Abstract
Background
Escherichia coli O157:H7 is a highly virulent human pathogen with severe consequences following infection, which claims many lives worldwide. A suggested method for controlling this bacterium is the competitive elimination through using probiotic bacteria that prevent its colonization. Some nonpathogenic E. coli strains that produce antibacterial colicins are among these probiotic bacteria. We aimed to isolate and characterize the colicinogenic E. coli strains from diarrheic and healthy sheep that inhibit E. coli O157:H7, which could be used as possible probiotic sources. A total of 292 E. coli isolates (146 from each diarrheic and healthy sheep) were screened for the presence of colicin and virulence genes. The phylogenetic group/subgroup determination was performed by PCR. In vitro evaluation of inhibitory effect of colicinogenic isolates on E. coli O157:H7 was done phenotypically.
Results
The frequency of diarrhea associated colicinogenic E. coli isolates was significantly higher than those isolated from healthy sheep. An association between ETEC and the genes coding for colicin-V & colicin-Iab in diarrheic E. coli isolates was observed. Moreover, there was an association between ipaH and Colicin-V encoding genes. Furthermore, E. coli isolates showing bacteriocinogeny while possessing no virulence genes had a frequency of 97.67 and 11.94% in healthy and diarrheic isolates, respectively. Of these strains, five isolates (3.42%) from diarrheic and twenty-five isolates (17.12%) from healthy sheep inhibited O157:H7 strain. Additionally, colicin E1 and colicin Iab genes were more prevalent in B1 phylogroup.
Conclusions
These results signified that healthy sheep could be considered as a potential source for anti-O175:H7 bacterial isolates.
Similar content being viewed by others
Background
E. coli O157: H7 is well known as a severely virulent foodborne bacterial pathogen that claims many lives annually worldwide [1]. The clinical symptoms of E. coli O157:H7 vary wildly from mild diarrhea to hemolytic uremic syndrome (HUS) leading to kidney failure. The severity of these symptoms depends on the immune status of the patient and also the infection dose of the bacteria [2, 3]. It has been shown that the main reservoir of E. coli O157:H7 are ruminants [4].
However, one proposed approach for the control of E. coli O157:H7 is through the competitive elimination, which is described as the use of other probiotic bacteria to prevent the growth and colonization of the pathogenic bacterium [5]. The reduction of E. coli O157:H7 could be achieved through the use of other nonpathogenic E. coli strains that produce colicins [6]. Colicins are antimicrobial proteins synthesized by E. coli for inhibiting other E. coli strains as well as other closely related bacteria [7]. These proteins have evolved to provide the carrying bacterium an advantage when competing with the closely related bacteria [8]. The ability to produce colicins is common in the Enterobacteriacea family, and studies have revealed that approximately 30% of E. coli isolates are capable of producing at least one type of colicin [9].
Host animal species can effect on the type of produced colicin and bovine isolates would be expected to show low rate of resistance against to non-cattle Colicinogenic E. coli [5]. Zhao et al., [10] developed the first competitive- exclusion system against E. coli O157:H7 by isolating colicinogenic E. coli isolates from cattle, but little information was provided about the types of colicins produced by these isolates and the extent of resistance among naturally present E. coli O157:H7 isolates [11].
For determination of the pathogenic or non-pathogenic nature of E. coli isolates, assessment of the evolutionary origins via phylogenetic analysis has been helpful [12]. Based on the phylogenetic studies, E. coli isolates can be allocated to one of the four main phylogenetic groups A, B1, B2 and D, which can fall into seven subgroups [13].
Previously published studies have only provided partial insight into the association between colicin production and virulence factors, as they were mainly focused upon uropathogenic Escherichia coli (UPEC) isolates and differed in the number of detected colicin and virulence genes [14]. The present project was designed to genotypic analysis of colicinogenic E. coli isolates from diarrheic and healthy sheep that inhibit serotype O157:H7. Additionally, this research was set to characterized phylogenetic groups/subgroups of the isolates.
The work presented here is a continuation of our previous study. 146 of the diarrheic strains that are used in the current study were also used in a previous study where their virulence genes profiles were studied [15]. Here we aimed to further expand our investigations through analyzing these strains in terms of determination of colicin genes, phylogrouping and the inhibitory effect of them on serotype O157:H7 and comparing them with strains from healthy once.
Methods
Sample collection and isolation of E. coli strains
This section of the study was performed from March to Oct 2015. 167 fecal samples from healthy sheep were collected in Kerman province, Iran. Each sample was belonged to one animal which were between 1 to 12 weeks old. Swab samples were placed directly in tubes containing the Amies medium (Becton Dickinson, BBL, and USA) and sent to the laboratory for immediate processing. Samples were streaked on MacConkey agar (Merck, Germany) and incubated at 37 °C for 24 h. Colonies showing E. coli features were sent to Gram staining and were confirmed to be E. coli by using the biochemical API 20E identification system (BioMérieux, Marcy l’Etoile, France). One confirmed isolate was chosen from each plate. Finally, 146 E.coli strains from healthy sheep were stored in Luria- Bertani broth (Invitrogen, Paisley, Scotland) with 30% sterile glycerol at − 80 °C and as the above-mentioned, 146 of the diarrheic strains that are used in the current study were also used in a previous study [15]. Therefore, the assays were done on 292 isolates (146 from each diarrheic and healthy sheep).
Ten E. coli strains were used as positive controls, including H10407 (LT-I +, ST-I+); 1404 (cdtIII +, cnf2 +, f17A+); 510 (F5 +, F41+); 28C (cdtIV +, cnf1+); 31A (f17c-A +); Sakaï (stx1 +, stx2+, eaeA+); 85b (ipaH+); 25KH9 (f17a-A+), S5 (f17b-A+); and ECOR62 (chuA +, yjaA + and Tspe4 C2+). The laboratory nonpathogenic E. coli MG1655 was used as a negative control. All the above-mentioned reference strains had been taken from the bacterial collection section of the Microbiology Department of Ecole Nationale Vétérinaire Toulouse, France. colicin reference strains were provided from the Pasture Institute of Iran.
Molecular detection of the virulence genes and phylogenetic groups
DNA extraction of the overnight E. coli cultures (current isolates and the reference isolates) was performed through the boiling method [15]. The virulence genes had been amplified and analyzed as described in details before. A complete list of the primers used for this virulence genotyping is published previously [15]. Additionally, the absence or presence of yjaA, chuA, and TSPE4.C2 sequences [13] in E. coli isolates (see Table 1), which were determined by multiplex PCR, was used to identify the phylogenetic groups.
Molecular detection of the colicin genes
All E. coli isolates were examined by several PCR protocols for the presence of the colicin encoding genes including colicin-Iab, V, ANS4, E1, Mix E, 5, 10, K and colicin YU [16] PCR Amplification was performed in 25 μL volumes containing: 2.5 μL of 10× PCR buffer, 3 μL prepared DNA, 2 mM MgCl2, 0.3 M of each oligonucleotide primer, 0.2 mM dNTP mix, 1 U Taq DNA polymerase (Cinnagen, Iran), and PCR grade water up to 25 μL. The PCR conditions included 1 cycle of denaturation at 94 °C for 2 min, annealing at the Tm specific for each primer (see Table 2) for 1 min, and elongation for 1 min at 72 °C; subsequently 35 cycles of denaturation at 94 °C for 1 min and then a final 5 min at 72 °C hold temperature. The PCR products were electrophoresed on 1.5% agarose gel for 90 min at 85 v. The DNA bands were visualized through staining of gels with DNA Fluorescent Loading Dye (Sigma, Germany).
Evaluation of the inhibitory activity of E. coli strains
The sensitivity of the pathogenic E. coli O157:H7 Sakai (EHEC) to the colicins was examined here. The E. coli O157:H7 colonies and the colicin positive strains, which have been identified in the previous steps, were inoculated into Lysogeny Broth (LB; Merck, Germany) and were grown overnight (12 to 16 h). The overnight grown strain of E. coli O157:H7 was streaked and spread thoroughly on the surface of LB agar plates containing 0.025 μg of Mitomycin C (Sigma, USA) and were then allowed to dry. Mitomycin C’s role here was to induce the colicins production in the isolated strains. Fresh overnight cultures (7 μL from each colicinogenic isolates were spotted on the same LB plates that had been in the previous step streaked with E. coli O157:H7. These bacterial test plates were afterwards stored at 37 °C for 12 to 16 h. Finally, the clearance zones around the spotted E. coli isolates were measured [16]. The clear zone indicates no growth of E. coli O157:H7.
Statistical analysis
SPSS software (version 17. SPSS Inc., USA) was used for the data analysis. P value was measured using Chi-square and Fisher’s exact tests to find any significant relationship. P values less than 0.05 were considered statistically meaningful.
Results
PCR assay showed that sixty-seven E. coli isolates (45.89%) from diarrheic samples contained colicin encoding genes. Twenty-five (17.12%) isolates of them harbored colicin Iab encoding genes and most of these isolates (15.06%) contain the virulence genes related to ETEC pathotype. Eighteen (12.32%) isolates possessed colicin E1 encoding genes and were widely distributed among STEC (9.58%) pathotype. Fifteen E. coli isolates (10.27%) were positive for colicin V encoding genes and were distributed among ETEC (6.16%) and EIEC (0.68) pathotypes, respectively. Colicin-Iab/ E1/ V were the most prevalent colicin encoding genes and colicin Mix E and colicin 5, 10, k were not detected in any of the diarrheic E. coli isolates.
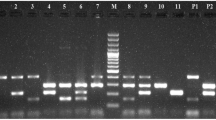
From one hundred and forty-six E. coli isolates of healthy sheep, eleven isolates (7.53%) contained stx1 and/or stx2 in combination with the eae or/and ehly genes and classified as STEC pathotype. Three isolates (2.05%) were positive for cnf1 gene which categorized into NTEC pathotype (Fig. 1). One isolate (0.68%) was positive for ipaH gene that is an EIEC virulence gene and one hundred twenty E. coli isolates from healthy sheep did not have any virulence genes (Fig. 2). The frequency of bacteriocinogeny was 29.45% among healthy isolates and all types of colicin ending genes were detected in these isolates. The figures of colicin genes are shown in Figs. 3, 4.
Statistical analysis showed that the frequency of colicinogenic isolates in diarrhea associated E. coli was significantly higher than E. coli isolates from healthy sheep (p < 0.05). A specific association between ETEC and colicin V/ colicin Iab encoding genes in diarrheic E. coli isolates was observed (p < 0.05). In addition, a specific association between ipaH gene and Colicin V encoding gene was found (p < 0.05). Totally, E. coli isolates possessing no virulence determinant had (42/43, 97.67%) and (8/67, 11.94%) frequency of bacterocingeny in healthy and diarrheic isolates, respectively. Altogether, 110 colicin producer strains (37.67%) were identified of 292 E. coli isolates.
Approximately 28% of the 146 E. coli isolates from diarrheic sheep produced inhibitory activity against E. coli O157:H7 Sakai (EHEC). Five of these isolates (12.19%) possess more than one of the mentioned colicin target genes. The largest zone of inhibition belonged to col. E1 producer (12.32%) and col. V producer (10.27%) isolates. No zone of inhibition was obtained from diarrheic isolates which were positive for col. Iab and col. yu encoding genes. Thirty two (21.9%) healthy isolates had the inhibitory effect on E. coli O157:H7 Sakai (EHEC). All types of colicin in healthy isolates had the inhibitory effect on E. coli O157:H7 with the exception of col. Iab. Details of detected colicin encoding genes in relation to the different pathotypes and the inhibitory effect of the colininogenic isolates on E. coli O157:H7 Sakai (EHEC) are shown in Table 3.
Phylogenetic analysis of diarrheic and healthy isolates showed that the highest frequency of colicin encoding genes were found in B1 phylogroup (65.45%) followed by A (26.36%), D (7.27%) and B2 (0.9%) phylogroups. In B1 phylogroup, colicin E1 (17/45, 37.7%) and colicin Iab (11/27, 40.74%) encoding genes were more common in diarrheic and healthy E. coli isolates, respectively. Producer of colicin V belonged more frequently to A phylogroup in diarrheic and healthy E. coli isolates (22.38, 7.46%, respectively; p < 0.0001) when compared to other colicin producer isolates (Table 4).
Discussion
In this study, the average prevalence of colicinogenic E. coli isolates from sheep was 37.67%. Surprisingly our data lies well within the recently published results from a different source for probiotic E. coli strains, humans [11, 14, 17, 18]. In those studies, Gordon et al., [19] detected 19 types bacteriocin genes when screening 266 E. coli strains that had been isolated from human fecal (38% of these isolates were bacteriocinogenic), and Smaj et al., [20] detected 29 bacteriocin types in 441 human fecal E. coli isolates (55% of which were bacteriocin-encoding isolates). Based on our presented data and the previously published results, sheep could also be considered as a potential source of the probiotic E. coli strains.
The results presented here showed that the frequency of bacteriocinogeny in E. coli isolates positively correlates with the number of detected virulence factors. To the best of our knowledge, this is the first time that the correlation between frequency of colicin encoding genes and the number of virulence factors in E. coli isolates from sheep have been investigated.
This study also showed that the prevalence of colicin E1 genes among diarrheic isolates was relatively high (10.27%) in comparison to the other colicin genes. Micenkova et al., [20] reported that colicin E1, which is a pore-forming bacteriocin, had a high prevalence in fecal and also in human UPEC isolates.
The prevalence of colicing Iab genes of E. coli isolates from diarrheic and healthy sheep (17.12 and 25.56%, respectively) were significantly higher than other colicin genes. This is in accordance with the study of Micenková., et al., [11, 14]. Another study showed that colicin Ia/Ib was one of the most frequent colicin types among ETEC isolates and also 38.2% of Shigella sonnei isolates produced colicin Ia/Ib [17]. In the present study, 15.06% of 292 of fecal isolates were positive for colicin Ia/Ib encoding gene and also categorized in ETEC pathotype.
The prevalence of colicin V encoding gene in healthy and diarrheic isolates was equal and there was a specific association between ipaH virulence gene and this type of colicin. This finding is in accordance with the previous studies, which showed that colicin V encoding plasmids are more commonly carried by the virulent E. coli isolates [7].
Furthermore, colicin Y is related to the colicin U that was isolated around 20 years ago in Europe from Shigella boydii. Colicin Y is rarely detected colicin type in Europe, but high frequency of colicin Y producers were detected in E. coli isolated in Amazonia [11]. In our study the prevalence of colicin Y/U genes was relatively low in both healthy and diarrheic isolates. Moreover, productions of nucelase colicins E2-E9 are also very rare in E. coli isolates. [18, 19, 21]. In this study, none of the diarrheic isolates were positive for colicin E2-E9 encoding genes.
Association of E. coli phylogroups and bacteriocin types has been tested in several previous publications, which produced different results. Gordon and O’Brien [19] tested a set of 266 fecal E. coli isolates and failed to find remarkable differences in the prevalence of bacteriocin determinants among the phylogroups. They reported phylogroup A and B1 tended to encode more colicin types, while in phylogroup A, genes encoding E1, Ia and V, were most common [21]. In this study, phylogenetic analysis of diarrheic and healthy isolates showed that the highest frequency of colicin encoding genes were found in B1 phylogroup (65.45%) followed by A (26.36%), D (7.27%) and B2 (0.9%) phylogroups. In phylogroup B1, colicin E1 and colicin Iab encoding genes were more common.
Setia et al., [16] showed that the colicin produced by 26.8% of E. coli isolates without known toxin genes recovered from healthy cattle were able to inhibit the growth of pathogenic E. coli K88. The present study showed that fifty E. coli isolates (50/292, 17.12%) from diarrheic and healthy sheep were able to produce different types of colicin, while they didn’t have any virulence factors. Thirty six isolates of them (12.32%) were able to inhibit the growth of E. coli O157:H7 Sakai (EHEC) in vitro. So, colicin produced by these isolates may potentially provide an effective strategy to reduce E. coli O157:H7 in food animals. In the recent study, all types of colicin were able to inhibit E. coli O157:H7 except for colicin Ia/Ib which is in agreement with the previous studies [22, 23]. Bradley et al., [24] considered that O157 isolates were most likely resistant due to masking of the colicin Ia/Ib receptor by O-antigen side-chains of LPS. Another explanation for this resistance is that some E. coli O157:H7 isolates are colicinogenic and produce specific concomitant immunity proteins [23], they can be resistant to certain colicins or even a broad classes of colicins. [25]. On the other hand, this possibility should be considered that O157:H7 Sakai (EHEC) of human could have resistance against the colicins of strains isolated from sheep. Therefore, simultaneous administration of a mixture of several classes of colicins should be considered as a treatment to reduce E. coli O157:H7 (and other EHEC) in the gastrointestinal tract of food animals. [23]
In this study, all types of colicin encoding genes in E. coli isolated from healthy sheep were detected. Whereas, E. coli isolates from diarrheic sheep didn’t harbored colicin Mix E/ANS4 encoding genes. Previous studies found colicinogenic isolates from all sources, but the greatest types of colicin were from cats and sheep [9, 26]. Tahamtan et al., [27] reported that Ia/Ib and ANS4 colicin encoding genes had the highest frequency among E.coli strains isolates from healthy and diarrheic cattle in Shiraz, Iran. It can be concluded that many factors may influence the types of detected colicin genes including species, illness, diet and geographical conditions [28].
Conclusion
The results and data presented in our report showed that the healthy sheep could be considered as a source of colicinogenic E. coli strains. These E. coli strains that do not harbor any virulent factors but are carrying the colicin genes are a potentially valuable source for anti-O175:H7 bacterial isolates. Further work is granted to study the in vivo inhibitory effects of these colicinogenic isolates of E. coli on the biocontrol of pathogenic O175:H7. On the other hand, it can be concluded that there is a correlation between presence of colicin genes and virulence genes in E. coli isolates which could be determined in the future studies.
Abbreviations
- a EPEC:
-
atypical Entropathogenic E. coli
- chuA :
-
E. coli haem-utilization gene
- cnf :
-
cytotoxic necrotizing factor
- eae :
-
attaching and effacing gene of E. coli
- EHEC:
-
Enterohemorrhagic E. coli
- EIEC:
-
Entroinvasive E. coli
- ETEC:
-
Entrotoxigenic E. coli
- HUS:
-
Hemolytic Uremic Syndrome
- ipaH :
-
invasion plasmid antigen
- LT:
-
heat labile toxin.
- NTEC:
-
Necrotoxic E. coli
- PCR:
-
Polymerase Chain Reaction
- SPSS:
-
Statistical Package for the Social Sciences
- ST:
-
heat-stable enterotoxin
- STEC:
-
Shiga Toxin producing E. coli
- Stx :
-
Shiga toxin
- TspE4.C2:
-
an anonymous DNA fragment in E. coli
- yjaA :
-
E. coli K12 gene
References
Abdissa R, Haile W, Fite AT, Beyi AF, Agga GE, Edao BM, et al. Prevalence of Escherichia coli O157: H7 in beef cattle at slaughter and beef carcasses at retail shops in Ethiopia. BMC Infect Dis. 2017;17:277.
Ferens WA, Hovde CJ. Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathog Dis. 2011;8:465–87.
Rahal EA, Kazzi N, Nassar FJ, Matar GM. Escherichia coli O157:H7—clinical aspects and novel treatment approaches. Front Cell Infect Microbiol. 2012;2:138.
Elder RO, Keen JE, Siragusa GR, Barkocy-Gallagher GA, Koohmaraie M, Laegreid WW. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc Natl Acad Sci. 2000;97:2999–3003.
Schamberger GP, Diez-Gonzalez F. Assessment of resistance to Colicinogenic Escherichia coli by E. coli O157:H7 strains. J Appl Microbiol. 2005;98:245–52.
Schamberger GP, Diez-Gonzalez F. Characterization of Colicinogenic Escherichia coli strains inhibitory to enterohemorrhagic Escherichia coli. J Food Prot. 2004;67:486–92.
Jeziorowski A, Gordon DM. Evolution of microcin V and Colicin Ia plasmids in Escherichia coli. J Bacteriol. 2007;189:7045–52.
Inglis RF, Bayramoglu B, Gillor O, Ackermann M. The role of bacteriocins as selfish genetic elements. Biol Lett. 2013;9 20121173–20121173.
Diez-Gonzalez F. Applications of bacteriocins in livestock. Curr Issues Intest Microbiol. 2007;8:15–23.
Zhao T, Doyle MP, Harmon BG, Brown CA, Mueller POE, Parks AH. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J Clin Microbiol. 1998;36:641–7.
Micenková L, Bosák J, Štaudová B, Kohoutová D, Čejková D, Woznicová V, et al. Microcin determinants are associated with B2 phylogroup of human fecal Escherichia coli isolates. Microbiology. 2016;5:490–8.
Chandran A, Mazumder A. Prevalence of diarrhea-associated virulence genes and genetic diversity in Escherichia coli isolates from fecal material of various animal hosts. Appl Environ Microbiol. 2013;79:7371–80.
Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8.
Micenková L, Bosák J, Vrba M, Ševčíková A, Šmajs D. Human extraintestinal pathogenic Escherichia coli strains differ in prevalence of virulence factors, phylogroups, and bacteriocin determinants. BMC Microbiol. 2016;16.
Ghanbarpour R, Askari N, Ghorbanpour M, Tahamtan Y, Mashayekhi K, Afsharipour N, et al. Genotypic analysis of virulence genes and antimicrobial profile of diarrheagenic Escherichia coli isolated from diseased lambs in Iran. Trop Anim Health Prod. 2017;49:591–7.
Setia A, Bhandari SK, House JD, Nyachoti CM, Krause DO. Development and in vitro evaluation of an Escherichia coli probiotic able to inhibit the growth of pathogenic Escherichia coli K88. J Anim Sci. 2009;87:2005–12.
Micenková L, Štaudová B, Bosák J, Mikalová L, Littnerová S, Vrba M, et al. Bacteriocin-encoding genes and ExPEC virulence determinants are associated in human fecal Escherichia coli strains. BMC Microbiol. 2014;14:109.
Šmajs D, Čejková D, Micenková L, Lima-Bittencourt CI, Chartone-Souza E, Šmarda J, et al. Human Escherichia coli strains of different geographical and time source: Bacteriocin types and their gene sequences are population-specific. Environ Microbiol Rep. 2012;4:459–66.
Gordon DM, O’Brien CL. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology. 2006;152:3239–44.
ŠŠmajs D, Micenková L, ŠŠmarda J, Vrba M, Ševčíková A, Vališová Z, et al. Bacteriocin synthesis in uropathogenic and commensal Escherichia coli: Colicin E1 is a potential virulence factor. BMC Microbiol. 2010;10:288.
Kohoutova D, Smajs D, Moravkova P, Cyrany J, Moravkova M, Forstlova M, et al. Escherichia coli strains of phylogenetic group B2 and D and bacteriocin production are associated with advanced colorectal neoplasia. BMC Infect Dis. 2014;14:733.
Toshima H, Yoshimura A, Arikawa K, Hidaka A, Ogasawara J, Hase A, et al. Enhancement of Shiga toxin production in enterohemorrhagic Escherichia coli serotype O157:H7 by DNase Colicins. Appl Environ Microbiol. 2007;73:7582–8.
Callaway TR, Stahl CH, Edrington TS, Genovese KJ, Lincoln LM, Anderson RC, et al. Colicin concentrations inhibit growth of Escherichia coli O157:H7 in vitro. J Food Prot. 2004;67:2603–7.
Bradley DE, Howard SP, Lior H. Colicinogeny of O157:H7 enterohemorrhagic Escherichia coli and the shielding of Colicin and phage receptors by their O-antigenic side chains. Can J Microbiol. 1991;37:97–104.
Alonso G. How bacteria protect themselves against channel-forming Colicins. Int Microbiol. 2000;3:81–8.
Garrett WS, Gallini CA, Yatsunenko T, Michaud M, Dubois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300.
Tahamtan Y, Shirazi Z, Pourbakhsh A, Kargar A. Detection of Colicin genes by PCR in Escherichia coli isolated from cattle in shiraz-Iran. Archives of Razi Institute. 2012:63–7.
Murinda SE, Liu SM, Roberts RF, Wilson RA. Colicinogeny among Escherichia coli serotypes, including O157 : H7, representing four closely related diarrheagenic clones. J Food Prot. 1998;61:1431–8.
Acknowledgments
The authors would like to thank Dr. Eric Oswald (Ecole Nationale Vétérinaire Toulouse, France) and Pasture Institute of Iran for providing the reference strains.
Funding
This work was financially supported by a Grant from the Iran National Science Foundation grant number 93027693.
Availability of data and materials
All data generated or analyzed during this study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
NA and RG designed the study, NA performed the experiments, NA and RG analyzed the data and NA and RG wrote and drafted the manuscript. NA and RG read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All investigations were conducted in accordance with the Guiding Principles for the Care and Use of Animals was approved by the Ethics Committee of Faculty of Veterinary Medicine, Shahid Bahonar University of Kerman, Iran.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Askari, N., Ghanbarpour, R. Molecular investigation of the colicinogenic Escherichia coli strains that are capable of inhibiting E. coli O157:H7 in vitro. BMC Vet Res 15, 14 (2019). https://doi.org/10.1186/s12917-018-1771-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-018-1771-y