within the majority of the STs, but some STs were also strongly linked with specific STs, such as ST2118, ST19, and ST734 with PI-1 + PI-2. The distribution of biofilm formation in different CCs of S. agalactiae strains is presented in Fig. 1. Notably, all CC1 and CC12, three-fourths of CC19, and half of CC22 isolates were confirmed as biofilm producers, while all CC17 and CC28 isolates were found to be nonproducers. Strong biofilm formation was exclusive to ST1 (30%; 9/30), ST7 (23.3%; 7/30), ST2118 (16.6%; 5/30), ST918 (10%; 3/30), ST19 (6.7%; 2/30), ST40 (6.7%; 2/30), and ST1372 (6.7%; 2/30). As stated earlier, among 61 S. agalactiae strains isolated from blood infection, seven virulence patterns were identified.
The phylogenetic analysis of 11 STs of S. agalactiae strains. The consensus trees were constructed by the RAxML analysis of the combined seven loci adhP, pheS, atr, glnA, sdhA, glcK, and tkt. The phylogeny was constructed with 1,000 bootstrap replications. Bootstrap branch supports above 75% are shown. Also, the distribution of biofilm formation and the distribution of different virulence genes have been shown in STs of S. agalactiae isolated from blood infection
Phylogenetic analysis
The consensus tree was constructed by the Maximum Likelihood Analysis from the combined seven loci adhP, pheS, atr, glnA, sdhA, glcK, and tkt of 11 STs of S. agalactiae isolated from blood infection. We chose one representative strain from each ST for phylogenetic analysis. All STs in this study were placed in separate clades. The ST2118 clade takes a basal position relative to the other clades. Bootstrap branch supports above 75% are shown (Fig. 1). Also, Fig. 1 indicates that cylE, lmb, rib, alp3 was the most prevalent profile among ST1 and ST7 isolates; cylE, lmb, bac, rib in ST918 isolates; cylE, lmb, alp2/3 in ST1372, ST40, and ST22 isolates; cylE, lmb, bca, rib in ST19 isolates; and cylE, lmb, bca, alp2 in ST48 isolates.
Discussion
To our knowledge, the molecular characteristics of S. agalactiae associated with blood infection, as well as its biofilm production, virulence, and adhesion factors, remain unexplored in Iran. Based on our current knowledge, this is the first document to reveal the genetic variability, biofilm production, and virulence detection of S. agalactiae strain isolates from blood infections in Iran.
The S. agalactiae isolates with the CCR phenotype (42.6%) were higher than isolates with the ICR phenotype (32.7%). Our findings, supported by evidence from other studies [13], indicate an increase in resistance to clindamycin in S. agalactiae worldwide. Possible explanations for this increased prevalence are the genetic diversity of bacteria, excessive consumption of this antibiotic, abuse of antibiotics, self-treatment, and the transmission of resistant strains from hospitals and communities, and vice versa.
In line with some previous studies that indicated PI-1 and PI-2a as the most prevalent PI family genes in S. agalactiae strains, we noticed a high percentage of PI-1 (83.6%), followed by PI-2a (65.6%), and PI-2b (36.1%) among our strains [9, 16]. We showed that the combination pattern of PI genes was greater than that of PI genes alone. In similar surveys conducted in China [9] and Ireland [23], a high prevalence of PI genes in combination compared to PI genes alone was also reported. In a study conducted in Africa, Madzivhandila et al. found similar results, reported that PI1 + PI2b, PI2a, PI1 + PI2a, and PI2b were observed in 45.1%, 29.8%, 24.8%, and 0.2% of isolates, respectively. Our research indicated that all isolates harbored at least one of the PI genes. Based on our findings, all the CC19 isolates were found to carry PI-1 + PI-2a. Additionally, the majority of CC22 (66.7%), CC17 (75%), and CC283 (50%) isolates harbored PI-1 + PI-2b. This result is in agreement with Khan et al.’s study, which reported the presence of at least one of the PI genes in S. agalactiae, with the majority of them being PI-1 + PI-2a. This study also reported that the PI-1 + PI-2b combination was exclusively found in CC17 isolates [3].
The main finding of the present research, based on the phylogenetic analysis, was the genetic variability of the tested strains with a high prevalence of CC1 (49.2%), which corresponded to ST1, ST7, and ST2118. All the strains of this clone were biofilm producers, had ICR phenotypes, and were positive for PI-1 + PI-2a (70%), PI-1 + PI-2b (16.6%), PI-2a (6.7%), and PI-2b (6.7%). According to our data, the tet(M) gene, as the most widespread tet family gene among CC1 isolates, was detected in 86.7% of isolates, followed by tet(L) (30%), tet(O) (10%), and tet(K) (6.79%). This clone was also reported in S. agalactiae strains obtained from Iran (5.3%) [24], China (13%) [17], and Sweden (10.2%) [11]. According to the evidence, there is an expected correlation between the virulence gene and specific STs. Our observations indicated that alp3 was only detected in CC1 strains. All of the CC1 isolates were found to carry cylE and lmb genes. This finding is in line with research conducted in the UK by Khan et al. that reported the predominance of alp3 in CC1 isolates [3]. In the research conducted by Rojo-Bezares et al. on 375 S. agalactiae strains isolated from pregnant women, alp2/3 and lmb genes were detected in all of the CC1 isolates [25]. In a study conducted in Spain, 16.8% of S. agalactiae from postmenopausal women exhibited CC1/ST1 and were all resistant to erythromycin and clindamycin [12]. Recently, a study conducted in the USA reported the presence of CC1 in 12% of the examined isolates, which was identified to have PI-1 + PI-2a as the most frequent PI type, followed by PI-1 + PI-2b, PI-2a, and PI-2b [16]. A similar survey conducted by Khan et al. in the UK on 192 S. agalactiae isolates (179 invasive and 13 non-invasive) showed that 91.4% of CC1 isolates were tetracycline resistant, and 82.9% carried tet(M), and 3 isolates carried tet(O), tet(S) and tet(W) each [3]. Overall, we found that 63.3% and 53.3% of CC1 S. agalactiae isolates harbored erm(A) and erm(B), respectively. In similar surveys conducted in UK, the carriage rates of erm(A) and erm(B) among S. agalactiae isolated from various clinical specimens were found to be 45.4% and 54.5%, respectively [3]. This controversy about the carriage of erm family genes may be due to the irrational prescribing or consumption of these antibiotics, public health policy, different sources of recovery of resistant S. agalactiae, or the possible exchange of erm and mef(A/E) genes among different S. agalactiae strains through the transfer of mobile genetic elements. Further investigation is required to validate the claims that the high prevalence of PIs, biofilm formation, and adhesion genes in the CC1 clone may play a significant role in its ability to colonize human cells. These assertions are currently speculative and necessitate verification. Therefore, additional research is necessary to comprehend the underlying factors that contribute to the adherence and colonization of human cells exhibited by the CC1 clone. However, it was hypothesized that the CC1 clone would have a higher likelihood of being identified in BSIs related to S. agalactiae in Iranian hospitals.
In the present study, CC12, which corresponded to ST918 and ST1372, was the second most frequently detected clone with simultaneous resistance to erythromycin and clindamycin and biofilm production. This confirms the previous results of Emaneini et al. in Iran, who showed CC12 in 10.5% of S. agalactiae isolated from newborns in intensive care units with full resistance to erythromycin, tetracycline, and clindamycin and carriage of scpB, hlyB, bca, and bac virulence genes [24]. A recent report by Moltó-García from Spain showed the emergence of ST12 in S. agalactiae from postmenopausal women at a low level (1.9%) [12]. These results coincide with those produced by Jiang et al., who reported ST12 as the third most predominant type of S. agalactiae causing community-onset and nosocomial infections in nonpregnant adults. They indicated that all the isolates were resistant to both erythromycin and clindamycin and showed the CCR phenotype [8]. Additionally, in a study conducted by Liu and colleagues in China in 2021, it was reported that CC12 was the most common clone of S. agalactiae isolated from blood infections, accounting for 86.7% of cases. They found that CC12 exhibited similarities to CC10 and CC1 in terms of virulence gene profiles. Liu et al. demonstrated that CC12 is an emerging clone of S. agalactiae that is both highly virulent and resistant to multiple drugs. This clone was found to be responsible for causing invasive and often fatal infections in pediatric patients [26]. In the current study, the high prevalence of bac in CC12 strains is in line with a study from Spain that indicated the presence of alphaC + bac in 88.9% of ST12 strains [25]. Consistent with earlier research, we found that CC12 isolates were linked to a high percentage of adhesion, PIs, and antibiotic resistance-related genes. This characteristic can enhance its ability to survive in the hospital setting and potentially lead to more severe illnesses in patients.
The current research highlights the emergence of CC19, which corresponded to two STs, including ST19 and ST734. This CC has been the predominant clone in many areas of the world. For example, a report on 60 S. agalactiae from Ethiopia highlighted CC19 (18.8%) as the second predominant type among tested isolates [20]. Similarly, Emaneini et al. in Iran reported CC19 as the most prevalent CC among S. agalactiae isolated from neonates (68.4%) [24]. Current findings in relation to the detection of rib in CC19 strains (50%) are in line with the study of Khan et al. in the UK, which indicated the rib gene was detected in the majority of CC19 (79.3%) isolates [3]. This result is similar to that of the study by Rojo-Bezares et al., who indicated that the rib gene was present in all CC19 strains (ST19, ST28, ST110, ST335, and ST601) [25]. This CC with a high frequency of the CCR phenotype and full resistance to tetracycline with a different carriage rate of the tet(K) (100%), tet(M) (25%), tet(S) (75%), and erm(B) (75%) genes was also reported in studies from different countries, including Poland (17%), Sweden (18.7%), Malaysia (8%), China (30.6%), and Taiwan (7.1%) [11, 18, 21, 27]. A study conducted by Khan et al. from the UK reported a prevalence rate of 78.6% of tet(M) among tetracycline-resistant S. agalactiae [3]. The differences in tet prevalence might be due to differences in the expression or transmissibility of these genes, but further investigation is needed to elucidate the underlying reasons for these differences. In similar surveys conducted in Portugal, an association between macrolide resistance and the CC19 lineages was noted [28]. These discrepancies among CC19 may reflect differences in the horizontal transfer of macrolide- and tetracycline resistance genes. Since CC19 might be highly pathogenic, careful attention is required to avoid possible problems and the spread of this strain.
The fourth known predominant genotype in the present research was CC22 (ST22 and ST0). Approximately 70% of these strains had PI-1 + PI-2b and CCR phenotypes; half of the strains produced biofilms, and all isolates carried tetracycline and erythromycin resistance genes. In concordance with our analysis, the presence of this clone, along with tetracycline resistance genes and CCR phenotypes, has been described by several investigators [29, 30]. To date, there have been few reports on the detection of CC22, and this is the first report to present clinically relevant data. This clone is suspected to be prevalent in a limited area.
According to published data, CC17 is a successful invasive hypervirulent clone that is distributed in different parts of the world. In the present survey, this clone was detected in bloodstream infections at a low level (6.6%), which is in agreement with Strakova et al.’s study that reported ST17 in CSF (85%) and blood (32%) isolates [31]. Likewise, a study by Bohnsack et al., conducted in the USA, displayed a strong correlation between CC17 and invasive infections, while this correlation was not detected in CC23 strains [32]. According to the evidence, ST17 and ST19 strains were most frequently detected in invasive infections, which confirms our results in the present research [33, 34]. We detected rib gene in 50% of CC17 isolates. This result is similar to that of the UK study by Khan et al., who indicated that the rib gene was the predominant virulence gene in S. agalactiae strains (100%) [3]. According to the study results of Rojo-Bezares et al., in Spain, the rib gene was detected in all of CC17 S. agalactiae isolates [25]. In line with findings reported from China, Poland, Kenya, Ethiopia, and Taiwan [10, 18,19,20], we noticed a high antibiotic resistance rate and no biofilm formation ability in CC17 strains. For instance, out of 4 CC17 isolates, none were biofilm producers; approximately 75% of isolates carried tet(M), 25% carried tet(O), and tet(K), while ICR and CCR phenotypes and carriage of erm family genes were not detected in any of the tested isolates. Khan and colleagues conducted an epidemiological study on 193 S. agalactiae strains obtained from clinical sources. They discovered that all CC17 isolates showed resistance to tetracycline. Among these isolates, 90.6% carried the tet(M) gene, while 9.3% carried the tet(O) gene [3]. The findings from the aforementioned studies align with our research, which confirms the diversity of these strains.
For the first time in Iran, we explored the emergence of CC/ST283 S. agalactiae-related bacteremia (3.3%). This study also found that CC/ST283 isolates did not have any ability to form biofilms, while PI-1 + PI-2b (50%) and aPI-2b (50%) were detected among the isolates. Although the first report of the emergence of this clone dates from 1985, the human outbreak of fish-borne S. agalactiae was first reported in 2015 in Singapore, with the CC/ST283 clone accounting for 70% of the outbreak [35]. Subsequent reports from China, France, and Thailand confirmed an increasing prevalence of this CC in Asia due to the consumption of undercooked aquaculture foods. According to the evidence, CC283 has been widespread in Southeast Asia for over two decades in humans [35, 36]. Therefore, it is predicted that this clone would be more likely to be frequently detected in Asia since the consumption of raw seafood, especially tilapia, is common in this region. Syuhada and colleagues conducted an epidemiological study on 256 S. agalactiae strains obtained from tilapia in Malaysia. The researchers examined the virulence gene profiles of these isolates. Their findings revealed that ST7 and ST283 were the most prevalent clones. They indicated that ST283 had higher virulence compared to ST7, as it carried a greater number of virulence genes, including cspA, bac, fbsB, lmb, bca, pavA, cyl, scpB, and others [37].
Contrary to expectations, the detection of the CC283 clone in Iran was rare; however, the results of this study suggest that further studies should be performed to complete the epidemiological picture of this clone.
The complex and diverse carriage of adhesion and alpha-protein-like genes in different CCs of S. agalactiae strains may be linked to the variability in the expression of the aforementioned factors, the possibility of transfer through mobile genetic elements, and their exclusive presence in different clones. Regarding the presence of alp genes in S. agalactiae strains, both the present and prior studies suggest the vital role of these genes in the pathogenesis of bacteria and their potential to be promising vaccine candidates. To our knowledge, this study is the first report showing the genetic diversity of S. agalactiae isolated from bloodstream infections. Overall, our findings indicate that there is genetic diversity with a strong ability for biofilm formation and the simultaneous carriage of resistance genes in clonal lineages. The observed genetic variability accentuates the potentially significant health risk posed by S. agalactiae in patients with bacteremia. Since CC1 and CC12, as successful clones in patients with bacteremia, are highly pathogenic and could cause a worse prognosis, careful attention to the spread of these clones is needed.
Materials and methods
Study design and identification of bacterial isolates
This cross-sectional study was conducted at the Department of Microbiology, Shahid Beheshti University of Medical Sciences (SBMU) over a duration of two years from February 2021 to January 2023. This study was performed at the Affiliated Hospitals of Shahid Beheshti University of Medical Sciences, Tehran, Iran. The Medical Ethics Committee of Shahid Beheshti University of Medical Sciences approved the study. All methods were performed in accordance with the relevant guidelines and regulations. BSI was defined as the presence of at least one positive blood culture for S. agalactiae in patients exhibiting signs of probable infection. For patients with persistent BSIs caused by the same organism, only the initial episode was included. If patients experienced multiple episodes of BSIs during the same admission period, only the first infection episode was counted for prevalence analysis, but subsequent episodes were included for analysis of microbiological characteristics and clinical outcomes. A total of 61 S. agalactiae strains causing blood infection were included in the present study. These samples were streaked on blood agar containing 5% sheep blood (BA, Oxoid, UK) for S. agalactiae isolation. Briefly, all suspected isolates from blood agar were gram-stained and tested for standard biochemical assays (sodium hippurate hydrolysis, catalase, and CAMP test) [8, 9]. All phenotypically confirmed S. agalactiae isolates underwent polymerase chain reaction (PCR) assay for atr gene detection and final confirmation. The sequences (5′ to 3′) of the oligonucleotide primers were: Forward-CAACGATTCTCTCAGCTTTGTTAA and reverse-TAAGAAATCTCTTGTGCGGATTTC [38]. The protocol of this study received approval from the Ethics Committee of the Shahid Beheshti University of Medical Sciences in Tehran, Iran (IR.SBMU.MSP.REC.1399.623). Informed consent was obtained from all of the participants in the study.
Determination of minimum inhibitory concentrations (MICs) and macrolide resistance phenotypes
For antimicrobial susceptibility testing of the isolated S. agalactiae, the microbroth dilution method was applied using commercially available antibiotic powders, including erythromycin, vancomycin, linezolid, penicillin, clindamycin, tetracycline, chloramphenicol, and levofloxacin (Sigma‒Aldrich, St. Louis, Mo), according to the Clinical and Laboratory Standards Institute (CLSI) methodology. The results were interpreted as recommended by the CLSI guidelines (CLSI 2022). The identification of ICR in S. agalactiae isolates was performed using a broth microdilution test (0.5 µg/mL clindamycin and 1 µg/mL erythromycin). Isolates with resistance to both clindamycin and erythromycin were considered CCR strains. Any growth in wells containing erythromycin and clindamycin was considered a positive test. MDR of S. agalactiae strains was specified as resistance to ≥ 3 classes of antibacterial agents, as previously explained [13, 39, 40]. The experiment was controlled using the Streptococcus pneumoniae ATCC 49,619 reference strain.
Evaluation of biofilm formation
Biofilm formation of S. agalactiae was performed by MtP assay, according to Goudarzi et al.’s previous research [39]. First, the S. agalactiae isolates were cultured in trypticase soy broth (TSB, Merck, Darmstadt, Germany) with 1% glucose and subsequently incubated at 37 °C overnight. The broth dilution was then performed at a ratio of 1:100. Next, two hundred microliters of the diluted cultures were inoculated in a sterile 96-well flat bottom polystyrene microtiter plate (Greiner Bio-One International, Kremsmunster, Austria) and subsequently incubated for 24 h at 37 °C without shaking. Then, the plate was air dried, and the wells were washed three times with 200 µl of phosphate-buffered saline (PBS; pH 7.2) to discard all nonadherent bacteria. Bacterial cells adhering to the slides were stained with 0.1% safranin after being fixed with 99% methanol. To remove the excess stain, the plate was rinsed under running tap water. After drying, 1 mL of 95% ethanol was added per well to solubilize the adherent stain. TSB containing 1% glucose was used as a negative control. An ELISA plate reader (plate reader, model–A4, serial no.-1910, Das, Italy) was used to measure the optical density (OD) at 490 nm. A cutoff value (ODc) was determined for each microtiter plate according to the following formula: ODc = average OD of negative control + (3× standard deviation (SD) of negative control). The resulting OD value of a tested strain was calculated by subtracting the ODc value from the average OD value of the strain (OD = average OD of a strain - ODc) [41]. In the present study, negative- and positive-biofilm controls were the wells that initially contained medium without any bacteria (non-inoculated) and wells with a biofilm-producing bacterium (S. agalactiae ATCC 13813), respectively.
Extraction of genomic DNA and detection of resistance determinants
According to a prior description, the phenol‒chloroform assay was used to extract the total genomic DNA of all the isolates. All isolates underwent a PCR assay for the presence of resistance determinants, erm(A), mef(A/E), and erm(B), as macrolide-mediated resistance genes and tet(S), tet(K), tet(M), tet(L), and tet(O) as tetracycline resistance genes, using specific oligonucleotides described by Palmeiro et al. [24, 42, 43]. The amplification products were electrophoresed on a 1.2–1.8% agarose gel.
Identification of alpha-protein-like and pilus island genes
All isolates were screened for the presence of alp2/3 (C alpha-like protein 2/3), alp4, bca (alpha C protein), eps (epsilon protein-encoding gene), and rib (cell surface rib protein) genes by multiplex PCR to detect alpha-protein-like and pilus island genes using specific oligonucleotide primer sequences described by Creti et al. [44]. To evaluate the presence of adhesion factors, a PCR assay was performed to detect lmb (human laminin-binding protein), cylE (beta-hemolysin), and bac genes (cell surface beta C protein), as previously described [8]. PI-1-, PI-2a-, and PI-2b-mediated virulence was detected by using PCR as described elsewhere [13].
MLST
To determine the clonal lineage and genetic diversity of the isolates, MLST was performed as described earlier. In this technique, seven S. agalactiae housekeeping genes (adhP, pheS, atr, glnA, sdhA, glcK, and tkt) were amplified by PCR assay. Afterwards, amplified fragments were subjected to bidirectional DNA sequencing. In the next step, STs were determined by comparing the aforementioned sequences of all seven housekeeping genes with standard alleles listed on the online MLST website (http://pubmlst.org/sagalactiae/). STs were further grouped into different CCs based on similar allelic profiles.
Phylogenetic analysis
The bioinformatics data were analyzed for 11 STs (a combination of seven genes) in this study. The sequence dataset was aligned using the MEGA7.0.21 software. The program RAxML version 8.2 (Miller et al., 2010; Stamatakis, 2014) was run on the CIPRES Science Gateway. Optimization in RAxML was carried out using the GTRCAT option. Bootstrap values for maximum likelihood were 1,000 replicates, with one search replicate per bootstrap replicate.
Data availability
All data are presented in manuscript.
Abbreviations
- MLST:
-
Multilocus sequence typing
- CC:
-
Clonal Complex
- SD:
-
Standard deviation
- MtP:
-
Microtiter plate
- CCR:
-
Constitutive clindamycin-resistant
- CLSI:
-
Clinical and laboratory standards institute
- MIC:
-
Minimum inhibitory concentration
- PCR:
-
Polymerase chain reaction
- ICR:
-
Inducible clindamycin resistance
- MDR:
-
Multidrug-resistant
- PI:
-
Pilus Island
- OD:
-
Optical density
References
Meroni G, Sora VM, Martino PA, Sbernini A, Laterza G, Zaghen F, et al. Epidemiology of antimicrobial resistance genes in Streptococcus agalactiae sequences from a public database in a one health perspective. Antibiotics. 2022;11(9):1236.
Hooven TA, Catomeris AJ, Bonakdar M, Tallon LJ, Santana-Cruz I, Ott S, et al. The Streptococcus agalactiae stringent response enhances virulence and persistence in human blood. Infect Immun. 2018;86(1). https://doi.org/10.1128/iai. 00612 – 17.
Khan UB. Detailed genomic and antimicrobial resistance comparison of UK Streptococcus agalactiae isolates from adults to those of diverse global origins. In.: Cardiff University; 2022.
Laupland KB, Pasquill K, Parfitt EC, Steele L. Bloodstream infection due to β-hemolytic streptococci: a population-based comparative analysis. Infection. 2019;47:1021–5.
Shelburne IIISA, Tarrand J, Rolston KV. Review of streptococcal bloodstream infections at a comprehensive cancer care center, 2000–2011. J Infect. 2013;66(2):136–46.
Ali M, Alamin MA, Ali A, Alzubaidi G, Ali K, Ismail B. Microbiological and clinical characteristics of invasive Group B streptococcal blood stream infections in children and adults from Qatar. BMC Infect Dis. 2022;22(1):881.
Morozumi M, Wajima T, Kuwata Y, Chiba N, Sunaoshi K, Sugita K, et al. Associations between capsular serotype, multilocus sequence type, and macrolide resistance in Streptococcus agalactiae isolates from Japanese infants with invasive infections. Epidemiol Infect. 2014;142(4):812–9.
Jiang H, Chen M, Li T, Liu H, Gong Y, Li M. Molecular characterization of Streptococcus agalactiae causing community-and hospital-acquired infections in Shanghai, China. Front Microbiol. 2016;7:1308.
Liu Z, Jiang X, Li J, Ji W, Zhou H, Gong X, et al. Molecular characteristics and antibiotic resistance mechanisms of clindamycin-resistant Streptococcus agalactiae isolates in China. Front Microbiol. 2023;14:1138039.
Lu B, Chen X, Wang J, Wang D, Zeng J, Li Y, et al. Molecular characteristics and antimicrobial resistance in invasive and noninvasive Group B Streptococcus between 2008 and 2015 in China. Diagn Microbiol Infect Dis. 2016;86(4):351–7.
Luan S-L, Granlund M, Sellin M, Lagergård T, Spratt BG, Norgren M. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J Clin Microbiol. 2005;43(8):3727–33.
Moltó-García B, del Carmen Liébana-Martos M, Cuadros-Moronta E, Rodríguez-Granger J, Sampedro-Martínez A, Rosa-Fraile M, et al. Molecular characterization and antimicrobial susceptibility of hemolytic Streptococcus agalactiae from post-menopausal women. Maturitas. 2016;85:5–10.
Nabavinia M, Khalili MB, Sadeh M, Eslami G, Vakili M, Azartoos N, et al. Distribution of Pilus island and antibiotic resistance genes in Streptococcus agalactiae obtained from vagina of pregnant women in Yazd, Iran. Iran J Microbiol. 2020;12(5):411.
Piccinelli G, Biscaro V, Gargiulo F, Caruso A, De Francesco MA. Characterization and antibiotic susceptibility of Streptococcus agalactiae isolates causing urinary tract infections. Infect Genet Evol. 2015;34:1–6.
Poyart C, Jardy L, Quesne G, Berche P, Trieu-Cuot P. Genetic basis of antibiotic resistance in Streptococcus agalactiae strains isolated in a French hospital. Antimicrob Agents Chemother. 2003;47(2):794–7.
Springman AC, Lacher DW, Waymire EA, Wengert SL, Singh P, Zadoks RN, et al. Pilus distribution among lineages of group b streptococcus: an evolutionary and clinical perspective. BMC Microbiol. 2014;14:1–11.
Zhang L, Ma L, Zhu L, Zhou X-H, Xu L-J, Guo C, et al. Molecular characterization of pathogenic group B streptococcus from a tertiary hospital in Shanxi, China: high incidence of sequence type 10 strains in infants/pregnant women. J Microbiol Immunol Infect. 2021;54(6):1094–100.
Kao Y, Tsai M-H, Lai M-Y, Chu S-M, Huang H-R, Chiang M-C, et al. Emerging serotype III sequence type 17 group B streptococcus invasive infection in infants: the clinical characteristics and impacts on outcomes. BMC Infect Dis. 2019;19(1):1–8.
Huber CA, McOdimba F, Pflueger V, Daubenberger CA, Revathi G. Characterization of invasive and colonizing isolates of Streptococcus agalactiae in east African adults. J Clin Microbiol. 2011;49(10):3652–5.
Gizachew M, Tiruneh M, Moges F, Adefris M, Tigabu Z, Tessema B. Molecular characterization of Streptococcus agalactiae isolated from pregnant women and newborns at the University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Infect Dis. 2020;20(1):1–9.
Brzychczy-Wloch M, Gosiewski T, Bulanda M. Multilocus sequence types of invasive and colonizing neonatal group B Streptococci in Poland. Med Principles Pract. 2014;23(4):323–30.
Tien N, Ho C-M, Lin H-J, Shih M-C, Ho M-W, Lin H-C, et al. Multilocus sequence typing of invasive group B Streptococcus in central area of Taiwan. J Microbiol Immunol Infect. 2011;44(6):430–4.
Meehan M, Cunney R, Cafferkey M. Molecular epidemiology of group B Streptococci in Ireland reveals a diverse population with evidence of capsular switching. Eur J Clin Microbiol Infect Dis. 2014;33:1155–62.
Emaneini M, Jabalameli F, Mirsalehian A, Ghasemi A, Beigverdi R. Characterization of virulence factors, antimicrobial resistance pattern and clonal complexes of group B streptococci isolated from neonates. Microb Pathog. 2016;99:119–22.
Rojo-Bezares B, Azcona-Gutiérrez J, Martin C, Jareño M, Torres C, Sáenz Y. Streptococcus agalactiae from pregnant women: antibiotic and heavy-metal resistance mechanisms and molecular typing. Epidemiol Infect. 2016;144(15):3205–14.
Liu J, Chen F, Guan H, Yu J, Yu J, Zhao J, et al. Emerging fatal Ib/CC12 hypervirulent multiresistant Streptococcus agalactiae in young infants with bloodstream infection in China. Front Microbiol. 2021;12:767803.
Ezhumalai M, Muthanna A, Suhaili Z, Dzaraly ND, Amin-Nordin S, Amal MNA, et al. Multilocus sequence typing analysis of invasive and non-invasive Group B Streptococcus of hospital origin in Malaysia. Malaysian J Med Sciences: MJMS. 2020;27(1):134.
Florindo C, Viegas S, Paulino A, Rodrigues E, Gomes JP, Borrego MJ. Molecular characterization and antimicrobial susceptibility profiles in Streptococcus agalactiae colonizing strains: association of erythromycin resistance with subtype III-1 genetic clone family. Clin Microbiol Infect. 2010;16(9):1458–63.
Jamrozy D, Gopal Rao G, Feltwell T, Lamagni T, Khanna P, Efstratiou A, et al. Population genetics of group B Streptococcus from maternal carriage in an ethnically diverse community in London. Front Microbiol. 2023;14:1185753.
Martins ER, Nascimento do OD, Marques Costa AL, Melo-Cristino J, Ramirez M. Characteristics of Streptococcus agalactiae colonizing nonpregnant adults support the opportunistic nature of invasive infections. Microbiol Spectr. 2022;10(3):e01082–22.
Straková L, Musílek M, Motlová J. Multilocus sequence types in Czech neonatal GBS strains from 2004 to 2008. Epidemiologie, Mikrobiologie, Imunologie: Casopis Spolecnosti pro Epidemiologii a Mikrobiologii Ceske Lekarske Spolecnosti. JE Purkyne. 2010;59(1):45–7.
Bohnsack JF, Whiting A, Gottschalk M, Dunn DM, Weiss R, Azimi PH, et al. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six US Academic Centers from 1995 to 1999. J Clin Microbiol. 2008;46(4):1285–91.
Björnsdóttir E, Martins E, Erlendsdóttir H, Haraldsson G, Melo-Cristino J, Kristinsson K, et al. Changing epidemiology of group B streptococcal infections among adults in Iceland: 1975–2014. Clin Microbiol Infect. 2016;22(4):379. e9-. e16.
Springman AC, Lacher DW, Waymire EA, Wengert SL, Singh P, Zadoks RN, et al. Pilus distribution among lineages of group b streptococcus: an evolutionary and clinical perspective. BMC Microbiol. 2014;14(1):1–11.
Barkham T, Zadoks RN, Azmai MNA, Baker S, Bich VTN, Chalker V, et al. One hypervirulent clone, sequence type 283, accounts for a large proportion of invasive Streptococcus agalactiae isolated from humans and diseased tilapia in Southeast Asia. PLoS Negl Trop Dis. 2019;13(6):e0007421.
Rothen J, Pothier JF, Foucault F, Blom J, Nanayakkara D, Li C, et al. Subspecies typing of Streptococcus agalactiae based on ribosomal subunit protein mass variation by MALDI-TOF MS. Front Microbiol. 2019;10:471.
Syuhada R, Zamri-Saad M, Ina-Salwany M, Mustafa M, Nasruddin N, Desa M, et al. Molecular characterization and pathogenicity of Streptococcus agalactiae serotypes Ia ST7 and III ST283 isolated from cultured red hybrid tilapia in Malaysia. Aquaculture. 2020;515:734543.
Arabestani MR, Mousavi SM, Nasaj M. Genotyping of clinical Streptococcus agalactiae strains based on molecular serotype of capsular (cps) gene cluster sequences using polymerase chain reaction. Archives Clin Infect Dis. 2017;12(1).
Goudarzi M, Mohammadi A, Amirpour A, Fazeli M, Nasiri MJ, Hashemi A, et al. Genetic diversity and biofilm formation analysis of Staphylococcus aureus causing urinary tract infections in Tehran, Iran. J Infect Developing Ctries. 2019;13(09):777–85.
Lo H-H, Nien H-H, Cheng Y-Y, Su F-Y. Antibiotic susceptibility pattern and erythromycin resistance mechanisms in beta-hemolytic group G Streptococcus dysgalactiae subspecies equisimilis isolates from central Taiwan. J Microbiol Immunol Infect. 2015;48(6):613–7.
Goudarzi M, Mohammadi A, Amirpour A, Fazeli M, Nasiri MJ, Hashemi A, et al. Genetic diversity and biofilm formation analysis of Staphylococcus aureus causing urinary tract infections in Tehran. Iran. 2019;13(09):777–85.
Palmeiro JK, Dalla-Costa LM, Fracalanzza SE, Botelho AC, da Silva Nogueira K, Scheffer MC, et al. Phenotypic and genotypic characterization of group B streptococcal isolates in southern Brazil. J Clin Microbiol. 2010;48(12):4397–403.
Maeda T, Tsuyuki Y, Fujita T, Fukushima Y, Goto M, Yoshida H, et al. Comparison with Streptococcus agalactiae isolates from humans reveals genotypic and phenotypic characteristics of isolates from Companion Animal-origin. Jpn J Infect Dis. 2020;JJID:2019–441.
Creti R, Fabretti F, Orefici G, von Hunolstein C. Multiplex PCR assay for direct identification of group B streptococcal alpha-protein-like protein genes. J Clin Microbiol. 2004;42(3):1326–9.
Acknowledgements
We would like to acknowledge Dr. Maryam Fazeli for editing this manuscript.
Funding
This work is financially supported by a research grant from the Research Deputy of Shahid Beheshti University of Medical Sciences, Tehran, Iran [Grant No. 25525].
Author information
Authors and Affiliations
Contributions
MG conceived and designed the study. MG, AM, CHA, and PB contributed to comprehensive research. BP, MG, and AM wrote the paper. ZS analyzed data and prepared figures. MG, AM, CHA, ZS and PB participated in manuscript editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Shahid Beheshti University of Medical Sciences in Tehran, Iran (IR.SBMU.MSP.REC.1399.623). Informed consent was obtained from all of the participants in the study. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no conflicts of interest directly relevant to this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mohammadi, A., Amini, C., Bagheri, P. et al. Unveiling the genetic landscape of Streptococcus agalactiae bacteremia: emergence of hypervirulent CC1 strains and new CC283 strains in Tehran, Iran. BMC Microbiol 24, 365 (2024). https://doi.org/10.1186/s12866-024-03521-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-024-03521-z