Abstract
Background
Moraxella catarrhalis is a leading cause of otitis media (OM) and chronic obstructive pulmonary disease (COPD). M. catarrhalis contains a Type III DNA adenine methyltransferase (ModM) that is phase-variably expressed (i.e., its expression is subject to random, reversible ON/OFF switching). ModM has six target recognition domain alleles (modM1–6), and we have previously shown that modM2 is the predominant allele, while modM3 is associated with OM. Phase-variable DNA methyltransferases mediate epigenetic regulation and modulate pathogenesis in several bacteria. ModM2 of M. catarrhalis regulates the expression of a phasevarion containing genes important for colonization and infection. Here we describe the phase-variable expression of modM3, the ModM3 methylation site and the suite of genes regulated within the ModM3 phasevarion.
Results
Phase-variable expression of modM3, mediated by variation in length of a 5′-(CAAC)n-3′ tetranucleotide repeat tract in the open reading frame was demonstrated in M. catarrhalis strain CCRI-195ME with GeneScan fragment length analysis and western immunoblot. We determined that ModM3 is an active N6-adenine methyltransferase that methylates the sequence 5′-ACm6ATC-3′. Methylation was detected at all 4446 5′-ACATC-3′ sites in the genome when ModM3 is expressed. RNASeq analysis identified 31 genes that are differentially expressed between modM3 ON and OFF variants, including five genes that are involved in the response to oxidative and nitrosative stress, with potential roles in biofilm formation and survival in anaerobic environments. An in vivo chinchilla (Chinchilla lanigera) model of otitis media demonstrated that transbullar challenge with the modM3 OFF variant resulted in an increased middle ear bacterial load compared to a modM3 ON variant. In addition, co-infection experiments with NTHi and M. catarrhalis modM3 ON or modM3 OFF variants revealed that phase variation of modM3 altered survival of NTHi in the middle ear during early and late stage infection.
Conclusions
Phase variation of ModM3 epigenetically regulates the expression of a phasevarion containing multiple genes that are potentially important in the progression of otitis media.
Similar content being viewed by others
Background
Moraxella catarrhalis is a human respiratory tract pathogen that is often carried asymptomatically in the nasopharynx [1], but frequently causes otitis media (OM) in infants and children, and exacerbations of chronic obstructive pulmonary disease (COPD) in adults. Along with Streptococcus pneumoniae and non-typeable Haemophilus influenzae (NTHi), M. catarrhalis is among the most prevalent bacterial causes of OM where it is detected by PCR in up to 56% of middle ear fluid associated with OM [2]. M. catarrhalis also causes approximately ~ 10% of exacerbations of COPD each year in the USA [3]. M. catarrhalis is also important as a co-pathogen in OM with S. pneumoniae and NTHi, as noted in both observational [4] and experimental studies [5, 6]. There is currently no vaccine available to prevent M. catarrhalis-mediated disease, and increased research is needed to identify stably-expressed vaccine antigens, correlates of protection, and to understand the progression from asymptomatic carriage to symptomatic disease.
Phase variation is the high frequency, reversible, random ON/OFF or graded switching of gene expression [7, 8]. Phase-variable gene expression is an important aspect of bacterial pathogenesis that aids in adaptation to changing host microenvironments, and which can aid immune evasion which has implications for vaccine development [9]. Several M. catarrhalis virulence factors and potential vaccine antigens, such as the outer membrane proteins UspA1 [10], UspA2 [11], UpsA2H [12] and Mid/Hag [13] (reviewed in [14]) exhibit phase-variable expression. Although phase variation is typically associated with genes encoding cell surface structures, phase variation of cytoplasmically located Restriction-Modification (R-M) systems has been observed in numerous host-adapted bacterial pathogens, as recently reviewed [15]. Phase-variable ON/OFF switching of DNA methyltransferase activity results in the presence or absence of methylation at a specific target sequence, leading to co-ordinated, epigenetically-regulated switching of expression of multiple genes across the genome. The suite of genes thus regulated are known as a phasevarion (phase-variable regulon) [15, 16]. Previously characterized phasevarions contain genes important for infection of the human host and potential vaccine candidates; for example lbpA and lbpB (encoding the lactoferrin binding proteins A and B, respectively) are regulated within the ModA11 phasevarion in Neisseria meningitidis [17], and the outer-membrane protein encoding gene hopG is regulated within the ModH5 phasevarion in Helicobacter pylori [18]. Switching of phasevarion expression has also been shown to modulate diverse phenotypes associated with virulence, such as biofilm formation [17], resistance to antimicrobial agents [19], resistance to oxidative stress [20], and survival within experimental models of infection [21].
We previously identified three phase-variable Type III DNA methyltransferases (modM, modN, and modO) that are variably distributed among M. catarrhalis isolates and strongly associated with phylogenetic lineage [22,23,24]. modM is the only phase-variable methyltransferase found in the disease-associated RB1 lineage and occurs in 76% of isolates from geographically and clinically diverse backgrounds [22]. Phase variation of modM is mediated at the translational level by a 5′-(CAAC)n-3′ tetranucleotide repeat tract present within its open reading frame (ORF) [23]. Analysis of the genomes of 51 M. catarrhalis strains identified six modM alleles (modM1–6) that vary in their target recognition domains [22]. Allelic variants of phase-variable Type III methyltransferases containing distinct target recognition domains methylate distinct target sequences, and regulate different suites of genes [17, 21]. In M. catarrhalis, only the most commonly occurring modM allele, modM2, has been investigated to date. ModM2 methylates the target sequence 5′-GARm6AC-3′, and ON/OFF switching of ModM2 results in the differential regulation of 34 genes, including genes involved in colonisation and protection against host defences [23]. modM3 is the second most frequently occurring modM allele in all strains analysed, and the most frequently occurring allele in strains belonging to the minor RB2/3 lineage [22]. Despite the RB2/3 lineage being less commonly associated with disease than the RB1 linage in several studies [reviewed in 14], the modM3 allele is also overrepresented in middle ear isolates from children with OM [22, 23]. Here we characterise the phase-variable expression of modM3, the ModM3 DNA methylation site and its distribution in the genome, the genes regulated in the ModM3 phasevarion, and phenotypes of the ModM3 ON versus ModM3 OFF variants.
Results
modM3 exhibits phase-variable expression
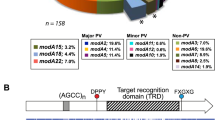
The modM3 gene contains a 5′-(CAAC)n-3′ tetranucleotide repeat tract in the N-terminal of its open-reading frame (ORF) (Fig. 1a). Different strains display varying numbers of modM3 repeat units (e.g. 24, 31, and 35 repeats are found in strains F24, BC1 and CCRI-195ME, respectively), which is suggestive of phase variation. The modM3 coding sequence downstream of the repeat tract is predicted to be in frame with the ATG start codon when 36 repeats are present, while a frameshift mutation resulting in a premature stop codon is predicted to occur when 37 repeats are present (Fig. 1a). To confirm modM3 is phase-variable, single colonies of M. catarrhalis strain CCRI-195ME (hereafter 195ME) were screened with GeneScan fragment length analysis to measure the length of the 5′-(CAAC)n-3′ repeat tract and to quantify the proportion of each repeat tract length present. From an initial mixed population with repeat tract lengths ranging from 34 to 37 repeats, separate colonies containing a majority (> 85%) of 36 or 37 repeats were isolated (Fig. 1b). Western immunoblot analysis of ModM3 protein expression confirmed that expression is correlated with the number of repeat units in the 5′-(CAAC)n-3′ repeat tract: ModM3 is expressed (ON) when 36 repeats units are present, whereas ModM3 expression is undetectable (OFF) when 37 repeats are present, confirming that ModM3 expression is phase-variable (Fig. 1c).
Phase-variable expression of modM3. a Schematic representation of the modM3 gene indicating the location of the 5′-(CAAC)n-3′ tetranucleotide repeat region and the central target recognition domain. Translation of the full length ModM3 is possible when the repeat tract contains 36 (ON) repeat units, but not when the tract contains 37 (OFF) repeat units (an asterisk indicates a premature stop codon). b GeneScan fragment length analysis of the of modM3 5′-(CAAC)n-3′ tetranucleotide repeat region. The area under each fluorescent peak is proportional to the percentage of a repeat length in the population. Separate populations were isolated containing ≥85% of 36 or 37 repeats. c) Western blot analysis confirmed ModM3 expression in 195ME ModM3 ON (36 repeats) and the absence of expression in the 195ME ModM3 OFF sample (37 repeats)
ModM3 is an active N6-adenine DNA methyltransferase that methylates 5′-ACm6ATC-3′
To determine the methylation specificity of ModM3, genomic DNA from our M. catarrhalis strain 195ME modM3 ON and modM3 OFF variants, as well as our ∆modM3 knockout mutant strain (generated by insertion of a kanamycin resistance cassette into the ORF of the gene), were subjected to Single-Molecule, Real-Time (SMRT) sequencing and methylome analysis. The closed genome sequence of 195ME was reported and deposited in GenBank at the time of sequencing (GenBank accession NZ_CP018059.1) [25]. Three methylated motifs were identified: one containing N6-adenine methylation and two containing 5-cytosine methylation (Table 1). N6-adenine methylation is present at the second adenine residue in the sequence 5′-ACm6ATC-3′ in the modM3 ON strain but not in the modM3 OFF or the ∆modM3 strains, indicating that this is the motif recognized and methylated by ModM3. The 195ME genome contains 4446 5′-ACATC-3′ sites in total, with 5′-ACm6ATC-3′ methylation observed at 100% of these sites in the modM3 ON variants, but at 0% of these sites in the modM3 OFF or ∆modM3 strains (Table 1).
In order to confirm the ModM3 methylation site, a restriction-inhibition assay was performed using the commercially available, methyl-sensitive restriction endonuclease BtsCI (5′-CATCC-3′) which partially overlaps the ModM3 recognition sequence (5′-ACm6ATC-5′) (Fig. 2a, b). Of the 2,491 BtsCI cleavage sites in the 195ME genome, 504 overlap a ModM3 recognition site and are sensitive to ModM3 methylation. There is no difference in the overall DNA digestion pattern on the DNA gel due to the majority (1,987) of BtsCI cleavage sites being cleaved in all samples. However, Southern blot analysis using a probe located adjacent to an overlapping BtsCI and ModM3 recognition sequence (and between two BtsCI sites that do not overlap with a ModM3 site; Fig. 2a) showed that genomic DNA isolated from modM3 ON was protected from digestion with BtsCI at the site investigated with an overlapping ModM3 site, whereas modM3 OFF and ∆modM3 genomic DNA was cleaved (Fig. 2c), validating that ModM3 is an active N6-methyladenine methyltransferase that methylates the target sequence 5′-ACm6ATC-3′. A quantitative real time PCR (qRT-PCR) based assay was designed to allow rapid, sensitive quantification of the relative frequency of methylation at a specific DNA site. Primers were designed to amplify a 99 bp amplicon spanning the overlapping BstCI and ModM3 recognition site (Fig. 2a), and BtsCI digested genomic DNA was used as the template. qRT-PCR analysis showed that there were 88-fold and 7-fold more copies of undigested DNA in the modM3 ON and modM3 OFF samples, respectively, relative to the ∆modM3 sample (set as 1 as a reference) (Fig. 2d). Minimal amplification was seen for the ∆modM3 sample, likely due to incomplete DNA digestion by the temperature sensitive BtsCI. The presence of methylated, undigested sites in the modM3 OFF population can be attributed to the 15% of cells having phase varied to modM3 ON, as indicated by GeneScan fragment length analysis (Fig. 1b).
Southern blotting confirms the ModM3 methylation target sequence 5′-ACm6ATC-3′. a Schematic representation of the restriction-inhibition assay used to confirm the ModM3 methylation site. The location of the Southern blot probe, and the BtsCI and ModM3 recognition sites are shown. The central BtsCI site overlaps the ModM3 recognition sequence and is sensitive to overlapping N6-methyladenine methylation. b Restriction-inhibition assay of modM3 ON, modM3 OFF and ∆modM3 genomic DNA using the methyl-sensitive restriction endonuclease BtsCI. The restriction endonuclease HindIII is not sensitive to methylation and is included as a control for digestion. c Southern blot of BtsCI digested genomic DNA isolated from modM3 ON, modM3 OFF, and ∆modM3 strains. Methylated DNA in the ModM3 ON strain is protected from BtsCI digestion resulting in a 1.5 kb band. All BtsCI sites are cleaved in the modM3 OFF and ∆modM3 strains as ModM3 methylation is absent, resulting in a 0.5 kb band. d qRT-PCR indicating the relative abundance of methylated, undigested genomic DNA in modM3 ON and modM3 OFF relative to ∆modM3 following digestion with BtsCI (Ct values of 18.32, 21.97, 24.77, respectively, normalised to copB reference)
ModM3 phase variation regulates the expression of multiple genes in a phasevarion
To determine whether phase variation of ModM3 alters expression of a phasevarion, the transcriptomes of modM3 ON and modM3 OFF variants were compared using RNASeq analysis. This identified 31 genes that are differentially regulated between the modM3 ON and modM3 OFF variants when grown to mid-log phase in aerated culture (≥1.25 fold expression ratio, P ≤ 0.05, Table 2). Seventeen genes were up-regulated and fourteen genes down-regulated in modM3 ON versus modM3 OFF. The modM3 gene and downstream res gene displayed the greatest difference in expression (9.03 and 5.49 fold increased expression in modM3 ON versus modM3 OFF, respectively), and ddc gene downstream of res also had increased expression (RS065760, 1.27 fold) relative to the modM3 OFF variants. Four genes involved in the nitrosative stress response (aniA, norB, narL, and narX), one gene potentially involved in the response to oxidative stress (RS03200), and three genes belonging to a Type I-F CRISPR-cas system operon (csy1, csy2 and csy3) were upregulated in modM3 ON. In addition, two genes (traW and RS09480) located on the large unnamed 195ME conjugative plasmid (GenBank accession NZ_CP018060.1 [25]) were also upregulated in modM3 ON. Three genes that encode part of the NADH:ubiquinone oxidoreductase (nuoB, nuoK and RS05045) were downregulated in modM3 ON. These results demonstrate that expression status of ModM3 affects the expression of multiple genes that have diverse functions and subcellular locations (Table 2). This confirms that ModM3 regulates a phasevarion.
ModM3 phase variation does not result in phenotypic differences in vitro
To investigate the phenotypic effects of ModM3 phase variation, M. catarrhalis ON and OFF variants were compared in in vitro assays, intended to simulate conditions relevant to infection of the human host. Growth curve analysis showed no significant difference in growth rate or final optical density between the ModM3 ON and OFF variants (Additional file 1: Figure S1a). The modM3 ON and modM3 OFF variants showed similar levels of survival under conditions of oxidative stress (hydrogen peroxide assays; Additional file 1: Figure S1b), and similar levels biofilm formation over 24, 48 and 72 h (Additional file 1: Figure S1c). Expression of ModM3 also did not significantly affect capacity of M. catarrhalis to adhere to or invade A549 cells (Additional file 1: Figure S1d).
ModM3 expression affects infection dynamics of M. catarrhalis and NTHi in the chinchilla model of experimental otitis media
We assessed whether differences in gene expression between M. catarrhalis modM3 ON and modM3 OFF variants impacts colonisation and/or virulence in vivo, using a chinchilla model of experimental OM [26]. Chinchillas were challenged by transbullar injection with modM3 ON or modM3 OFF populations and the number of M. catarrhalis colony forming units (CFU) in middle ear fluids and mucosal membrane homogenates were counted at days + 1 and + 2 post challenge. At all time points, a greater number of M. catarrhalis CFU were isolated from middle ear fluids and mucosal membranes of ears challenged with the modM3 OFF input pool than from chinchillas challenged with the modM3 ON population. This difference was statistically significant for middle ear mucosa samples on day + 1, middle ear fluid samples on day + 2, and total CFU per ear on day + 2 (P < 0.05, Mann-Whitney test) (Fig. 3a). Due to differential gene regulation between modM3 ON vs OFF, bacterial load may not necessarily be linked to disease severity. The severity of disease caused by M. catarrhalis phase variants was assessed using tympanometry and imaging of dissected bullae to monitor for signs of OM. On day + 1 post challenge small regions of erythema were observed in the middle ears of chinchillas from both cohorts, with an apparent slight increase in the ears challenged with the modM3 ON population (Fig. 3b). At Day + 2 post challenge, obvious erythema was observed in all middle ears with no clear difference seen in the relative amount of erythema present between cohorts. However, the development of a greater number of submucosal pockets of bacteria was observed in bullae of chinchillas challenged with modM3 OFF compared to modM3 ON (Fig. 3b). Middle ear pressure reduction is characteristic of Eustachian tube dysfunction in OM which hinders equilibration of the middle ear space against barometric pressure in the nasopharynx. The mean middle ear pressure on day + 1 and day + 2 post challenge was below the normal range for chinchillas (+/− 60 daPa), for both modM3 ON and modM3 OFF challenged cohorts, consistent with infection. A lower middle ear pressure was observed in chinchillas challenged with modM3 OFF compared to the cohort challenged with modM3 ON at day + 1 and day + 2 post challenge, however this difference was not significant (P > 0.05) (Additional file 1: Figure S2a). Expression of ModM did not affect tympanic membrane compliance, as values were within the normal range for chinchillas (0.75 to 1.5) and not significantly different between cohorts (P > 0.05) (Additional file 1: Figure S2b).
ModM3 expression alters survival of M. catarrhalis in chinchilla ears. a Number of M. catarrhalis CFU found in middle ear fluids (MEF), middle ear mucosa (MEM), or whole bullae (MEF + MEM) from chinchilla ears transbullarly inoculated with either M. catarrhalis modM3 ON or modM3 OFF. The median CFU and interquartile range is displayed for each cohort. * indicates P < 0.05. b Images of middle ears challenged with M. catarrhalis modM3 ON or modM3 OFF populations. Submucosal pockets of bacteria, characteristic of OM, are indicated by black circles
In order to determine whether modM3 phase variation also affects infections in polymicrobial challenge, chinchillas were co-challenged with NTHi strain 86-028NP (which contains a modA2 genes that only contains three DNA repeats and is therefore not phase variable) in combination with either M. catarrhalis modM3 ON, or modM3 OFF populations. Middle ear fluid and mucosal membrane homogenates were counted at days + 1, + 2 and + 7 post challenge. At days + 1 and + 2 post challenge the number of M. catarrhalis CFU was below the level of detection in the majority of mucosal membrane homogenates. However, M. catarrhalis was detected in mucosal membrane homogenates at day + 7 in 2/4 animals challenged with modM3 ON versus 4/4 animals challenged with modM3 OFF (Fig. 4a). Although more NTHi was recovered from mucosal membrane homogenates of ears co-challenged with M. catarrhalis modM3 ON at days + 1 and + 2 post challenge, and a greater number of NTHi CFU were retrieved from ears co-challenged with M. catarrhalis modM3 OFF on day + 7, these differences were not significant (Fig. 4b). M. catarrhalis and NTHi were not detected in middle ear fluids at any time point.
Discussion
M. catarrhalis is a leading bacterial pathogen of OM in children and exacerbations of COPD in adults. Despite the significant morbidity caused, factors contributing to M. catarrhalis virulence and their role in the progression of OM and exacerbations of COPD have not been fully elucidated, and there is currently no vaccine available to protect against M. catarrhalis infection. The presence of phasevarions in bacterial pathogens results in increased phenotypic diversity of these organisms. Where multiple allelic variants of phase-variable methyltransferases are present, each unique Mod allelic variant has been shown to methylate a unique target sequence, and regulate a different phasevarion [17, 21, 27]. Six modM alleles (modM1–6) have been identified in M. catarrhalis that vary in their central target recognition domain, and are therefore all expected to regulate distinct phasevarions [22]. Our previous analysis showed that ModM2 methylates the target sequence 5′-GARm6AC-3′ and regulates a phasevarion containing 34 genes [23]. In this study we have characterized the ModM3 phasevarion in M. catarrhalis strain 195ME, showing that phase variation of ModM3 is mediated by changes in the number of 5′-(CAAC)n-3′ repeat units present in the repeat tract of the modM3 ORF, and that the ModM3 protein methylates the sequence 5′-ACm6ATC-3′.
We demonstrated expression of a suite of genes differs in populations of ModM3 ON and OFF. Under aerobic growth, to mid-log phase, 29 genes were differentially expressed (1.27–1.61 fold). Five genes involved in the response to oxidative and nitrosative stress are upregulated in modM3 ON versus modM3 OFF: the nitrite and nitric oxide reductases aniA and norB, the two-component system narX/narL, and the predicted AhpC/TSA family peroxiredoxin RS03200. AniA and NorB are components of the truncated M. catarrhalis denitrification pathway required for survival in the presence of exogenously generated reactive nitrogen species [28, 29]. Detoxification of nitric oxide by NorB in N. meningitidis was shown to enhance intracellular survival within macrophages and nasopharyngeal mucosal cells [30]. Whether increased expression of NorB in modM3 ON confers greater protection against macrophage-generated nitric oxide warrants further investigation. Gene expression experiments [31] found that aniA and norB are highly upregulated during growth in biofilms versus planktonic growth in vitro, suggesting a role for both genes in the progression of OM as M. catarrhalis biofilms have been observed in middle ears of children with chronic OM [32]. However, we observed no difference in biofilm formation between our modM3 ON and modM3 OFF strain pair, suggesting that the role of ModM3 in regulation of aniA and norB is more subtle, and may only occur under specific in vivo conditions that we are unable to reproduce in the in vitro assay. The narX and narL operon form a two-component regulatory system that responds to nitrate and nitrite stimuli and modulates gene expression. The genes regulated by NarX-NarL have not been defined in M. catarrhalis, however in other organisms NarX-NarL induces expression of genes important for survival and growth in anaerobic environments [33, 34]. The middle ear is a microaerophilic environment [35] and anaerobic conditions may be reached in mature biofilms [36]. Upregulation of NarX/L in the modM3 ON strain may promote persistence in the middle ear through increased survival within biofilms under anaerobic conditions, where additional regulators may be induced and be acting in concert with ModM3. Intriguingly, several of these genes, or pathways, have previously been noted to be upregulated in M. catarrhalis when isolated from the nasopharynx of chinchillas, including aniA, norB, and other genes associated with denitrification pathway, while in the chinchilla nasopharynx, elements of energy production (NADH:ubiquinone oxidoreductases) are down regulated [37]. These in vivo expression changes have parallels to the expression changes we noted in aerobic, log phase modM3 ON strains, suggesting this population may have a pre-adapted advantage to in vivo growth in the nasopharynx. Although expression of the peroxiredoxin AhpC was increased 1.27 fold in our ModM3 ON strain, we observed no difference in survival between our modM3 ON and OFF variants following exposure to hydrogen peroxide. It is possible that the observed difference in AhpC expression was too small to have a noticeable impact on CFU counts in this assay. Alternatively, as AhpC is only minimally involved in H2O2 degradation in M. catarrhalis compared to catalase (KatA) [38], the effect of differential regulation of AhpC may have been undetectable in this assay. The lack of detectable in vitro phenotypic differences between the modM3 ON and OFF strains may be due to the narrow range of conditions able to be replicated in vitro compared to in vivo. Our gene expression analysis was also only carried out under aerobic culture conditions, and thus the present characterization may not encompass the entire suite of genes that are differentially regulated by ModM3 phase variation. In order to observe the differential regulation of genes that are not constitutively expressed, further characterization experiments need to be performed with cells grown in vivo or under a range of in vitro conditions that simulate the physiological environment encountered upon infection of the human host and where various regulatory proteins mediated expression. DNA methylation typically affects DNA interactions of regulatory proteins involved in transcription, rather than acting directly on transcriptional machinery (reviewed in [39]). As such, ModM3 may impact genes expression either directly by methylation changes at the promoter region of the regulated gene that affect interactions with active regulators, or indirectly by altering expression of regulatory proteins [40].
We have previously examined the role of phasevarions in animal models using other causative agents of OM: it was noted that the modA2 ON state was selected in NTHi in chinchilla [21, 41]. This indicated that a particular expression state of a phase-variable methyltransferase, and the genes they regulate, gives an advantage in this anatomical niche. We investigated M. catarrhalis infection dynamics using a chinchilla infection model and observed differences between the M. catarrhalis modM3 ON and modM3 OFF strain variants despite not detecting any in vitro phenotypic differences between this strain pair. Expression profiles of our aerobically grown modM3 ON more closely resembled the previously reported expression profile of M. catarrhalis isolated from chinchilla nasopharynx [37], including changes in oxidative stress, and denitrification pathways. Given this, it was a surprise that infection with M. catarrhalis strain 195ME modM3 ON resulted in a decreased bacterial load in chinchilla ears compared to modM3 OFF infected animals, as we predicted the modM3 ON mediated phasevarion would provide a survival advantage. This highlights that the nasopharynx and middle ear are distinct compartments presenting different challenges and selective pressures, and that in vivo models reflect the complexity of human carriage and disease.
The chinchilla model of OM has also been used to study polymicrobial interactions between the three major bacterial OM pathogens: M. catarrhalis survival increased when co-infected with H. influenzae compared to infection with M. catarrhalis alone [42]. The impact of switching of expression of the ModM3 phasevarion on survival in polymicrobial OM was investigated by co-infecting chinchillas transbullarly with either M. catarrhalis modM3 ON or modM3 OFF variants in combination with NTHi. Although the observed difference of M. catarrhalis and NTHi CFU retrieved from bullar mucosal membrane homogenates was not statistically significant, it would seem that co-infection with NTHi reduced the level of infection by M. catarrhalis, compared to when the animals were challenged with M. catarrhalis alone. However, co-infection with the M. catarrhalis modM3 ON variant provided an advantage to survival to NTHi during early infection (days + 1 and + 2), while modM3 OFF enhances survival of NTHi at later stages of OM. Phenotypic analysis of bacteria that contain several phase-variable genes, such as M. catarrhalis, can be confounded by phase variation of genes that are not the subject of investigation. It is unlikely that genes other than modM3 have phase varied during isolation of the ModM3 ON and OFF populations, as none of the genes identified as differentially regulated by RNASeq are known to be phase variable. However, the low bacterial recovery from in vivo chinchilla experiments prevented analysis of the expression status of other known phase variable genes (i.e., uspA1, uspA2, and mid/hag) and we cannot definitively say that the phenotypes seen in the animal model were solely due to the ON vs OFF status of ModM3. Furthermore, we were also unable to determine whether there was selection of ModM3 ON or OFF variants during in vivo infection. It is important to note that there are several limitations to the animal model data presented due to the small number of animals tested in addition to the low numbers of bacteria recovered in each experiment. Although the chinchilla model is considered the gold standard for investigating OM [43], M. catarrhalis is a human-host adapted bacteria that does not naturally infect any known animal model. Therefore, investigations in animals are challenging and further experimentation is required to validate these in vivo observations, ideally from samples collected from humans with OM.
Conclusions
In this study we have confirmed that ModM3 is phase variable, identified the ModM3 methylation target sequence, and characterized the ModM3 phasevarion. ModM3 regulates the expression of multiple genes (15 gene upregulated and 14 genes downregulated 1.27–1.61 fold between the ModM3 ON vs OFF states), belonging to pathways important for infection of the human host, including those involved in biofilm formation and in the response to oxidative and nitrosative stress. Our observations that the modM3 OFF state may confer a survival advantage to M. catarrhalis in vivo, and that phase variation of modM3 may alter survival of NTHi at different stages of infection has important implications in OM progression and treatment. The observed modulation of gene expression may facilitate adaptation of M. catarrhalis to changing host microenvironments, potentially promoting persistence of M. catarrhalis in the middle ear and contributing to the ability of M. catarrhalis to cause OM.
Methods
Bacterial strains and growth conditions
M. catarrhalis strains used in this study include American Type Culture Collection (ATCC) 25,239 [23] and CCRI-195ME, an OM isolate recovered from the middle ear of a 16-month-old otitis-prone child, at the Columbus Children’s Research Institute (CCRI), Nationwide Children’s Hospital, Columbus, Ohio [25]. M. catarrhalis strains were routinely cultured on brain heart infusion (BHI) agar (Oxoid, Basingstoke, UK) at 37 °C with 5% CO2 overnight (~ 16 h). For use in all biological assays, cells cultured overnight on BHI agar were re-suspended in 20 mL of BHI broth (Oxoid, Basingstoke, UK) to an optical density at 600 nm (OD600) = 0.05 and grown for an additional 4 h to mid log phase at 37 °C with orbital shaking at 200 rpm.
Construction of the M. catarrhalis modM3 knockout mutant strain
To generate the 195ME modM3-knockout mutant (∆modM3), 195ME was transformed with 1 μg of BsaI linearized plasmid pMcrepI::kan [23] by co-incubation on BHI agar at 37 °C, 5% CO2 for 3 h. Transformants were selected on BHI agar containing 20 μg/mL kanamycin and confirmed by PCR analysis.
Fragment length analysis
The length of the modM 5′-(CAAC)n-3′ tetranucleotide repeat tract in M. catarrhalis 195ME, and the proportion of each fragment length present was determined using GeneScan fragment analysis as per [23]. Briefly, the modM repeat region was amplified with the 6-carboxyfluorescein (6-FAM) labelled forward primer Mcmod2F-6FAM (5′-[6FAM]-TTACTTGACACTCTGAATGGA-3′) and the unlabelled reverse primer McmodrepR2 (5′-GTATTATGGGCAGTTTTTAAG-3′) using GoTaq Flexi DNA polymerase (Promega) in 20 μL reaction volumes as per manufacturer’s instructions. Analysis of fluorescent traces and quantification of fragment lengths was performed using PeakScanner v1.0 (Applied Biosystems, Grand Island, NY, USA). M. catarrhalis subpopulations containing different modM3 repeat tract lengths were selected by fragment length analysis of individual colonies, as previously described [20].
Western blot analysis
M. catarrhalis 195ME modM3 ON, modM3 OFF were grown to mid log phase as described above. Cultures were then centrifuged at 6000 x g for 5 min to pellet, and the pellets were re-suspended in water to an OD600 of 5. Cell lysates were prepared by boiling for 10 min and 20 μg of protein was electrophoresed on a 4–12% Bis-Tris NuPAGE polyacrylamide gel at 180 V for 60 min. Protein was transferred to a nitrocellulose membrane using the XCell II Blot Module at 30 V for 70 min. Western blot analysis was performed using a mouse polyclonal anti-Mod antibody at 1:1000 dilution, followed by a 1:5000 dilution of a rabbit anti-mouse alkaline phosphatase conjugated antibody. Western blots were developed using the AP Conjugate Substrate Kit (Bio-Rad) as per manufacturer’s instructions.
Methylome determination by single molecule real-time (SMRT) sequencing
The 195ME modM3 ON (36 repeats), modM3 OFF (37 repeats), and ∆modM3 strains were grown to mid-log phase as described above. Genomic DNA was extracted using the Qiagen Genomic-tip 20/G kit (Qiagen) and SMRTbell libraries were prepared as described previously [23, 44]. Sequencing was performed on the PacBio RS II (Menlo Park, CA, USA) using standard protocols for short and long insert libraries.
Confirmation of the ModM3 methylation target sequence
Restriction inhibition assay
Genomic DNA was extracted from mid log cultures of 195ME modM3 ON, modM3 OFF, and ∆modM3 strains using GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich). Genomic DNA was digested with the restriction enzyme BtsCI (NEB) at 50 °C or HindIII (NEB) at 37 °C for 60 min. Restriction enzymes were heat inactivated at 80 °C for 20 min. Digested and undigested genomic DNA was electrophoresed on a 0.8% agarose gel at 110 V for 50 min.
Southern blotting
Electrophoresis of BtsCI digested DNA performed as above and was then transferred to a charged nylon membrane by the capillary transfer method [45]. The DNA probe for Southern hybridization was amplified using the primer pair M3ResInh-F (5′- TTTAGCAAAGGGTAACCACC-3′) and M3ResInh-R (5′- ACAATGACACGCTCAACTCG-3′) and labelled with Digoxigenin (DIG) using the DIG-High Prime DNA Labelling and Detection Starter Kit II (Roche) as directed by the manufacturer.
Quantitative real-time PCR
Quantitative real-time PCR was performed using Sybr Green Master Mix (Bio-Rad) in 20 μL reaction volumes with 1 ng of BtsCI digested genomic DNA as template and 250 nmol of each primer. qRT-PCR was performed on the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) with the following cycle conditions: polymerase activation at 98 °C for 2 min, followed by 40 cycles of denaturation at 98 °C for 15 s, and annealing/extension at 60 °C for 30 s. The primers M3Met_RT_F (5′-CAGGTTCGCCATCAACAAAC-3′) and M3Met_RT_R (5′-TGCCTGCCATCGCTGAATC-3′) were designed to amplify a 99 bp amplicon spanning an overlapping BstCI and ModM3 recognition sequence. The primers CopB_RT-F (5′- GTGAGTGCCGCTTTTACAACC-3′) and CopB_RT-R (5′- TGTATCACCTGCCAAGACAA − 3′) were used in control reactions and amplify a 72 bp amplicon in the copB gene that does not contain a BtsCI cleavage site. Relative quantitation of BtsCI digested sites in each sample was performed using the ∆∆Ct method.
RNA sequencing
M. catarrhalis strains 195ME ModM3 ON and OFF were cultured in triplicate on BHI agar at 37 °C with 5% CO2 overnight (16 h). Each strain was then re-suspended in 20 mL BHI broth to an optical density of OD600 = 0.05 and grown for a further 3 h at 37 °C with orbital shaking at 200 rpm. 2 mL of culture was added directly to 4 mL of RNAprotect Bacteria Reagent (Qiagen), and incubated at room temperature for 5 min. Cultures were pelleted by centrifugation at 5000 x g for 10 min at 4 °C and the supernatant was discarded. Pellets were resuspended in 200 μL of lysis buffer (30 mM Tris·Cl, 1 mM EDTA, pH 8.0) containing 15 mg/mL lysozyme and 80 units/mL proteinase K. Total RNA was extracted using RNeasy Mini Kit (Qiagen) and residual DNA digested using RNase-Free DNase Set (Qiagen) as per manufacturer’s instructions. RNASeq was performed by the Australian Genome Research Facility (Melbourne, VIC, Australia). RNA libraries were prepared using the Illumina Ribo-Zero Gold protocol. Pooling and clustering of libraries was performed using the Illumina cBot system with TruSeq PE Cluster kit v3 reagents. Sequencing (150 bp paired-end reads) was performed on the Illumina MiSeq system with TruSeq SBS Kit v3 reagents.
Growth curve analysis
M. catarrhalis overnight plate cultures were re-suspended in 1 mL BHI broth and equalized to an OD600 of 0.05 and 100 μL volumes of each bacterial suspension was transferred to a 96 well plate in triplicate. Automated growth curve analysis was performed using a Tecan Infinite M1000 Pro microplate reader. Growth curves were performed at 37 °C with orbital shaking at an amplitude of 4 mm, and spectroscopy readings were taken at a wavelength of 600 nm at 1 h intervals.
H2O2 killing assay
M. catarrhalis strains were grown to mid log phase as described above. Mid-log phase cultures were diluted in BHI broth to 1 × 106 CFU/mL and 90 μL of each suspension was added to wells of a 96 well plate in triplicate. The killing assay was started by addition of 10 μL H2O2 in BHI broth to a final concentration of either 5 or 10 mM H2O2 and plates were incubated at 37 °C, 5% CO2. Samples were taken at 15, 30, 45 and 60 min, 10 fold serial dilutions were performed in BHI broth containing catalase, and 5 μL of each dilution was plated onto BHI agar and incubated overnight at 37 °C, 5% CO2.
Biofilm formation assay
M. catarrhalis biofilm assays were performed as previously described [46]. Briefly, strains were standardized to OD600 = 1.0 in BHI broth and diluted 1:10 in chemically defined media (final OD600 = 0.1). Biofilm assays were performed in 0.5 mL volumes in 48-well tissue culture plates, and grown without agitation for 24, 48, or 72 h at 37 °C, 5% CO2. Wells were stained with 0.05% crystal violet (Sigma-Aldrich) for 15 mins, washed three times with PBS, and crystal violet was solubilised with 90% ethanol. Quantification was performed at 600 nm using the Tecan Infinite M1000 Pro microplate reader.
Adherence and invasion assays
The A549 human lung epithelial cell line (ATCC, CCL185) was maintained in Minimal Essential Media (MEM) supplemented with 10% Foetal Bovine Serum at 37 °C, 5% CO2. A549 cells were grown to confluent monolayers in 24-well tissue culture plates (1 × 105 cells/well) and infected with 1 × 106 M. catarrhalis 195ME cells. Plates were incubated for 3 h at 37 °C, 5% CO2. For adherence assays, non-adherent bacteria were removed by washing the wells three times with 1 mL PBS. For invasion assays, extracellular bacteria were removed by treating wells with 500 μL of 100 μg/mL gentamycin for 15 min at 37 °C, 5% CO2 and then washing with PBS as above. Cells were detached from the plate by treatment with 200 μL of 0.25% trypsin for 10 min at 37 °C, 5% CO2, and lysed by adding 300 μL of 1% saponin and thorough vortexing. To enumerate adherent and intracellular bacteria, 10 fold serial dilutions of each sample were performed in BHI broth and 5 μL of each dilution was plated onto BHI agar and incubated overnight at 37 °C, 5% CO2.
Chinchilla model of OM
Due to anatomical similarities of the chinchilla (Chinchilla lanigera) and human middle ear, and immunological similarities with children in the response to middle ear infection, the chinchilla model has been extensively used to study prominent otopathogens, including M. catarrhalis, in the pathogenesis of OM [43].
Housing of chinchillas and experimental procedures
Adult, outbred, chinchillas of mixed sex were acquired from Rausher Chinchilla Ranch (LaRue, Ohio) and allowed to acclimate to the vivarium for 7 days prior to the beginning of the study. Animals were housed individually in clear cages with autoclaved corn cobb bedding and access to autoclaved water and certified feed ad libitum. Racks of cages were maintained in negative air flow isolation units with a 12 h light cycle. All animals were examined by a veterinarian upon arrival for general health and were examined twice each day by veterinary staff and laboratory personnel in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines. No deviations were observed in health or behaviour over the course of the study. Cohort sizes for each study were determined based on extensive prior experience with this animal model and in keeping with the principles of Replacement, Refinement and Reduction in animal numbers. Animals were randomly clustered into cohorts such that the mean weight of each cohort was comparable. Animals were examined in numerical order based on their ear tag, not by cohort clustering. Based on American Veterinary Association (AVMA) Guidelines for Euthanasia of Animals, the primary method for euthanasia was cardiac injection of Euthosol (10 mg ketamine-HCl plus 2 mg xylazine per kg body weight) to anesthetized animals followed by decapitation as the secondary physical method for euthanasia.
Transbullar infection of M. catarrhalis
Eight chinchillas were weighed and divided into two cohorts of four animals with equal mean weight (609 +/− 76 g). On Day + 0, cohorts were challenged with either M. catarrhalis 195ME modM3 ON or modM3 OFF variants. Challenge doses consisted of 2800 CFU in 300 μL of saline delivered directly into both the left and right bulla for each animal. Tympanometry was performed on a daily basis to assess progression of otitis media. On day + 1 and day + 2 post challenge, two animals from each cohort were sacrificed, and the bullae were dissected from the skull and aseptically opened and imaged. CFU enumeration was performed by plating dilutions of middle ear effusates and mucosal membrane homogenates on chocolate agar.
Transbullar co-infection of M. catarrhalis and NTHi
Twelve chinchillas were divided into two cohorts of six animals with equal mean weight (663 +/− 21 g). On Day + 0 cohorts were challenged with NTHi strain 86-028NP in combination with either M. catarrhalis 195ME modM3 ON or modM3 OFF variants. Transbullar challenge was performed as previously stated, with challenge doses consisting of 1000 CFU for NTHi and 3000 CFU for M. catarrhalis being co-administered into each bulla. Two animals from each cohort were sacrificed on days + 1, + 2, and + 7 post challenge and the bullae were dissected from the skull and aseptically opened and imaged. In order to discriminate between M. catarrhalis and NTHi and permit enumeration of each bacterial species, dilutions of middle ear effusates and mucosal membrane homogenates were plated on chocolate agar to quantitate NTHi, and chocolate agar with 10 μg/mL vancomycin and 5 μg/mL trimethoprim to select for and quantitate M. catarrhalis, as previously reported [47, 48].
Statistical analysis
Each chinchilla ear was counted as a separate experimental unit. Exact P values were calculated with the Mann-Whitney U test using GraphPad Prism version 7.02.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. All supplementary material is available online at BMC Genomics. The CCRI-195ME sequence is available in GenBank under the accession number CP018059. The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO accession number GSE140417.
Abbreviations
- BHI:
-
Brain Heart Infusion
- CFU:
-
Colony forming units
- COPD:
-
Chronic obstructive pulmonary disease
- NTHi:
-
Non-typeable Haemophilus influenzae
- OM:
-
Otitis media
- qRT-PCR:
-
Quantitative real time PCR
References
Vaneechoutte M, Verschraegen G, Claeys G, Weise B, Van den Abeele AM. Respiratory tract carrier rates of Moraxella (Branhamella) catarrhalis in adults and children and interpretation of the isolation of M. catarrhalis from sputum. J Clin Microbiol. 1990;28(12):2674–80.
Pettigrew MM, Alderson MR, Bakaletz LO, Barenkamp SJ, Hakansson AP, Mason KM, Nokso-Koivisto J, Patel J, Pelton SI, Murphy TF. Panel 6: Vaccines. Otolaryngol Head Neck Surg. 2017;156(4_suppl):S76–87.
Murphy TF, Brauer AL, Grant BJ, Sethi S. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am J Respir Crit Care Med. 2005;172(2):195–9.
Ruohola A, Pettigrew MM, Lindholm L, Jalava J, Raisanen KS, Vainionpaa R, Waris M, Tahtinen PA, Laine MK, Lahti E, et al. Bacterial and viral interactions within the nasopharynx contribute to the risk of acute otitis media. J Infect. 2013;66(3):247–54.
Krishnamurthy A, McGrath J, Cripps AW, Kyd JM. The incidence of Streptococcus pneumoniae otitis media is affected by the polymicrobial environment particularly Moraxella catarrhalis in a mouse nasal colonisation model. Microbes Infect. 2009;11(5):545–53.
Perez AC, Pang B, King LB, Tan L, Murrah KA, Reimche JL, Wren JT, Richardson SH, Ghandi U, Swords WE. Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog Dis. 2014;70(3):280–8.
Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet. 2006;40:307–33.
van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17(3):581–611 table of contents.
Tan A, Atack JM, Jennings MP, Seib KL. The capricious nature of bacterial pathogens: Phasevarions and vaccine development. Front Immunol. 2016;7:586.
Lafontaine ER, Wagner NJ, Hansen EJ. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J Bacteriol. 2001;183(5):1540–51.
Attia AS, Hansen EJ. A conserved tetranucleotide repeat is necessary for wild-type expression of the Moraxella catarrhalis UspA2 protein. J Bacteriol. 2006;188(22):7840–52.
Wang W, Pearson MM, Attia AS, Blick RJ, Hansen EJ. A UspA2H-negative variant of Moraxella catarrhalis strain O46E has a deletion in a homopolymeric nucleotide repeat common to uspA2H genes. Infect Immun. 2007;75(4):2035–45.
Mollenkvist A, Nordstrom T, Hallden C, Christensen JJ, Forsgren A, Riesbeck K. The Moraxella catarrhalis immunoglobulin D-binding protein MID has conserved sequences and is regulated by a mechanism corresponding to phase variation. J Bacteriol. 2003;185(7):2285–95.
Blakeway LV, Tan A, Peak IRA, Seib KL. Virulence determinants of Moraxella catarrhalis: distribution and considerations for vaccine development. Microbiology (Reading, England). 2017;163(10):1371–84.
Atack JM, Tan A, Bakaletz LO, Jennings MP, Seib KL. Phasevarions of bacterial pathogens: Methylomics sheds new light on old enemies. Trends Microbiol. 2018;26(8):715–26.
Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci U S A. 2005;102(15):5547–51.
Srikhanta YN, Dowideit SJ, Edwards JL, Falsetta ML, Wu HJ, Harrison OB, Fox KL, Seib KL, Maguire TL, Wang AH, et al. Phasevarions mediate random switching of gene expression in pathogenic Neisseria. PLoS Pathog. 2009;5(4):e1000400.
Srikhanta YN, Gorrell RJ, Steen JA, Gawthorne JA, Kwok T, Grimmond SM, Robins-Browne RM, Jennings MP. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One. 2011;6(12):e27569.
Jen FE, Seib KL, Jennings MP. Phasevarions mediate epigenetic regulation of antimicrobial susceptibility in Neisseria meningitidis. Antimicrob Agents Chemother. 2014;58(7):4219–21.
Seib KL, Pigozzi E, Muzzi A, Gawthorne JA, Delany I, Jennings MP, Rappuoli R. A novel epigenetic regulator associated with the hypervirulent Neisseria meningitidis clonal complex 41/44. FASEB J. 2011;25(10):3622–33.
Atack JM, Srikhanta YN, Fox KL, Jurcisek JA, Brockman KL, Clark TA, Boitano M, Power PM, Jen FE, McEwan AG, et al. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat Commun. 2015;6:7828.
Blakeway LV, Tan A, Lappan R, Ariff A, Pickering JL, Peacock CS, Blyth CC, Kahler CM, Chang BJ, Lehmann D, et al. Moraxella catarrhalis restriction-modification systems are associated with phylogenetic lineage and disease. Genome Biol Evol. 2018;10(11):2932–46.
Blakeway LV, Power PM, Jen FE, Worboys SR, Boitano M, Clark TA, Korlach J, Bakaletz LO, Jennings MP, Peak IR, et al. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J. 2014;28(12):5197–207.
Seib KL, Peak IR, Jennings MP. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol Med Microbiol. 2002;32(2):159–65.
Tan A, Blakeway LV, Bakaletz LO, Boitano M, Clark TA, Korlach J, Jennings MP, Peak IR, Seib KL. Complete Genome Sequence of Moraxella catarrhalis Strain CCRI-195ME, Isolated from the Middle Ear. Genome Announc. 2017;5(21):e00384-17.
Novotny LA, Mason KM, Bakaletz LO. Development of a chinchilla model to allow direct, continuous, biophotonic imaging of bioluminescent nontypeable Haemophilus influenzae during experimental otitis media. Infect Immun. 2005;73(1):609–11.
Seib KL, Jen FE, Tan A, Scott AL, Kumar R, Power PM, Chen LT, Wu HJ, Wang AH, Hill DM, et al. Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N (6)-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res. 2015;43(8):4150–62.
Wang W, Richardson AR, Martens-Habbena W, Stahl DA, Fang FC, Hansen EJ. Identification of a repressor of a truncated denitrification pathway in Moraxella catarrhalis. J Bacteriol. 2008;190(23):7762–72.
Wang W, Kinkel T, Martens-Habbena W, Stahl DA, Fang FC, Hansen EJ. The Moraxella catarrhalis nitric oxide reductase is essential for nitric oxide detoxification. J Bacteriol. 2011;193(11):2804–13.
Stevanin TM, Moir JW, Read RC. Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infect Immun. 2005;73(6):3322–9.
Wang W, Reitzer L, Rasko DA, Pearson MM, Blick RJ, Laurence C, Hansen EJ. Metabolic analysis of Moraxella catarrhalis and the effect of selected in vitro growth conditions on global gene expression. Infect Immun. 2007;75(10):4959–71.
Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. Jama. 2006;296(2):202–11.
Stewart V. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol Microbiol. 1993;9(3):425–34.
Schreiber K, Krieger R, Benkert B, Eschbach M, Arai H, Schobert M, Jahn D. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J Bacteriol. 2007;189(11):4310–4.
Okubo J, Noshiro M. Analysis of middle ear cavity gas composition by mass spectrometry. Nihon Jibiinkoka Gakkai kaiho. 1994;97(7):1181–90.
Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother. 2004;48(7):2659–64.
Hoopman TC, Liu W, Joslin SN, Pybus C, Sedillo JL, Labandeira-Rey M, Laurence CA, Wang W, Richardson JA, Bakaletz LO, et al. Use of the chinchilla model for nasopharyngeal colonization to study gene expression by Moraxella catarrhalis. Infect Immun. 2012;80(3):982–95.
Hoopman TC, Liu W, Joslin SN, Pybus C, Brautigam CA, Hansen EJ. Identification of gene products involved in the oxidative stress response of Moraxella catarrhalis. Infect Immun. 2011;79(2):745–55.
Casadesus J, Low DA. Programmed heterogeneity: epigenetic mechanisms in bacteria. J Biol Chem. 2013;288(20):13929–35.
Phillips ZN, Husna AU, Jennings MP, Seib KL, Atack JM. Phasevarions of bacterial pathogens - phase-variable epigenetic regulators evolving from restriction-modification systems. Microbiology (Reading, England). 2019;165(9):917–28.
Brockman KL, Jurcisek JA, Atack JM, Srikhanta YN, Jennings MP, Bakaletz LO. ModA2 phasevarion switching in nontypeable Haemophilus influenzae increases the severity of experimental otitis media. J Infect Dis. 2016;214(5):817–24.
Armbruster CE, Hong W, Pang B, Weimer KE, Juneau RA, Turner J, Swords WE. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio. 2010;1(3):e00102-10.
Bakaletz LO. Chinchilla as a robust, reproducible and polymicrobial model of otitis media and its prevention. Expert Rev Vaccines. 2009;8(8):1063–82.
Clark TA, Murray IA, Morgan RD, Kislyuk AO, Spittle KE, Boitano M, Fomenkov A, Roberts RJ, Korlach J. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 2012;40(4):e29.
Southern E. Southern blotting. Nat Protoc. 2006;1(2):518–25.
Tan A, Li WS, Verderosa AD, Blakeway LV, Tsitsi DM, Totsika M, Seib KL. Moraxella catarrhalis NucM is an entry nuclease involved in extracellular DNA and RNA degradation, cell competence and biofilm scaffolding. Sci Rep. 2019;9(1):2579.
Brockson ME, Novotny LA, Jurcisek JA, McGillivary G, Bowers MR, Bakaletz LO. Respiratory syncytial virus promotes Moraxella catarrhalis-induced ascending experimental otitis media. PLoS One. 2012;7(6):e40088.
Mokrzan EM, Novotny LA, Brockman KL, Bakaletz LO. Antibodies against the Majority Subunit (PilA) of the Type IV Pilus of Nontypeable Haemophilus influenzae Disperse Moraxella catarrhalis from a Dual-Species Biofilm. mBio. 2018;9(6):e02423-18.
Acknowledgements
The authors wish to thank Mathew Boitano, Tyson A. Clark, and Jonas Korlach for their continued support with SMRT sequencing and methylome analysis. The authors wish to thank Laura A. Novotny for her assistance with the chinchilla model of OM.
Funding
This work was supported by the Australian National Health and Medical Research Council (NHMRC) [Career Development Fellowship to KLS; Project Grant 1099279 to KLS, JMA and LOB], the Garnett Passe and Rodney Williams Memorial Foundation [Fellowship to AT and Grant in Aid Supplementation to KLS and JMA], and an Australian Government Research Training Program Scholarship to LVB. The funders had no role in study design, data collection and analysis, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
The study was conceived and co-ordinated by KLS and IRP. Significant contributions to experimental design were made by LVB, AT, JMA. All experimentation and statistical analysis was performed by LVB and AT, with the exception of in vivo chinchilla experiments which were designed and performed by JAJ and LOB. All authors contributed to writing and have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Written consent for the acquisition and use of chinchillas was obtained from Rausher Chinchilla Ranch (LaRue, Ohio). Animal care and all procedures were performed under a protocol approved by the Nationwide Children’s Hospital Research Institute Animal Care and Use Committee (Protocol #01304AR) and in compliance with the U.S. Department of Health and Human Services Guide for the Care and Use of Laboratory Animals. Strain CCRI-195ME was recovered from a patient undergoing surgery for tympanostomy tube insertion, after obtaining written Informed Consent from patient’s guardian. Consent forms and study protocols were approved by the Nationwide Children’s Hospital Institutional Review Board (protocol #0312HS196).
Consent for publication
Not applicable.
Competing interests
KLS is a member of the editorial board (Associate Editor) of BMC Microbiology. The authors declare that they have no other competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
In vitro phenotypic assays of M. catarrhalis 195ME modM3 ON and modM3 OFF variants. a) Automated microtiter plate growth curve analysis of optical density at 600 nm (OD600) over 10 h; b) hydrogen peroxide (H2O2) killing assay showing colony forming units/ml (CFU/ml) after exposure to 0, 5 or 10 mM (H2O2) over 60 min; c) biofilm formation assay with biofilm biomass quantified after 24, 48 or 72 h by analysis of OD600 after crystal violet staining; d) adherence and invasion of A549 human lung epithelial cells assay measures as CFU/ml. For all assays, the mean of three replicates is shown and error bars indicate +/− 1 standard deviation of the mean. Figure S2. Tympanometry and imaging of bullae in the chinchilla model of otitis media. a) Mean middle ear pressure measured as decapascals (daPa); and b) mean tympanic membrane compliance measured as middle ear volume (mL) in two cohorts of chinchillas challenged with either M. catarrhalis modM3 ON or modM3 OFF populations. The mean of four ears is shown and error bars indicate +/− 1 standard deviation of the mean.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Blakeway, L.V., Tan, A., Jurcisek, J.A. et al. The Moraxella catarrhalis phase-variable DNA methyltransferase ModM3 is an epigenetic regulator that affects bacterial survival in an in vivo model of otitis media. BMC Microbiol 19, 276 (2019). https://doi.org/10.1186/s12866-019-1660-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-019-1660-y