Abstract
Oxygen-induced retinopathy (OIR) animal model is widely used for retinopathy of prematurity (ROP) researches. The purpose of this study was to identify proteins and related pathways of OIR with or without anti-vascular endothelial growth factor (VEGF) treatment, for use as biomarkers in diagnosing and treating ROP. Nine samples were subjected to proteomic analysis. Retina specimens were collected from 3 OIR mice, 3 OIR mice with anti-VEGF treatment and 3 normal mice (control group). Liquid chromatography-tandem mass spectrometry analysis was performed using the 4D label-free technique. Statistically significant differentially expressed proteins, gene ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway representations, InterPro (IPR) and protein interactions were analyzed. In total, 4585 unique proteins were identified as differentially expressed proteins (DEPs). Enrichment analysis of the GO and KEGG indicated functional clusters related to peptide biosynthetic and metabolic process, cellular macromolecule biosynthetic process and nucleic acid binding in OIR group. For anti-VEGF treatment group, DEPs were clustered in DNA replication, PI3K/Akt signaling pathway and Jak/STAT signaling pathway. Proteomic profiling is useful for the exploration of molecular mechanisms of OIR and mechanisms of anti-VEGF treatment. These findings may be useful for identification of novel biomarkers for ROP pathogenesis and treatment.
Similar content being viewed by others
Introduction
Preterm birth is a significant global health problem. With the decreasing total fertility rate, which is most pronounced in developing countries, a growing number of studies have focused on the development and quality of life of preterm infants [1, 2]. Further, retinopathy of prematurity (ROP), with an overall estimated incidence of 50,000 infants annually, is a leading cause of childhood blindness in industrialized countries [3].
To ablate the peripheral avascular retina and reverse neovascularization, the gold standard treatment of ROP has been laser photocoagulation [4]. On the other hand, laser therapy might cause visual impairment, such as peripheral visual field defect and refractive error [5]. Because neovascularization in ROP is largely vascular endothelial growth factor (VEGF) driven and anti-VEGF therapy appears to have fewer ocular side effects, various intravitreal anti-VEGF medications have substituted or been in conjunction with traditional laser therapy [6, 7].
Because of the special physiological condition of premature infants, animal models are widely used in ROP researches. The mouse oxygen-induced retinopathy (OIR) model is a well-established animal model for ROP researches, which is characterized by retinal vessel change from hyperoxia-induced vessel loss to neovascular in normoxia [8, 9]. OIR model is analyzed and studied mainly in angiogenesis disease field, but some researches have also reported neuronal damage and pathological effects on retina in OIR model [10]. Furthermore, VEGF plays an extremely important role in OIR model, as well as pathogenesis of preterm infants. Because of multiple interactions among vascular, neuronal, glial, and immune cells in retinal vascular pathology, it is necessary to conduct researches in whole retinal tissue to gain more information about the mechanism of pathogenesis.
Proteomics has been widely used in ophthalmic research and proved to be reliable. Because of the advancement in the mass spectrometry technology, proteomics could facilitate accurately identification of potential disease markers for subsequent diagnosis and prognosis, as verified by many previous studies [11, 12]. Therefore, label-free quantitative proteomics strategy was applied to comprehensively characterize the retinal proteome in a model of VEGF-induced retinal insult as well as to evaluate the efficacy of anti-VEGF treatment.
In this study, we identified proteins and related pathways examined in OIR model and examined effects of anti-VEGF treatment. There is a possibility for revealing alterations in the major metabolic functions, cellular stress, and corresponding dysfunctions, because of oxygen-mediated insult and protective characteristics of anti-VEGF in retina.
Results
Analysis of significantly differentially expressed proteins
For the details of the data, 159,155 matched spectra, 35,450 peptides, 4585 identified and quantifiable proteins were obtained from 9 samples. The number of up-regulated proteins and down-regulated proteins was listed in the Table 1 and Fig. 1. Hierarchical clustering analysis was performed on dysregulated proteins of three groups, and the heatmaps generated from the analysis showed a clear distinction in protein expression levels among the ranibizumab, OIR and normal groups (Fig. 2). Retinal images of immunohistochemistry and OCTA was present in Fig. 3.
Volcano plot of differentially expressed proteins. A Comparisons of the volcano plot of the differential proteins expressed between the ranibizumab group and the OIR group. B Comparisons of the volcano plot of the differential proteins expressed between the OIR group and the normal group. C Comparisons of the volcano plot of the differential proteins expressed between the ranibizumab group and the normal group. The X-axis represents protein difference (log2-transformed fold changes), and the Y-axis represents the corresponding − log10-transformed P values. Red dots indicate significantly up-regulated proteins, green dots denote significantly down-regulated proteins, and gray dots symbolize proteins with no significant change
Comparisons of the heatmaps of the differential proteins expressed in three groups. A Comparisons of the heatmaps of the differential proteins expressed between the ranibizumab group and the OIR group. B Comparisons of the heatmaps of the differential proteins expressed between the OIR group and the normal group. C Comparisons of the heatmaps of the differential proteins expressed between the ranibizumab group and the normal group. Higher red and blue intensities indicate higher degrees of upregulation and downregulation, respectively
VEGFA and HIF-1α double-stained confocal images of retinas and OCTA images in three groups. Group A: the ranibizumab group; Group B: the OIR group; Group C: the normal group. At P17, neovascularization emerges on the surface of the retina (arrows). Avascular area and tortuous neovascularization could be directly observed on the enface images. Abnormal vascularity also caused inhomogeneous distribution of blood flow signals on OCTA images
GO functional analysis of the DEPs
We conducted GO analysis to enrich and cluster the differentially expressed proteins (DEPs) of the ranibizumab, OIR, and normal groups. Details of the cellular components (CC), molecular functions (MF), and biological processes (BP) are elaborated in Fig. 4. GO annotation analysis suggested that the DEPs of the three groups mainly participate in MF, such as protein binding, ATP binding, nucleic acid binding, and GTP binding. In BP, oxidation − reduction process, metabolic process and protein phosphorylation had many DEPs aggregation. Using GO annotation-enriched analysis, it indicated how anti-VEGF affects vascular changes. The GO enrichment of the ranibizumab group was involved in regulation of response to cellular macromolecule metabolic process, nucleic acid metabolic process and cellular macromolecule biosynthetic process (p < 0.05). On the other hand, the GO enrichment of the OIR group showed that a large number of up-regulated DEPs were enriched in terms related to MF, such as organic cyclic compound binding, heterocyclic compound binding, nucleic acid binding, et al. (p < 0.05). The GO enrichment analysis of the ranibizumab group was similar with the OIR group when compared with normal group.
GO annotation and GO enrichment analyses of the DEPs. A GO enrichment analysis of the DEPs (Top 20) between the ranibizumab group and the OIR group. B GO enrichment analysis of the DEPs (Top 20) between the OIR group and the normal group. C GO enrichment analysis of the DEPs (Top 20) between the ranibizumab group and the normal group. D GO annotation analysis of the DEPs (Top 10) of the three groups
KEGG pathway analysis functional analysis of the DEPs
For further physiological characterization of samples from different groups, we conducted annotation and enrichment analysis of KEGG pathway to identify the main biochemical metabolic pathways involving the differential metabolites. In total, all differential metabolites of three groups were enriched in 34 pathways (Fig. 5). KEGG pathway analysis revealed that the most obvious pathway affected by intravitreal injection of ranibizumab was “PI3K/Akt signaling pathway”. A large number of DEPs were detected in this pathway.
For OIR group, spliceosome, ribosome and other pathway associated with RNA-transport had significant difference compared to control group. The KEGG analysis of ranibizumab. vs. normal group presented similar result.
IPR enrichment analysis of the DEPs
We conducted IPR analysis to enrich and cluster the DEPs of the ranibizumab, OIR, and normal groups (Fig. 6). IPR enrichment analysis showed that 21 significantly enriched functional domains were identified between the ranibizumab and OIR groups, especially mini-chromosome maintenance (MCM), DNA-dependent ATPase and WD40 repeat domain. On the other hand, 46 domains were identified between the OIR and normal groups. The most significant enriched functional domains were RNA recognition motif domain, K homology domain (KHD) and C-terminal domain (CTD). The IPR analysis of ranibizumab. vs. normal group presented similar result.
PPI analysis of the DEPs
Using the STRING online database and Cytoscape software, we established a protein–protein interaction network of DEPs (Fig. 7). In comparison of the ranibizumab and OIR group, the network consisted of 411 nodes and 5273 edges (ranibizumab group vs. OIR group). Compared with the OIR group, DEPs up-regulated in the ranibizumab group were mainly enriched in pathways related to cell responses, migration and growth, transcription of DNA, GTP-binding, nucleotide-binding, protein homeostasis, neuronal migration, and promotes neurogenesis. DEPs down-regulated in the ranibizumab group were enriched in pathways related to nucleotide excision repair, modulation of proteasomal degradation, mRNA processing and splicing, and tRNA modification.
PPI analysis of the DEPs. A DEPs interaction networks of the DEPs between the ranibizumab group and the OIR group. B DEPs interaction networks of the DEPs between the OIR group and the normal group. C DEPs interaction networks of the DEPs between the ranibizumab group and the normal group. Red node: up-regulated. Blue node: down-regulated
In comparison of the OIR and normal group, the interaction network of DEPs contained 1274 nodes and 88,453 edges. Compared with the normal group, DEPs up-regulated in the OIR group were mainly enriched in pathways related to correct protein translation and folding, heat shock protein formation, protein sumoylation, RNA helicase, and folding of actin and tubulin. DEPs down-regulated in the OIR group were enriched in pathways related to cell protection against oxidative stress, neuroprotective mechanisms, cell growth and transformation, glutathione biosynthesis, and protein phosphorylating. In comparison of the ranibizumab and normal group, the interaction network of DEPs contained 1515 nodes and 112,066 edges. The result of regulation was similar with comparison of the OIR and normal group.
Another smaller-scale simplified interactome map was present in Fig. 8. The nodes with high scores were selected for display.
Simplified interactome map of PPI analysis. A DEPs interaction networks of the DEPs between the ranibizumab group and the OIR group. B DEPs interaction networks of the DEPs between the OIR group and the normal group. C DEPs interaction networks of the DEPs between the ranibizumab group and the normal group. Red node: up-regulated. Blue node: down-regulated
Discussion
This study implemented 4D label-free proteomics and analyses of functional protein network and pathways to elaborate the global and specific proteomics alterations in the OIR and anti-VEGF treatment animal models. Because of the characteristic of ROP and physiological structure of infants, it’s extremely difficult to obtain pathological eye contents, such as aqueous humor, vitreous humor, or retinal tissue [13]. Nevertheless, full-fledged OIR model could maturely simulate pathological mechanism of ROP, which is characterized by abnormal avascular area and neovascularization. Up to now, some proteomics studies have been conducted on OIR model [14,15,16]. Although there were few researches focus on the influence caused of treatment on the model. At the same time, anti-VEGF treatment takes on increasing importance in retinal ischemic diseases [17, 18]. In this study, we introduced anti-VEGF treatment to OIR model for proteomics analysis. We thought that revelation of the complex proteome changes happening after anti-VEGF treatment would be an important source for detecting ROP-specific biological targets. And it will help to find biomarkers for diagnosis and disease treatment, and for the better knowledge of the pathogenic mechanisms of the disease.
Neovascularization in OIR
ROP is constituted by two oxygen-dependent stage. The first stage is induced by the hyperoxic environment and activated immediately after the premature birth; the increase of oxygen initiates a decrease in VEGF, which causes the stagnation of retinal vascular development. At this stage, the retinal vessels are so vulnerable and ineffective that retina becomes hypoxic [19]. When oxygen concentration recovers to a normal level, the second stage starts with an increase in the levels of VEGF, which promotes an excessive growth of abnormal retinal vessels that extend into the vitreous and even retinal disease [20]. When the mouse is born, its retinal development is not yet mature, and the hyperoxic environment would further inhibit retinal vascular development [21]. After the pups return to normal room air environment, aberrant retinal vessels further evolve to the formation of neovascular tufts [22]. The pathological mechanism of OIR model is extremely similar to human ROP [23]. The retinal angiogenic changes under hypoxia are stimulated by up-regulated VEGF [24], while the Müller cells release VEGF for vessel growth in the deep vascular plexus [25].
In this study, we used OCTA to visualize retinal vascular structure. Compared with control group, abnormal avascular zone and neovascularization area could be clearly observed in OIR mice.
The result of proteomic analysis displayed that a lot of proteins participated in pathological angiogenesis. Heat shock proteins (HSPs) were significantly upregulated proteins, which plays an important role in angiogenesis [26]. The expression of HSPB1 is increased in DR and cause retinal injury by enhancing VEGF expression [27, 28]. HSPC could induce retinal neovascularization by regulating HIF-1 and VEGF [29]. α-crystallins, which are small heat shock proteins, were detected to be expressed severely. Their expression is dramatically upregulated in numerous retinal diseases, such as mechanical injury, ischemic insults, age-related macular degeneration (AMD), and DR [30].
Erythropoietin (EPO) production can be stimulated by hypoxia, and have a direct relationship, and both are stimulated by HIF-1 [31]. EPO have promotional functions in vascular proliferation and endothelial cell proliferation [32]. We also detected a significantly differential protein, U4/U6.U5 tri-snRNP-associated protein 1, which is one of the building blocks of the spliceosome and plays a role in hypoxia-induced regulation of EPO gene expression [33].
Neuronal tissue in OIR
The results revealed that in addition to the changes detected on proteins responding to hypoxia and inducing pathological neovascularization, some changes in protein expression took place in the neuronal tissue of the retina. As previous researches showed, not only angiogenesis, but also neurodevelopment was affected by abnormal oxygen environment [16]. The effect of hyperoxia-hypoxia induction is harmful to the retinal neurons due to the presence of oxidative stress [34].
ROP can also be considered one of the neurodegeneration diseases. The hypoxic damage could induce the production of free radicals, inadequate blood supply and other inflammatory actions, along with the apoptotic effect in neuronal cells [35]. It has been reported that persistent ectopic synapses, prolonged cellular apoptosis, and gliosis exist in the OIR retina [36], which is similar with ROP pathogenesis. Many children with a history of ROP show persistent vision impairment, and there is evidence of an association between ROP and neurosensory disabilities [37]. A study shows that preterm infants with severe ROP have a significantly poorer outcome at 11 years than preterm infants without ROP, with neurosensory impairments detected in 50% of the infants who had suffered from severe ROP [38]. We found that OIR showed an up-regulated expression of 14–3-3 proteins, which have proved the importance of the 14–3-3 protein family in the development of the nervous system [39].
Glial cells participate in neuroinflammation and synaptic homeostasis, the latter being essential for maintaining the physiological function of the central nervous system (CNS) [40]. Retinal microglia have been noted to be involved in the OIR model while their role in the OIR process is still unclear. In mouse OIR model, the number of retinal microglia was increased and associated with a loss of deep retinal vessels [10]. In addition, it has been observed that microglia are the predominant myeloid cells in the neovascularization area in mouse OIR [41]. Further study showed that microglia might cooperate with astrocytes, promote the regrowth of blood vessels after vascular occlusion in OIR, and thus result in less neovascularization afterward in the mouse OIR model [42]. In our study, we detected some proteins associated with the differentiation and metabolism of glial cell. Up-regulated expression of these proteins corresponded to the activity of glial cells in OIR. These mediators might play a potential protective role in the inflammatory and apoptotic conditions of ischemic injury.
Proteins involved in ncRNAs in OIR
GO analysis demonstrated that peptide biosynthetic and metabolic process, cellular macromolecule biosynthetic process and nucleic acid binding had the major role in significant enrichment. KEGG enrichment analysis indicated that a large number of differential proteins were mainly expressed in upregulation and involved in non-coding RNAs (ncRNAs), including spliceosome, RNA transport, rRNA processing and mRNA surveillance. NcRNAs are functional RNAs that are not translated into proteins and regulate various retinal diseases [43]. Based on their molecular weight, they are classified into microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) [44]. In existing research, miRNAs are the most extensively explored ncRNAs about ROP. MiR-18a-5p and miR-145, which are up-regulated in mouse OIR model, could enhance pathological neovascularization [45, 46]. Some studies have uncovered the effect of partial lncRNAs in ROP, which alleviates retinal neovascularization by dampening the Akt/VEGF pathway [47,48,49]. Some circRNAs in ROP have been reported to regulating retinal neovascularization [50, 51]. In this study, we detected a large number of proteins that are involved in ncRNA regulation of OIR. We identified some DEPs encoded by serine/arginine-rich splicing factors (SRSF) family. SRSF family plays an essential role in the progression of neurodevelopment [52], and has been reported in mouse OIR model [53]. In addition, we detected some of small nuclear ribonucleoproteins (snRNPs). It has been proved that snRNP contributes ischemia-hypoxia regulation [54]. RNA-binding proteins (RBPs) were also significantly differential proteins detected in this study. RBPs are critical effectors of gene expression, and as such their malfunction could induce many diseases [55]. It has been proved that RBPs also participate in diabetic retinopathy (DR) and affect retinal neovascularization through binding to VEGF and increasing its stability [56,57,58]. Protein splicing factors play key roles in mediating the progression of the spliceosome pathway [59], and we identified plenty of differential splicing factor subunits expressed significant differently in OIR model.
Proliferation and repairment after anti-VEGF treatment
Via OCTA image, we observed that eyes taking anti-VEGF injection had less abnormal retinal area, comparing to those without treatment. As shown in previous animal studies and clinical trials, intravitreal injection of anti-VEGF displayed a beneficial effect on reducing neovascular area and promoting vessels growth toward the peripheral retina after treatment [60, 61].
GO enrichment analysis demonstrated that among the cellular macromolecule and nucleic acid metabolic process showing strong positive correlations with the ranibizumab group, DNA replication and its correlative metabolites displayed the most significant enrichment. The IPR enrichment analysis showed that a large number of proteins about MCM was detected in the ranibizumab group, including MCM3, MCM4, MCM5, MCM7 and their fragments. Past research has shown that MCM played a necessary role in the biologic processes and pathways of DNA replication [62, 63], DNA repair [64], cell cycle [65, 66], et al. MCM5 has been prove to be an indispensable role in the zebrafish retina for ensuring efficient genomic duplication and cell-cycle progression [67].
KEGG enrichment analysis indicated that Jak/STAT and HIF-1 were major down-regulated signaling pathways expressed in treatment group. It has been reported that activated STAT3 to lead to neovascularization by activation of NADPH oxidase [68]. And HIF-1 is well-known as a stimulus that leads to an increase in VEGF and angiogenesis [69]. EPO could also be stimulated by HIF-1 in a hypoxia environment [31], and promote vascular proliferation. From the view of result, intravitreal injection of anti-VEGF could control abnormal angiogenesis by these pathways.
Among the up-regulated proteins, we detected a great deal of collagens and fibronectins enriched in PI3K/Akt signaling pathway. It might be related to degeneration of neovascularization. The studies have shown that collagen and laminin-entactin enhances the stability of vessels [70]. In addition, low concentrations of type IV collagen promote elongation, and high concentrations of type IV collagen stabilize microcapillaries [71].
Conclusion
In conclusion, our study revealed that the proteomic characteristics of retinal tissue in mouse OIR model were significantly different from the proteomic characteristics of the control group. Further analysis between the ranibizumab and OIR group indicated partial signaling pathways and DEPs were affected by intravitreal injection of anti-VEGF. These findings may be useful for identification of novel biomarkers for ROP pathogenesis and treatment.
Material and methods
Animals
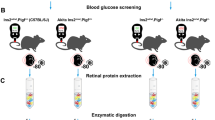
In this study, SPF C57BL/6 J mice (purchased from Hangzhou hangsi Biotechnology Co., Ltd. (Hangzhou, China)) were housed under standard conditions with 12-h dark/12-h light cycle and fed with standard laboratory pellets and water ad libitum. The OIR model was generated as described in previous study [8]. To sum up, the newborn mice at postnatal day 7 (P7) and their nursing mothers were exposed to 75% ± 1% oxygen environment for 5 days until P12. After that, they were returned to normal room air to induce OIR. On the other hand, OIR mice were divided into two groups. Intraocular injections of 5 μg /0.5 µL of ranibizumab into the vitreous body (group A) were performed at P13 using 33-gauge needles (Hamilton, Bonaduz, Switzerland). Group B didn’t take any therapeutic intervention but intraocular injections of 0.5µL phosphate-buffered saline (PBS; Proteintech Inc, USA). Moreover, another group of normal newborn mice (control group, group C) were housed under normal room air conditions. All mice were humanely euthanized with an intraperitoneal injection of saline-diluted pentobarbital sodium (200 mg/kg) and eyeballs collected at P17 (late hypoxic phase and the peak of neovascularization) to assess the effect of OIR and anti-VEGF treatment on retinal proteome. The study design is described in Fig. 9. In previous study, it demonstrated that postnatal weight would affect outcome in the OIR model [72]. In this study, only the pups weighing between 6.3 and 7.5 g at P17 were included in the study.
OCTA Examination
Optical coherence tomography angiography (OCTA) examination of the fundus in each group of mice was performed using Beiming-Kun (400,000 ultrawide-angle, full-field sweep OCT) from Toward Pi (Beijing) Medical Technology Ltd. (Beijing, China). All pups were pharmacologically dilated after receiving intraperitoneal anesthesia (1.25% tribromoethanol) according to the manufacturer’s operating instructions to capture high resolution scan imaging images of the various retinal layers.
We picked enrolled mice by OCTA images. For ranibizumab and OIR group, the inclusion criteria are abnormal fundus vascular, including pathological avascular area and neovascularization. After OCTA examination, pups were decapitated and eyes enucleated.
Proteomics
Detailed methods of protein extraction, trypsin digestion, HPLC fractionation, modification enrichment and LC–MS analysis were described in Supplementary Data.
Immunohistochemistry
Enucleated eyes were fixed with 4% paraformaldehyde (PFA) for 1 h, and processed for paraffin embedding. Five-micrometer thick sections were subjected to antigen retrieval (20xsodium citrate antigen retrieval solution, pH6.0), blocked and incubated either with HIF-1α antibody (66,730–1-Ig; 1:50; Proteintech, Rosemont, IL, USA) or VEGFA polyclonal antibody (19,003–1-ap; 1:200; Proteintech, Rosemont, IL, USA) followed by horseradish peroxidase (HRP) conjugated secondary antibodies. Samples were imaged confocal laser microscope (KF-PRO-120; KFBIO; Ningbo, China).
Statistical analysis
All data were analyzed by SPSS 25.0 software (IBM, Chicago, IL, USA). The normal distribution of the data was confirmed by the Shapiro‒Wilk and Kolmogorov–Smirnov tests. T test and Mann–Whitney U test were used to compare differences between two groups. The differences at p < 0.05 were considered statistically significant.
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD045378.
References
Lee M, Hall ES, Taylor M, DeFranco EA. Regional contribution of previable infant deaths to infant mortality rates in the United States. Am J Perinatol. 2021;38(2):158–65.
Blencowe H, Krasevec J, de Onis M, Black RE, An X, Stevens GA, Borghi E, Hayashi C, Estevez D, Cegolon L, et al. National, regional, and worldwide estimates of low birthweight in 2015, with trends from 2000: a systematic analysis. Lancet Glob Health. 2019;7(7):e849–60.
Wood EH, Chang EY, Beck K, Hadfield BR, Quinn AR, Harper CA 3rd. 80 Years of vision: preventing blindness from retinopathy of prematurity. J Perinatol. 2021;41(6):1216–24.
Stahl A, Sukgen EA, Wu WC, Lepore D, Nakanishi H, Mazela J, Moshfeghi DM, Vitti R, Athanikar A, Chu K, et al. Effect of intravitreal aflibercept vs laser photocoagulation on treatment success of retinopathy of prematurity: the firefleye randomized clinical trial. JAMA. 2022;328(4):348–59.
Kang HG, Choi EY, Byeon SH, Kim SS, Koh HJ, Lee SC, Kim M. Intravitreal ranibizumab versus laser photocoagulation for retinopathy of prematurity: efficacy, anatomical outcomes and safety. Br J Ophthalmol. 2019;103(9):1332–6.
Graziosi A, Perrotta M, Russo D, Gasparroni G, D’Egidio C, Marinelli B, Di Marzio G, Falconio G, Mastropasqua L, Li Volti G, et al. Oxidative stress markers and the retinopathy of prematurity. J Clin Med. 2020;9(9):2711.
Murakami T, Okamoto F, Kinoshita T, Shinomiya K, Nishi T, Obata S, Ogura S, Nishihara Y, Tsukitome H, Jujo T, et al. Comparison of long-term treatment outcomes of laser and anti-VEGF therapy in retinopathy of prematurity: a multicentre study from J-CREST group. Eye. 2023;37:3589.
Connor KM, Krah NM, Dennison RJ, Aderman CM, Chen J, Guerin KI, Sapieha P, Stahl A, Willett KL, Smith LE. Quantification of oxygen-induced retinopathy in the mouse: a model of vessel loss, vessel regrowth and pathological angiogenesis. Nat Protoc. 2009;4(11):1565–73.
Zhou Y, Tan W, Zou J, Cao J, Huang Q, Jiang B, Yoshida S, Li Y. Metabolomics analyses of mouse retinas in oxygen-induced retinopathy. Invest Ophthalmol Vis Sci. 2021;62(10):9.
Vessey KA, Wilkinson-Berka JL, Fletcher EL. Characterization of retinal function and glial cell response in a mouse model of oxygen-induced retinopathy. J Comp Neurol. 2011;519(3):506–27.
Grotegut P, Perumal N, Kuehn S, Smit A, Dick HB, Grus FH, Joachim SC. Minocycline reduces inflammatory response and cell death in a S100B retina degeneration model. J Neuroinflammation. 2020;17(1):375.
Xue M, Ke Y, Ren X, Zhou L, Liu J, Zhang X, Shao X, Li X. Proteomic analysis of aqueous humor in patients with pathologic myopia. J Proteomics. 2021;234:104088.
Sugioka K, Saito A, Kusaka S, Kuniyoshi K, Shimomura Y. Identification of vitreous proteins in retinopathy of prematurity. Biochem Biophys Res Commun. 2017;488(3):483–8.
Xu M, Jiang Y, Su L, Chen X, Shao X, Ea V, Shang Z, Zhang X, Barnstable CJ, Li X, et al. Novel regulators of retina neovascularization: a proteomics approach. J Proteome Res. 2022;21(1):101–17.
Zasada M, Suski M, Bokiniec R, Szwarc-Duma M, Borszewska-Kornacka MK, Madej J, Bujak-Gizycka B, Madetko-Talowska A, Revhaug C, Baumbusch LO, et al. An iTRAQ-based quantitative proteomic analysis of plasma proteins in preterm newborns with retinopathy of prematurity. Invest Ophthalmol Vis Sci. 2018;59(13):5312–9.
Vähätupa M, Nättinen J, Jylhä A, Aapola U, Kataja M, Kööbi P, Järvinen TAH, Uusitalo H, Uusitalo-Järvinen H. SWATH-MS proteomic analysis of oxygen-induced retinopathy reveals novel potential therapeutic targets. Invest Ophthalmol Vis Sci. 2018;59(8):3294–306.
Uemura A, Fruttiger M, D’Amore PA, De Falco S, Joussen AM, Sennlaub F, Brunck LR, Johnson KT, Lambrou GN, Rittenhouse KD, et al. VEGFR1 signaling in retinal angiogenesis and microinflammation. Prog Retin Eye Res. 2021;84:100954.
Wallsh JO, Gallemore RP. Anti-VEGF-resistant retinal diseases: a review of the latest treatment options. Cells. 2021;10(5):1049.
Pesce NA, Canovai A, Plastino F, Lardner E, Kvanta A, Cammalleri M, André H, Dal Monte M. An imbalance in autophagy contributes to retinal damage in a rat model of oxygen-induced retinopathy. J Cell Mol Med. 2021;25(22):10480–93.
Higgins RD. Oxygen saturation and retinopathy of prematurity. Clin Perinatol. 2019;46(3):593–9.
Chan-Ling T, Gole GA, Quinn GE, Adamson SJ, Darlow BA. Pathophysiology, screening and treatment of ROP: a multi-disciplinary perspective. Prog Retin Eye Res. 2018;62:77–119.
Hartnett ME. Pathophysiology and mechanisms of severe retinopathy of prematurity. Ophthalmology. 2015;122(1):200–10.
Fulton AB, Hansen RM, Moskowitz A, Akula JD. The neurovascular retina in retinopathy of prematurity. Prog Retin Eye Res. 2009;28(6):452–82.
Provis JM. Development of the primate retinal vasculature. Prog Retin Eye Res. 2001;20(6):799–821.
Gariano RF, Gardner TW. Retinal angiogenesis in development and disease. Nature. 2005;438(7070):960–6.
Bellini S, Barutta F, Mastrocola R, Imperatore L, Bruno G, Gruden G. Heat shock proteins in vascular diabetic complications: review and future perspective. Int J Mol Sci. 2017;18(12):2709.
Du Y, Tang J, Li G, Berti-Mattera L, Lee CA, Bartkowski D, Gale D, Monahan J, Niesman MR, Alton G, et al. Effects of p38 MAPK inhibition on early stages of diabetic retinopathy and sensory nerve function. Invest Ophthalmol Vis Sci. 2010;51(4):2158–64.
Lee YJ, Lee HJ, Choi SH, Jin YB, An HJ, Kang JH, Yoon SS, Lee YS. Soluble HSPB1 regulates VEGF-mediated angiogenesis through their direct interaction. Angiogenesis. 2012;15(2):229–42.
Jo DH, An H, Chang DJ, Baek YY, Cho CS, Jun HO, Park SJ, Kim JH, Lee HY, Kim KW, et al. Hypoxia-mediated retinal neovascularization and vascular leakage in diabetic retina is suppressed by HIF-1α destabilization by SH-1242 and SH-1280, novel hsp90 inhibitors. J Mol Med (Berl). 2014;92(10):1083–92.
Kannan R, Sreekumar PG, Hinton DR. Novel roles for α-crystallins in retinal function and disease. Prog Retin Eye Res. 2012;31(6):576–604.
Watts KD, McColley SA. Elevated vascular endothelial growth factor is correlated with elevated erythropoietin in stable, young cystic fibrosis patients. Pediatr Pulmonol. 2011;46(7):683–7.
Chen J, Connor KM, Aderman CM, Smith LE. Erythropoietin deficiency decreases vascular stability in mice. J Clin Investig. 2008;118(2):526–33.
Gupta M, Mungai PT, Goldwasser E. A new transacting factor that modulates hypoxia-induced expression of the erythropoietin gene. Blood. 2000;96(2):491–7.
Subirada PV, Paz MC, Ridano ME, Lorenc VE, Fader CM, Chiabrando GA, Sánchez MC. Effect of autophagy modulators on vascular, glial, and neuronal alterations in the oxygen-induced retinopathy mouse model. Front Cell Neurosci. 2019;13:279.
Lalkovičová M, Danielisová V. Neuroprotection and antioxidants. Neural Regen Res. 2016;11(6):865–74.
Mezu-Ndubuisi OJ, Macke EL, Kalavacherla R, Nwaba AA, Suscha A, Zaitoun IS, Ikeda A, Sheibani N. Long-term evaluation of retinal morphology and function in a mouse model of oxygen-induced retinopathy. Mol Vis. 2020;26:257–76.
Lucchesi M, Marracci S, Amato R, Filippi L, Cammalleri M, Dal Monte M. Neurosensory alterations in retinopathy of prematurity: a window to neurological impairments associated to preterm birth. Biomedicines. 2022;10(7):1603.
Farooqi A, Hägglöf B, Sedin G, Serenius F. Impact at age 11 years of major neonatal morbidities in children born extremely preterm. Pediatrics. 2011;127(5):e1247-1257.
Cornell B, Toyo-Oka K. 14-3-3 Proteins in brain development: neurogenesis, neuronal migration and neuromorphogenesis. Front Mol Neurosci. 2017;10:318.
Kim YS, Choi J, Yoon BE. Neuron-glia interactions in neurodevelopmental disorders. Cells. 2020;9(10):2176.
Boeck M, Thien A, Wolf J, Hagemeyer N, Laich Y, Yusuf D, Backofen R, Zhang P, Boneva S, Stahl A, et al. Temporospatial distribution and transcriptional profile of retinal microglia in the oxygen-induced retinopathy mouse model. Glia. 2020;68(9):1859–73.
Dorrell MI, Aguilar E, Jacobson R, Trauger SA, Friedlander J, Siuzdak G, Friedlander M. Maintaining retinal astrocytes normalizes revascularization and prevents vascular pathology associated with oxygen-induced retinopathy. Glia. 2010;58(1):43–54.
Karali M, Banfi S. Non-coding RNAs in retinal development and function. Hum Genet. 2019;138(8–9):957–71.
Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20(8):629–51.
Liu CH, Wang Z, Huang S, Sun Y, Chen J. MicroRNA-145 regulates pathological retinal angiogenesis by suppression of TMOD3. Mol Ther Nucleic Acids. 2019;16:335–47.
Guan JT, Li XX, Peng DW, Zhang WM, Qu J, Lu F, D’Amato RJ, Chi ZL. MicroRNA-18a-5p administration suppresses retinal neovascularization by targeting FGF1 and HIF1A. Front Pharmacol. 2020;11:276.
Di Y, Wang Y, Wang X, Nie QZ. Effects of long non-coding RNA myocardial infarction-associated transcript on retinal neovascularization in a newborn mouse model of oxygen-induced retinopathy. Neural Regen Res. 2021;16(9):1877–81.
Xia F, Xu Y, Zhang X, Lyu J, Zhao P. Competing endogenous RNA network associated with oxygen-induced retinopathy: expression of the network and identification of the MALAT1/miR-124-3p/EGR1 regulatory axis. Exp Cell Res. 2021;408(1):112783.
Wang Y, Wang X, Wang YX, Ma Y, Di Y. Effect and mechanism of the long noncoding RNA MALAT1 on retinal neovascularization in retinopathy of prematurity. Life Sci. 2020;260:118299.
Deng Y, Li S, Li S, Yu C, Huang D, Chen H, Yin X. CircPDE4B inhibits retinal pathological angiogenesis via promoting degradation of HIF-1α though targeting miR-181c. IUBMB Life. 2020;72(9):1920–9.
Liu C, Yao MD, Li CP, Shan K, Yang H, Wang JJ, Liu B, Li XM, Yao J, Jiang Q, et al. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7(11):2863–77.
Bustos F, Segarra-Fas A, Nardocci G, Cassidy A, Antico O, Davidson L, Brandenburg L, Macartney TJ, Toth R, Hastie CJ, et al. Functional diversification of srsf protein kinase to control ubiquitin-dependent neurodevelopmental signaling. Dev Cell. 2020;55(5):629-647.e627.
Zhou H, Song H, Wu Y, Liu X, Li J, Zhao H, Tang M, Ji X, Zhang L, Su Y, et al. Oxygen-induced circRNA profiles and coregulatory networks in a retinopathy of prematurity mouse model. Exp Ther Med. 2019;18(3):2037–50.
Schmidt-Kastner R, Yamamoto H, Hamasaki D, Yamamoto H, Parel JM, Schmitz C, Dorey CK, Blanks JC, Preising MN. Hypoxia-regulated components of the U4/U6.U5 tri-small nuclear riboprotein complex: possible role in autosomal dominant retinitis pigmentosa. Mol Vision. 2008;14:125–35.
Gebauer F, Schwarzl T, Valcárcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22(3):185–98.
Amadio M, Pascale A, Cupri S, Pignatello R, Osera C, V DA, AG DA, Leggio GM, Ruozi B, Govoni S et al. Nanosystems based on siRNA silencing HuR expression counteract diabetic retinopathy in rat. Pharmacol Res. 2016;111:713-720.
Chang SH, Hla T. Post-transcriptional gene regulation by HuR and microRNAs in angiogenesis. Curr Opin Hematol. 2014;21(3):235–40.
Chen X, Wu J, Li Z, Han J, Xia P, Shen Y, Ma J, Liu X, Zhang J, Yu P. Advances in the study of RNA-binding proteins in diabetic complications. Mol Metab. 2022;62:101515.
Chen HC, Cheng SC. Functional roles of protein splicing factors. Biosci Rep. 2012;32(4):345–59.
Sankar MJ, Sankar J, Chandra P. Anti-vascular endothelial growth factor (VEGF) drugs for treatment of retinopathy of prematurity. Cochrane Database Syst Rev. 2018;1(1):Cd009734.
Mintz-Hittner HA, Kennedy KA, Chuang AZ. Efficacy of intravitreal bevacizumab for stage 3+ retinopathy of prematurity. N Engl J Med. 2011;364(7):603–15.
Ishimi Y, Komamura-Kohno Y, Karasawa-Shimizu K, Yamada K. Levels of MCM4 phosphorylation and DNA synthesis in DNA replication block checkpoint control. J Struct Biol. 2004;146(1–2):234–41.
Ishimi Y, Komamura-Kohno Y, Kwon HJ, Yamada K, Nakanishi M. Identification of MCM4 as a target of the DNA replication block checkpoint system. J Biol Chem. 2003;278(27):24644–50.
Casey JP, Nobbs M, McGettigan P, Lynch S, Ennis S. Recessive mutations in MCM4/PRKDC cause a novel syndrome involving a primary immunodeficiency and a disorder of DNA repair. J Med Genet. 2012;49(4):242–5.
Fujita M, Yamada C, Tsurumi T, Hanaoka F, Matsuzawa K, Inagaki M. Cell cycle- and chromatin binding state-dependent phosphorylation of human MCM heterohexameric complexes. A role for cdc2 kinase. J Biol Chem. 1998;273(27):17095–101.
Komamura-Kohno Y, Karasawa-Shimizu K, Saitoh T, Sato M, Hanaoka F, Tanaka S, Ishimi Y. Site-specific phosphorylation of MCM4 during the cell cycle in mammalian cells. FEBS J. 2006;273(6):1224–39.
Ryu S, Holzschuh J, Erhardt S, Ettl AK, Driever W. Depletion of minichromosome maintenance protein 5 in the zebrafish retina causes cell-cycle defect and apoptosis. Proc Natl Acad Sci USA. 2005;102(51):18467–72.
Byfield G, Budd S, Hartnett ME. The role of supplemental oxygen and JAK/STAT signaling in intravitreous neovascularization in a ROP rat model. Invest Ophthalmol Vis Sci. 2009;50(7):3360–5.
Lajko M, Cardona HJ, Taylor JM, Shah RS, Farrow KN, Fawzi AA. Hyperoxia-induced proliferative retinopathy: early interruption of retinal vascular development with severe and irreversible neurovascular disruption. PLoS One. 2016;11(11):e0166886.
Nicosia RF, Bonanno E, Smith M, Yurchenco P. Modulation of angiogenesis in vitro by laminin-entactin complex. Dev Biol. 1994;164(1):197–206.
Bonanno E, Iurlaro M, Madri JA, Nicosia RF. Type IV collagen modulates angiogenesis and neovessel survival in the rat aorta model. In Vitro Cell Dev Biol Anim. 2000;36(5):336–40.
Stahl A, Connor KM, Sapieha P, Chen J, Dennison RJ, Krah NM, Seaward MR, Willett KL, Aderman CM, Guerin KI, et al. The mouse retina as an angiogenesis model. Invest Ophthalmol Vis Sci. 2010;51(6):2813–26.
Supplemental data
This article contains supplemental data.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceptualization, L.S.; methodology, Z.X., J.M., Y.C., Y.W., S.Z., H.C., and L.S..; software, Z.X., J.M., Y.C. and X.D.; validation, Z.X., Y.W., S.Z. and H.C.; formal analysis, Z.X., J.M., Y.C. and X.D.; investigation, Z.X., H.C., S.Z. and Y.W.; resources, X.D., and L.S..; data curation, Z.X., J.M. and Y.C.; writing—original draft preparation, Z.X. and L.S..; writing—review and editing, Z.X., Y.W., S.Z., H.C., J.Y., and L.S..; visualization, Z.X. and J.M.; supervision, J.Y., X.D., and L.S.. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All animal protocols were in accordance with the ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments). and approved and monitored by the ethics committee of Animal Care Committee of the Zhejiang Provincial People’s Hospital (protocol code ZH20220915087).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xu, Z., Wu, Y., Mao, J. et al. 4D label-free proteomics analysis of oxygen-induced retinopathy with or without anti-VEGF treatment. BMC Genomics 25, 415 (2024). https://doi.org/10.1186/s12864-024-10340-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10340-z