Abstract
Background
To screen whether E. coli strains encoding type 1 fimbriae, isolated from fecal microflora, produce bacteriocins more often relative to fimA-negative E. coli strains of similar origin.
Methods
PCR assays were used to detect presence of genes encoding 30 bacteriocin determinants (23 colicin- and 7 microcin-encoding genes) and 18 virulence determinants in 579 E. coli strains of human and animal origin isolated from hospitals and animal facilities in the Czech and Slovak Republic. E. coli strains were also classified into phylogroups (A, B1, B2 and D).
Results
fimA-negative E. coli strains (defined as those possessing none of the 18 tested virulence determinants) were compared to fimA-positive E. coli strains (possessing fimA as the only detected virulence determinant). Strains with identified bacteriocin genes were more commonly found among fimA-positive E. coli strains (35.6 %) compared to fimA-negative E. coli strains (21.9 %, p < 0.01) and this was true for both colicin and microcin determinants (p = 0.02 and p < 0.01, respectively). In addition, an increased number of strains encoding colicin E1 were found among fimA-positive E. coli strains (p < 0.01).
Conclusions
fimA-positive E. coli strains produced bacteriocins (colicins and microcins) more often compared to fimA-negative strains of similar origin. Since type 1 fimbriae of E. coli have been shown to mediate adhesion to epithelial host cells and help colonize the intestines, bacteriocin synthesis appears to be an additional feature of colonizing E. coli strains.
Similar content being viewed by others
Background
Escherichia coli (E. coli) is a common, variable, aerobic bacterial species that inhabits the gut of vertebrates [1]. Strains of E. coli differ in a number of important characteristics including genome size [2], gene content and virulence [3]. E. coli strains are classified into four phylogroups; E. coli strains of phylogroups A and B1 contain smaller genomes and are frequently non-pathogenic, while strains of phylogroups B2 and D encode more genes and are more often pathogenic [2, 4–6].
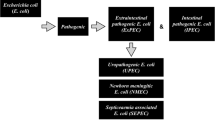
Flow chart of the E. coli isolates investigated in this study. *Fecal E. coli strains used in this study (n = 579) either tested negative for all 18 virulence determinants (pCVD432, α-hly, afaI, aer, cnf1, sfa, pap, ial, lt, st, bfpA, eaeA, ipaH, iucC, fimA, stx1, stx2 and ehly) and were, therefore, considered fimA-negative, or the strains were fimA-positive, while still testing negative for all other virulence determinants
Approximately 80 % of all E. coli strains of fecal origin are able to produce type 1 fimbriae, which are encoded by the chromosomal fim operon [7]. This operon consists of fimAICDFGH genes [8], where fimA encodes the major fimbrial subunit (FimA). FimA is arranged in a helical manner along with minor components, one of which includes FimH (an adhesin that mediates attachment of type 1 fimbriae to mannose-containing receptors) [9]. E. coli type 1 fimbriae have been shown to mediate adhesion to a number of host cell types (e.g. epithelial and endothelial cells), thus potentially increasing virulence of fimA-positive E. coli strains [10].
E. coli strains are able to synthesize two types of bacteriocins - colicins and microcins. While microcins are low molecular weight oligopeptides, colicins are proteins with molecular weights between 30–70 kDa. Colicins and microcins differ in a number of additional parameters including operon organization, regulation of gene expression, export from producer cells, presence of post-translational modifications, antimicrobial activity, etc. [11–13]. However, this classification is not strict, since colicin Js [14, 15] is known to share features of both bacteriocin types. Several bacteriocins (i.e. colicins E1, Ia and S4, and microcins B17, E492, H47, I47, M and V) have been found to be associated with virulence factors (i.e. with aer, cnf1, fyuA, hlyA, iroCDN, iucC, papCG, sfa, tcpC and usp determinants) in E. coli strains [16–20].
Previous studies have found associations between several genes encoding bacteriocin types and several virulence determinants, however, no association between type 1 fimbriae and bacteriocin genes has been identified [16–20]. The role of fimbriae type 1 in the E. coli virulence is not clear [21, 22]. This situation is likely a result of very frequent presence of type 1 fimbriae among E. coli strains (over 80 %) and the fact that in most E. coli strains the type 1 fimbriae are combined with other virulence determinants. Therefore we collected a set of fecal E. coli strains encoding type 1 fimbriae as the only detected virulence determinant (out of 18 tested) and a set of E. coli strains with no virulence determinants (out of 18 tested).
In this communication, we studied prevalence of bacteriocin production and prevalence of bacteriocin types in both sets of E. coli strains to assess association of fimbriae type I encoding genes and bacteriocin determinants in E. coli strains. In addition, we also tested association between type 1 fimbriae determinants and other factors including biochemical profiles and E. coli phylogroups.
Results
Characteristics of E. coli strains
Fecal E. coli strains used in this study (n = 579) (Fig. 1) either tested negative for all 18 virulence determinants (pCVD432, α-hly, afaI, aer, cnf1, sfa, pap, ial, lt, st, bfpA, eaeA, ipaH, iucC, fimA, stx1, stx2 and ehly) and were, therefore, considered fimA-negative, or the strains were fimA-positive, while still testing negative for all other virulence determinants. To assess presence of other genes of the fimA cluster, a fimH determinant, encoding adhesin mediating attachment of type 1 fimbriae, was tested on all fimA-positive strains. Except of 10 isolates, fimA-positive isolates were also positive for fimH determinants (98.3 % showed both determinants for type 1 fimbriae). Within groups of strains with different origins (Table 1), a relatively small proportion (≈17 %) of E. coli strains had no detected virulence determinants. Therefore, all strains were analyzed without regard for their origin.
Biochemical analysis
E. coli isolates (n = 579) were positive for the following biochemical reactions: TRE (99.1 %), MAN (99.0 %), SOR (97.8 %), ONP (97.6 %) IND (97.4 %), LYS (93.6 %), SUC (51.8 %), ORN (50.4 %), ESL (14.2 %), ADO (8.8 %), H2S (3.6 %), CEL (2.2 %), SCI (2.1 %), MAL (1.0 %), URE (0.7 %), INO (0.7 %) and PHE (0.2 %). The utilization of substrate showed variable results in ORN and SUC reactions between fimA-positive and fimA-negative E. coli strains. Biochemical analysis showed that the production of ornithine decarboxylase was significantly higher among fimA-positive E. coli strains (n = 254; 52.6 %) compared to fimA-negative strains (n = 38; 39.6 %, p = 0.03). In addition, production of succinate dehydrogenase was significantly higher among fimA-positive E. coli strains (n = 269; 55.7 %) compared to fimA-negative E. coli strains (n = 31; 32.3 %, p < 0.01).
Detection of phylogenetic groups in E. coli strains
Phylogenetic analysis of 579 E. coli strains revealed that fimA-negative E. coli strains contained a significantly higher prevalence of phylogenetic group A (66.7 %) compared to fimA-positive E. coli strains (39.1 %; p < 0.01). Phylogroups B1 and B2 were found less frequently among fimA-negative E. coli strains (p = 0.01 and p = 0.04, respectively), while the prevalence of phylogroup D was similar in both groups of E. coli strains (Table 2). Frequency of bacteriocin production in E. coli strains belonging to phylogenetic groups A, B1, B2 and D, respectively, was not significantly different between fimA-positive and fimA-negative E. coli strains (Additional file 1: Table S1).
Detection of bacteriocin-encoding determinants
Genetic determinants encoding 30 bacteriocin types including 23 colicins (A, B, D, E1, E2-9, Ia, Ib, Js, K, L, M, N, S4, U, Y and 5/10) and 7 microcins (H47, M, B17, C7, J25, L and V) were tested in all 579 E. coli strains used in this study. Strains with identified bacteriocin genes were more frequently found among fimA-positive E. coli strains (35.6 %) compared to fimA-negative E. coli strains (21.9 %, p < 0.01). Altogether, 3 microcin types and 8 colicin types were identified among fimA-negative E. coli strains while all 7 tested microcin types and 14 of the colicin types were found among fimA-positive E. coli strains (Tables 2 and 3). All of the identified bacteriocin determinants found among fimA-negative E. coli strains were also found among the fimA-positive E. coli strains.
A higher number of microcin determinants was found among fimA-positive E. coli strains (22.4 %) compared to fimA-negative E. coli strains (9.4 %; p < 0.01). Similarly, a higher number of detected colicin determinants was also found among fimA-positive E. coli strains (38.7 %) compared to fimA-negative E. coli strains (26.0 %; p = 0.02). In addition, an increased number of strains encoding colicin E1 was found among fimA-positive E. coli strains (8.1 %) compared to fimA-negative E. coli strains (1.0 %; p < 0.01).
In silico analysis of E. coli genomes
A set of 1951 publicly available E. coli genomes including 121 completed genomes in the NCBI database was analyzed for the presence of virulence determinants tested in our study. Out of 1951 genomes, 490 genomes (25 %) met the criteria of our study (i.e. absence of tested virulence determinants or the sole presence of fimA determinant). In this set, 286 (58 %) genomes contained type 1 fimbriae as the only detected virulence determinant (fimA-positive E. coli strains) and 204 (42 %) genomes did not contain any of the 18 tested virulence factors (fimA-negative E. coli strains). No significant difference was found in the prevalence of bacteriocin determinants in the group of fimA-positive E. coli strains (15.4 %) compared to fimA-negative E. coli strains (14.7 %).
In addition, the subset of 121 complete genomes was analyzed. Out of them, 64 genomes were suitable for our study (i.e. contained no tested virulence determinants or contained only fimA determinant). While 50 (78 %) genomes belonged to fimA-positive group, 14 (22 %) genomes were fimA-negative. Bacteriocin genes were detected in only three fimA-positive genomes.
Discussion
The fimA-negative E. coli, as well as fimbriae type I-possessing E. coli strains, used in this study were found to have similar frequencies in both humans and animals. A relatively small proportion (≈17 %) of E. coli strains had no detected virulence factors. These results are in accordance with other published data where 83 – 100 % of fimA-positive E. coli strains were found [7, 23]. Detection of both fimA and fimH determinants in majority of strains suggests that the complete fimA cluster is present in most of the tested strains.
In silico analysis of 121 complete genomes identified fimA determinant in 78 % of E. coli complete genomes. This finding is in accordance to previous studies [7, 23] and also with the experimental results of this work, where fimA virulence determinant was identified in more than 80 % of isolates. On the other hand, the fimA determinant was identified only in 25 % of 1830 draft genomes suggesting that the fimA determinant likely remained unsequenced in a number of draft genomes. Similar underrepresentation was found also for bacteriocin determinants (24 % and 38 % of all complete and draft genomes contained bacteriocin determinants, respectively; data not shown). This is in contrast to experimentally determined prevalence of bacteriocinogeny among human E. coli isolates where over 50 % of E. coli strains produced bacteriocins [17, 24]. These findings suggest that in silico analysis of draft genomes is of limited value in this and similar studies.
To assess clonal character of isolates, the obtained data from biochemical screening and analysis of phylogenetic groups and bacteriocin determinants were analyzed using Paup* 4 (Phylogenetic Using Analysis Parsimony). Using this approach, 52 and 226 individual strain types (data not shown) were identified in the groups of 96 and 483 fimA-negative E. coli isolates and fimbriae type I-possessing E. coli isolates, respectively, indicating that E. coli isolates in this study were not predominantly of clonal character.
E. coli type 1 fimbriae mediate adhesion to a number of host cell types including epithelial, endothelial and lymphoid cells [10, 25, 26], where they recognize mannose-containing glycoproteins and activate epithelial cells via Toll-like receptor 4 [27]. Type 1 fimbriae are expressed by both uropathogenic and fecal E. coli strains. In murine models, type 1 fimbriae have been shown to be important in the persistence of E. coli urine infections and deletion of the fim gene cluster from the virulent E. coli strain O1:K1:H7 has been shown to decrease the virulence of this strain in the urinary tract infection model [22, 28]. However, several other studies have demonstrated that the presence of the fim gene cluster was not correlated with uropathogenicity in humans [21, 29–33]. Additionally, E. coli strain A0 34/86 (O83:K24:H31), which has been approved as live oral vaccine preparation for infants in the Czech and Slovak Republic, was shown to possess type 1 fimbriae [34, 35]. Another widely used probiotic strain, E. coli Nissle 1917, is known to possess type 1 fimbriae as well as other adhesins (e.g. F1C fimbriae) [36, 37]. These examples demonstrate that type 1 fimbriae are primarily important for attachment to eukaryotic cells and could be, in certain strains, important also with regard to virulence.
While phylogroup A (and also B1) consists mostly of commensal strains, phylogroup B2 (and also D) consists mainly of extraintestinal pathogenic E. coli strains [4–6, 38]. The majority of tested strains in this study (61 %) belonged to A and B1 phylogenetic groups, which was the direct result of sampling E. coli strains without a specific set of virulence factors or E. coli strains harboring fimA determinant as the only detected virulence gene. These findings are in agreement with the observation that non-pathogenic E. coli strains are usually in phylogroup A and B1 [1, 39]. Interestingly, our set of strains also contained strains from group B2 (17 %). Moreover, no virulence factors were detected in of the 9 strains from this phylogroup, which indicates that the relationship between E. coli phylogroup and the presence of virulence factors is not exclusive.
Since prevalence of phylogroups among fimA-negative strains differed from fimA-positive E. coli strains, it is possible that the observed differences in the prevalence of bacteriocin determinants between both groups of E. coli strains were in fact due to differences in the E. coli phylogroups. However, there is no clear association between bacteriocinogeny and E. coli phylogroups. Gordon and O'Brien (2006) detected 4 phylogenetic groups and 19 bacteriocin types in a set of 266 fecal E. coli strains and did not find significant differences in the frequency of bacteriocinogeny in different E. coli phylogroups [40]. In our previous study, we have found that prevalence of colicinogenic strains was higher in phylogroups A and D compared to phylogroups B2 [17]. In contrast, the study of Budič et al. (2011) revealed increased bacteriocinogeny in the phylogroup B2 among 105 uropathogenic strains [18]. In this study, no differences in the prevalence of bacteriocinogeny were found among fimA-negative E. coli, while an increased bacteriocinogeny was found in the phylogenetic group B2 compared to phylogenetic group A in the set of fimA-positive E. coli strains. Although more frequent phylogenetic group B2 could be the reason of increased prevalence of bacteriocinogeny among fimA-positive E. coli strains, increased prevalence of bacteriocin genes were found in all tested E. coli phylogroups (statistically not significant; Additional file 1: Table S1), suggesting the association between bacteriocinogeny and the fimA gene cluster.
In humans, two types of commensal E. coli strains (resident and transient) are known to exist. They differ in their ability to persist in the human intestine. While resident strains are present in the intestines of an individual for months at a time, transient strains only persist for days to weeks [41–43]. In addition, it has been shown that the ability of E. coli strains to persist in the human intestines is associated with several virulence factors, especially various fimbriae [44, 45]. Since the E. coli strains in phylogroup B2 are typical for resident flora [39, 46, 47] and E. coli of phylogroup A is typical for transient strains [39] the fimA-negative E. coli isolates in this study appear to be more frequently transient strains.
The fimA-positive E. coli strains were more often positive for activity of ornithine decarboxylase compared to fimA-negative E. coli. Activity of ornithine decarboxylase, which results in production of polyamines (e.g. putrescine), helps to cope with stress conditions, such as oxidative radicals [48] and low pH [49]. In addition, polyamines play an important role in biofilm formation [50]. There is a relationship between cellular adherence and biofilm formation in certain strains of E. coli [51].
This study has shown that fecal fimA-positive E. coli strains produced bacteriocins more often compared to similar, but fimA-negative, strains. Bacteriocin synthesis appears to be important in microbial communities because of its potential invasive and defensive roles [52]. Moreover, antimicrobial effect of individual bacteriocin types showed differences with respect to their activity on E. coli strains [18]. In previous studies, the occurrence of several bacteriocin genes was found to be associated with several genes encoding virulence factors [16–20] and the results of this study extends the original findings. Bacteriocin types and their sequences have been shown to be host population-specific [53], indicating that bacteriocin-encoding determinants mainly spread among and within hosts. Since virulence genes likely evolved and are being maintained to improve inter-host persistence of commensal bacteria [54, 55], bacteriocin synthesis may further promote stable colonization of the gut. Similar findings were published by Gillor et al. 2009 [56], in which they reported that bacteriocinogeny plays a significant role in the colonization of E. coli in the intestinal tract. As with type 1 fimbriae, which were shown to increase virulence in the urinary tract infection model [26, 27], synthesis of colicin E1 was found to be associated with uropathogenic strains [17].
Conclusions
In summary, fimA-positive E. coli strains of human and animal origin were found most often to be in phylogroup B2; additionally, fimA-positive E. coli strains tested positive for ornithine decarboxylase, succinate dehydrogenase and bacteriocin synthesis more frequently than fimA-negative E. coli strains. All these findings are consistent with increased adherence to intestinal epithelium, increased bacterial virulence, and increased ability to survive in the intestine.
Methods
Bacterial strains
The origins of E. coli strains used in this study are shown in Table 1. E. coli strains were collected between 2007 and 2012 from intestinal microflora of patients at two University Hospitals in Brno (n = 399) and one University Teaching Hospital in Hradec Králové (n = 44), Czech Republic. Strains were collected from feces of patients without bacterial gut infection. The patients were admitted for a number of concerns including infectious and parasitic diseases (n = 165); neoplasms (n = 60); blood diseases (n = 2); endocrine, nutritional and metabolic diseases (n = 42); mental and behavioral disorders (n = 4); diseases of the nervous system (n = 5); diseases of the circulatory system (n = 7); diseases of the respiratory system (n = 4); diseases of the digestive system (n = 73); diseases of the skin and subcutaneous tissue (n = 5); diseases of the musculoskeletal system and connective tissue (n = 3); diseases of the genitourinary system (n = 6); symptoms, signs and abnormal clinical and laboratory findings, not elsewhere classified (n = 30); injury, poisoning and certain other consequences of external causes (n = 5); and factors influencing health status and contact with health services (n = 32). An International Statistical Classification of Diseases and Related Health Problems, 10th Revision (ICD-10)-2015-WHO Version for 2015, was used for the classification of diseases. In addition, fecal E. coli strains of animal origin were isolated from pigs (n = 100) and non-human primates (NHP) (n = 36). The animal isolates were included into the study because of their availability and because there was no statistically significant difference in the ratio of fimA-negative to fimA-positive strains between human and animal E. coli strains. E. coli strains from pigs were isolated during 2010–2012 in Hradec Králové [57, 58]. E. coli strains isolated from NHP feces were collected in 2012 from 7 zoological gardens in the Czech and Slovak Republic (Zoological Garden Hodonín (48°51′52.06″N, 17°6′24.52″E), Zoological Garden Jihlava (49°23'51.834"N, 15°35'57.872"E), Zoological Garden Košice (48°47'00.8"N, 21°12'13.6"E), Zoological Garden Liberec (50°46′34.038″N, 15°4′32.655″E), Zoological and Botanical Garden Plzeň (49°45'27.85"N, 13°21'35.90″E), The Prague Zoological Garden (50°7'0.099"N, 14°24'39.676"E) and Zoological Garden Zlín - Lešná (49°16'20.048"N, 17°42'54.118"E)). From each patient or animal, a single E. coli strain was isolated using selective diagnostic ENDO agar. Metabolic profiles of isolates were obtained during determination of E. coli among isolates using commercial screening kit EnteroTest 16 (test for the presence of several metabolic reactions (H2S, LYS, IND, ORN, URE, PHE, ESL, SCI, MAL, INO, ADO, CEL, SUC, SOR, TRE and MAN) (Lachema, Brno, CZ) and ONP test for detection of β-galactosidase (Lachema, Brno, CZ). The obtained metabolic profiles were compared with the database (TNW ProAuto 7 software) for classification of isolates.
All human data used in the study were anonymized and the study was approved by the Joint Ethical Committee (Charles University in Praha, Faculty of Medicine at Hradec Králové & University Teaching Hospital Hradec Králové) and the ethics committee of the Faculty of Medicine, Masaryk University, Czech Republic. All clinical samples were collected after patients gave written informed consent for participation in the study and for their samples to be used for research. For children under the age of 18, consent was obtained from parents. The animal part of the study (i.e. E. coli strains isolated from pigs) was approved by the Institutional Review Board of the Animal Care Committee of the Institute of Experimental Biopharmaceutics, Academy of Sciences of the Czech Republic, Record Number 1492006. NHP fecal samples were collected after presentation of a preliminary research plan that specified the agreement between particular ZOO zoologists or veterinarians. We obtained all the required permits needed to collect the samples, which were collected during routine cages cleaning, without direct contact or interaction with animals.
Detection of virulence determinants
E. coli strains were tested for the presence of 18 virulence determinants (α-hly, afaI, aer, cnf1, sfa, pap, pCVD432, ial, lt, st, bfpA, eaeA, ipaH, iucC, fimA, stx1, stx2 and ehly). Primer pair sequences and PCR product lengths are shown in Additional file 2: Table S2; the PCR protocols were previously described [59–67]. Positive controls for detection of virulence genes were taken from the laboratory stock and comprised following strains: E. coli B2917 (pCVD432), E. coli B3428 (α-hly), E. coli B3406 (afaI), E. coli B3427 (aer), E. coli B3410 (cnf1), E. coli B3418 (sfa), E. coli B3406 (pap), E. coli B3430 (ial), E. coli B2541 (st), E. coli B2802 (lt), E. coli B1804 (bfpA), E. coli B2905 (eaeA), E. coli B2987 (ipaH), E. coli B3411 (iucC), E. coli B3404 (aer), E. coli B3423 (fimA) and E. coli B2871 (ehly). To assess presence of other genes of the fimA cluster, a fimH determinant, encoding adhesin mediating attachment of type 1 fimbriae, was tested on a set of fimA-positive strains. E. coli B3423 strain was used as a positive control for fimH gene. E. coli strains with none of the 18 tested virulence determinants were used as control strains (i.e. fimA-negative E. coli strains), while the experimental strains consisted of E. coli strains encoding only fimbriae type I (i.e. fimA-positive E. coli strains).
Detection of bacteriocinogeny and bacteriocin determinants
E. coli strains were cultivated (37 °C for 48 h) in parallel on (i) TY agar and (ii) nutrient broth agar plates. The TY agar consisted of yeast extract (Hi-Media, Mumbai, India) 5 gl−1, tryptone (Hi-Media) 8 gl−1, sodium chloride 5 gl−1, and a 1.5 % (w/v) of agar (Hi-Media). Nutrient broth agar contained a Nutrient Agar (HiMedia) 28 gl−1. The bacteria were then killed using chloroform vapors and each plate was then overlaid with a thin layer of soft TY agar (0.7 %; w/v) containing 107 cells ml−1 of an indicator strain. The plates were then incubated at 37 °C overnight and bacteriocin producers were identified [17, 20]. Indicator strains E. coli K12-Row, C6 (ϕ), B1, P400, and Shigella sonnei 17 and E. coli S40 were used to detect bacteriocin production [17, 20]. The set of these strains is capable to detect all known colicin types and most of the microcin types.
Altogether, 30 bacteriocin types were detected among tested strains (23 colicin and 7 microcin genes) using methods previously described [17, 20, 40]. Isolated DNA (using DNAzol reagent, Invitrogen, Carlsbad, CA, according to the manufacturer's protocol) was diluted 100-fold in sterile distilled water. Alternatively, one bacterial colony of each E. coli strain was resuspended in 100 μl of sterile distilled water and 1 μl of this suspension was added to the PCR mix. A list of primers is shown in Additional file 2: Table S2. Cycling conditions were 94 °C (2 min); 94 °C (30 s), 60 °C (30 s), 72 °C (1 min), 30 cycles; and 72 °C (7 min). For colony PCR, the initial step was set for 5 min. For identification of bacteriocin determinants among tested strains, known bacteriocin producers were used as positive controls: E. coli BZB2101pColA - CA31, BZB2102 pColB - K260, BZB2103 pColD - CA23, BZB2107 pColE4 - CT9, BZB2108 pColE5 - 099, BZB2150 pColE6 - CT14, BZB2120 pColE7 - K317, BZB2279 pColIa - CA53, BZB2202 ColIb - P9, BZB2116 pColK - K235, PAP1 pColM - BZBNC22, BZB2123 pColN - 284 (original source: A. P. Pugsley), E. coli 189BM pColE2 - P9 (B. A. D. Stocker), E. coli 385/80 pColE1, pColV (H. Lhotová), E. coli 185 M4 pColE3 - CA38 (P. Fredericq), E. coli W3110 pColE8, W3110 pColE9 (J. R. James), E. coli K-12 pColS4 (D. Šmajs), S. boydii M592 (serovar 8) pColU (V. Horák), E. coli K339 pColY (D. Friedman), Shigella sonnei (colicinotype 7) pColJs (J. Šmarda), E. coli pCol5 and E. coli pCol10 (H. Pilsl). As microcin control producers, the following bacterial strains were used: E. coli 449/82 pColX (microcin B17); E. coli 313/66 pColG (microcin H47); E. coli 363/79 pColV (microcin V, original source: H. Lhotová); E. coli TOP10F' pDS601 (microcin C7); E. coli D55/1 (microcin J25); E. coli B1239 (microcin L, D. Šmajs). E. coli B3423 strain was used as a positive control for fimH gene detection. Because of sensitivity of microcins H47 and M to chloroform vapours, all E. coli strains were tested by PCR method for the presence of mH47 and mM genes [36]. PCR products of related bacteriocin types (colicins E2-9, Ia-Ib, U-Y) were sequenced using dideoxy-terminator sequencing with amplification primers. Sequence analyses were carried out using Lasergene software (DNASTAR, Inc., Madison, WI).
Phylogenetic analysis of E. coli strains
A previously described triplex PCR method [68] was used to assign E. coli strains to one of four main phylogenetic groups (A, B1, B2 and D).
Statistical analyses
The statistical analyses of the prevalence bacteriocin and phylogroups used standard methods derived from the binomial distribution, including the two-tailed Fisher's exact test. STATISTICA software, version 8.0 (StatSoft, Tulsa, OK), was used for calculations.
In silico analysis of E. coli genomes
In total, 121 complete and 1830 draft genome sequences of E. coli strains were downloaded as FASTA files from ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria/ and ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria_DRAFT/ NCBI public databases, respectively, using ncbi_ftp_download script (available at: https://github.com/aleimba/bac-genomics-scripts/). For determination of the presence/absence of virulence determinants and bacteriocin encoding genes in downloaded genome sequences, DNA comparison using Smith-Waterman algorithm [69], implemented in a cross-match software (unpublished) was used. Identity scores higher than 75 % were used.
Availability of supporting data
The data set supporting the results of this article is included in the Additional file 1: Table S1. The data set of colicin gene sequences supporting the results of the article has been deposited in the GenBank/EMBL/DDBJ. Accession numbers for colicin sequences are shown in the Additional file 3: Table S3.
Abbreviations
- E. coli :
-
Escherichia coli
- TRE:
-
Trehalose
- MAN:
-
Mannitol
- SOR:
-
Sorbitol
- ONP:
-
Beta-galatosidase
- IND:
-
Indole
- LYS:
-
Lysine
- SUC:
-
Sucrose
- ORN:
-
Ornithine
- ESL:
-
Esculin
- ADO:
-
Adonitol
- H2S:
-
Hydrogen sulphide
- CEL:
-
Cellobiose
- SCI:
-
Simmons citrate
- MAL:
-
MAL
- URE:
-
Urease
- INO:
-
Inositol
- PHE:
-
Phenylalanine
References
Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–17.
Bergthorsson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16.
Dobrindt U, Agerer F, Michaelis K, Janka A, Buchrieser C, Samuelson M, et al. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J Bacteriol. 2003;185:1831–40.
Boyd EF, Hartl DL. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–65.
Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–53.
Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–72.
Kaczmarek A, Budzynska A, Gospodarek E. Prevalence of genes encoding virulence factors among Escherichia coli with K1 antigen and non-K1 E. coli strains. J Med Microbiol. 2012;61:1360–5.
Klemm P, Jørgensen BJ, van Die I, de Ree H, Bergmans H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol Gen Genet. 1985;199:410–4.
Jones CH, Pinkner JS, Roth R, Heuser J, Nicholes AV, Abraham SN, et al. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–5.
Khan NA, Kim Y, Shin S, Kim KS. FimH-mediated Escherichia coli K1 invasion of human brain microvascular endothelial cells. Cell Microbiol. 2007;9:169–78.
Braun V, Pilsl H, Gross P. Colicins: structures, modes of action, transfer through membranes, and evolution. Arch Microbiol. 1994;161:199–206.
Šmarda J, Šmajs D. Colicins--exocellular lethal proteins of Escherichia coli. Folia Microbiol (Praha). 1998;43:563–82.
Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, et al. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229.
Šmajs D, Weinstock GM. Genetic organization of plasmid ColJs, encoding colicin Js activity, immunity, and release genes. J Bacteriol. 2001;183:3949–57.
Šmajs D, Weinstock GM. The iron- and temperature-regulated cjrBC genes of Shigella and enteroinvasive Escherichia coli strains code for colicin Js uptake. J Bacteriol. 2001;183:3958–66.
Azpiroz MF, Poey ME, Laviña M. Microcins and urovirulence in Escherichia coli. Microb Pathog. 2009;47:274–80.
Šmajs D, Micenková L, Šmarda J, Vrba M, Ševčíková A, Vališová Z, et al. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: colicin E1 is a potential virulence factor. BMC Microbiol. 2010;10:288.
Budič M, Rijavec M, Petkovšek Z, Zgur-Bertok D. Escherichia coli bacteriocins: antimicrobial efficacy and prevalence among isolates from patients with bacteraemia. PloS One. 2011;6:e28769.
Petkovšek Z, Zgur-Bertok D, Starcic Erjavec M. Colicin insensitivity correlates with a higher prevalence of extraintestinal virulence factors among Escherichia coli isolates from skin and soft-tissue infections. J Med Microbiol. 2012;61:762–5.
Micenková L, Štaudová B, Bosák J, Mikalová L, Littnerová S, Vrba M, et al. Bacteriocin-encoding genes and ExPEC virulence determinants are associated in human fecal Escherichia coli strains. BMC Microbiol. 2014;14:109.
Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991;4:80–128.
Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–32.
Gonçalves A, Igrejas G, Radhouani H, Santos T, Monteiro R, Pacheco R, et al. Detection of antibiotic resistant enterococci and Escherichia coli in free range Iberian Lynx (Lynx pardinus). Sci Total Environ. 2013;456–457:115–9.
Šmarda J, Obdrzálek V. Incidence of colicinogenic strains among human Escherichia coli. J Basic Microbiol. 2001;41:367–74.
Abraham S, Shin J, Malaviya R. Type 1 fimbriated Escherichia coli-mast cell interactions in cystitis. J Infect Dis. 2001;183(s1):S51–5. doi:10.1086/318853.
Ponniah S, Abraham SN, Dockter ME, Wall CD, Endres RO. Mitogenic stimulation of human B lymphocytes by the mannose-specific adhesin on Escherichia coli type 1 fimbriae. J Immunol. 1989;142:992–8.
Hedlund M, Frendéus B, Wachtler C, Hang L, Fischer H, Svanborg C. Type 1 fimbriae deliver an LPS- and TLR4-dependent activation signal to CD14-negative cells. Mol Microbiol. 2001;39:542–52.
Mulvey MA, Schilling JD, Hultgren SJ. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun. 2001;69:4572–9.
Hagberg L, Jodal U, Korhonen TK, Lidin-Janson G, Lindberg U, Svanborg Edén C. Adhesion, hemagglutination, and virulence of Escherichia coli causing urinary tract infections. Infect Immun. 1981;31:564–70.
Leffler H, Svanborg-Edén C. Glycolipid receptors for uropathogenic Escherichia coli on human erythrocytes and uroepithelial cells. Infect Immun. 1981;34:920–9.
Edén CS, Freter R, Hagberg L, Hull R, Hull S, Leffler H, et al. Inhibition of experimental ascending urinary tract infection by an epithelial cell-surface receptor analogue. Nature. 1982;298:560–2.
Väisänen-Rhen V, Elo J, Väisänen E, Siitonen A, Orskov I, Orskov F, et al. P-fimbriated clones among uropathogenic Escherichia coli strains. Infect Immun. 1984;43:149–55.
Bergsten G, Wullt B, Svanborg C. Escherichia coli, fimbriae, bacterial persistence and host response induction in the human urinary tract. Int J Med Microbiol. 2005;295:487–502.
Lodinová R, Jouja V, Vinsová N, Vocel J, Melková J. New attempts and possibilities in prevention and treatment of intestinal coli-infections in infants. Czech Med. 1980;3:47–58.
Lodinová-Zádníková R, Tlaskalová H, Korych B, Bartáková Z. The antibody response in infants after oral administration of inactivated and living E. coli vaccines and their protective effect against nosocomial infections. Adv Exp Med Biol. 1995;371B:1431–8.
Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiol Read Engl. 2003;149:2557–70.
Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, et al. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol. 2004;186:5432–41.
Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, et al. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis. 1998;177:642–50.
Nowrouzian FL, Wold AE, Adlerberth I. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J Infect Dis. 2005;191:1078–83.
Gordon DM, O'Brien CL. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology. 2006;152:3239–44.
Sears HJ, Brownlee I, Uchiyama JK. Persistence of individual strains of Escherichia coli in the intestinal tract of man. J Bacteriol. 1950;59:293–301.
Sears HJ, Brownlee I. Further observations on the persistence of individual strains of Escherichia coli in the intestinal tract of man. J Bacteriol. 1952;63:47–57.
Sears HJ, Janes H, Saloum R, Brownlee I, Lamoreaux LF. Persistence of individual strains of Escherichia coli in man and dog under varying conditions. J Bacteriol. 1956;71:370–2.
Adlerberth I, Svanborg C, Carlsson B, Mellander L, Hanson LA, Jalil F, et al. P fimbriae and other adhesins enhance intestinal persistence of Escherichia coli in early infancy. Epidemiol Infect. 1998;121:599–608.
Nowrouzian F, Adlerberth I, Wold AE. P fimbriae, capsule and aerobactin characterize colonic resident Escherichia coli. Epidemiol Infect. 2001;126:11–8.
Zhang L, Foxman B, Marrs C. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J Clin Microbiol. 2002;40:3951–5.
Gordon DM, Stern SE, Collignon PJ. Influence of the age and sex of human hosts on the distribution of Escherichia coli ECOR groups and virulence traits. Microbiol Read Engl. 2005;151:15–23.
Tkachenko AG, Akhova AV, Shumkov MS, Nesterova LY. Polyamines reduce oxidative stress in Escherichia coli cells exposed to bactericidal antibiotics. Res Microbiol. 2012;163:83–91.
Viala JPM, Méresse S, Pocachard B, Guilhon AA, Aussel L, Barras F. Sensing and adaptation to low pH mediated by inducible amino acid decarboxylases in Salmonella. PloS One. 2011;6:e22397. doi:10.1371/journal.pone.0022397.
Wortham BW, Oliveira MA, Fetherston JD, Perry RD. Polyamines are required for the expression of key Hms proteins important for Yersinia pestis biofilm formation. Environ Microbiol. 2010;12:2034–47.
Puttamreddy S, Minion FC. Linkage between cellular adherence and biofilm formation in Escherichia coli O157:H7 EDL933. FEMS Microbiol Lett. 2011;315:46–53.
Riley MA, Wertz JE. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 2002;84:357–64.
Šmajs D, Čejková D, Micenková L, Lima-Bittencourt CI, Chartone-Souza E, Šmarda J, et al. Human Escherichia coli strains of different geographical and time source: bacteriocin types and their gene sequences are population-specific. Environ Microbiol Rep. 2012;4:459–66.
Levin BR. The evolution and maintenance of virulence in microparasites. Emerg Infect Dis. 1996;2:93–102.
Le Gall T, Clermont O, Gouriou S, Picard B, Nassif X, Denamur E, et al. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol Biol Evol. 2007;24:2373–84.
Gillor O, Giladi I, Riley MA. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol. 2009;9, e165.
Bureš J, Šmajs D, Květina J, Förstl M, Šmarda J, Kohoutová D, et al. Bacteriocinogeny in experimental pigs treated with indomethacin and Escherichia coli Nissle. World J Gastroenterol WJG. 2011;17:609–17.
Šmajs D, Bureš J, Šmarda J, Chaloupková E, Květina J, Förstl M, et al. Experimental administration of the probiotic Escherichia coli strain Nissle 1917 results in decreased diversity of E. coli strains in pigs. Curr Microbiol. 2012;64:205–10.
Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. 1995;12:85–90.
Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995;33:701–5.
López-Saucedo C, Cerna JF, Villegas-Sepulveda N, Thompson R, Velazquez FR, Torres J, et al. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerg Infect Dis. 2003;9:127–31.
Kuhnert P, Hacker J, Mühldorfer I, Burnens AP, Nicolet J, Fey J. Detection system for Escherichia coli-specific virulence genes: absence of virulence determinants in B and C strains. Appl Environ Microbiol. 1997;63:703–9.
Martínez JL, Herrero M, de Lorenzo V. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutA genes. J Mol Biol. 1994;238:288–93.
Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx 1, stx 2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. J Clin Microbiol. 1998;36:598–602.
Asadi KM, Oloomi M, Habibi M, Bouzari S. Cloning of fimH and fliC and expression of the fusion protein FimH/FliC from Uropathogenic Escherichia coli (UPEC) isolated in Iran. Iran J Microbiol. 2012;4:55–62.
Paciorek J. Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhoea. J Med Microbiol. 2002;51:548–56.
Bírošová E, Siegfried L, Kmetová M, Makara A, Ostró A, Gresová A, et al. Detection of virulence factors in alpha-haemolytic Escherichia coli strains isolated from various clinical materials. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2004;10:569–73.
Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8.
Smith TF, Waterman MS. Identification of Common Molecular Subsequences. J Mol Biol. 1981;147:195–7.
Acknowledgments
We thank Zoological Garden Hodonín, Zoological Garden Jihlava, Zoological Garden Košice, Zoological Garden Liberec, Zoological and Botanical Garden Plzeň, The Prague Zoological Garden and Zoological Garden Zlín - Lešná for kind collaboration and providing us with the collection of non-human primate samples. We thank to Prof. Thomas Secrest (Secrest Editing, Ltd.) for English editing of the manuscript. This work was supported by a grant from the Ministry of Health of the Czech Republic (NT13413-4/2012) to D.S.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DS designed the study and together with BS, LM and JB wrote the manuscript. BS, LM and JB performed bacteriocin and virulence testing of E. coli strains. DS and LM analyzed the data. KH, ES, MV, AS, DK, VW and JB contributed to isolation and characterization of the bacterial strains and gathered data. All authors read and approved the final manuscript.
Authors’ information
Not applicable.
Additional files
Additional file 1: Table S1.
Complete data set presented in this article. (DOCX 19 kb)
Additional file 2: Table S2.
DNA primers used for PCR detection of colicin and microcin encoding genes and genes encoding virulence factors. (XLSX 167 kb)
Additional file 3: Table S3.
Colicin gene sequences deposited in the GenBank/EMBL/DDBJ. (XLSX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Štaudová, B., Micenková, L., Bosák, J. et al. Determinants encoding fimbriae type 1 in fecal Escherichia coli are associated with increased frequency of bacteriocinogeny. BMC Microbiol 15, 201 (2015). https://doi.org/10.1186/s12866-015-0530-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-015-0530-5