Abstract
Background
Retinal gene expression pattern is severely altered after exposition to hyperoxia in mice with oxygen-induced retinopathy (OIR), a common model of retinopathy of prematurity. Gene ontology and signaling pathway analyses may add new insights into a better understanding of the pathogenesis of this disease.
Methods
Seven-day-old C57BL/6J mice (n = 60) were exposed to 75% oxygen for 5 days and then recovered in room air. The controls (n = 60) were kept in the normoxic conditions. Retinas were harvested immediately following hyperoxia, during the phase of maximal neovascularization, and at the time of neovascularization regression. The retinal RNA samples were evaluated for gene expression using mouse gene expression microarrays. DAVID annotation tools were used for gene ontology and pathway analyses.
Results
The most significantly enriched signaling pathways during the neovascularization phase of OIR were: focal adhesion; ECM–receptor interaction; PI3K-Akt; oxidative phosphorylation; and Alzheimer’s, Parkinson’s and Huntington’s disease signaling pathways. Genes involved in apoptosis, cell proliferation, cell differentiation, and immune responses were associated with neovascularization regression.
Conclusions
Performed analyses revealed the possible involvement of various signaling pathways in OIR pathomechanism, mostly specific to the OIR phase. Dysregulation of genes involved in oxidative phosphorylation may have an impact on neovascularization development.
Similar content being viewed by others
Introduction
Retinopathy of prematurity (ROP) is a vision-threatening disorder affecting the immature retina of nearly one-third of infants with birth weight <1500 g.1 ROP develops in two phases: suppression of growth factors due to excessive oxygen treatment and cessation of the maternal–fetal interaction causes an arrest of retinal vascularization (phase 1). Afterward, increased metabolic activity of the hypoxic retina triggers abnormal vasoproliferation (phase 2), which ultimately may lead to retinal detachment and blindness.2 Until now, the regulatory mechanisms of ROP have not been completely uncovered. Transcriptome analysis provides information about gene expression alterations associated with the disorder and helps to obtain a thorough picture of dysregulated genes and pathways. A better understanding of processes that regulate neonatal retina injury course may help us find pathomechanisms to be potentially targeted by new therapeutic interventions.
In our previous article, we described the short- and long-term impact of hyperoxia on retinal cells’ gene expression level in a mouse model of oxygen-induced retinopathy (OIR), a common model of ROP.3 In this animal model, which reiterates both ROP phases, 7-day-old mice pups are treated with 75% oxygen for 5 days, and on postnatal day 12 (P12) they present with the obliteration of retinal vessels. Five days later (P17), there is a time of maximal retinal neovascularization. Interestingly, mice with OIR characterize with a spontaneous regression of abnormal retinal neo-vessels till day P25.4 In our study, we showed that overall 1519 genes (556 downregulated and 963 upregulated) had different expression levels in the retinal cells from OIR mouse model when compared to the normal control animals at three examined time points. Right after the end of hyperoxia (P12), the majority of differentially expressed genes (DEGs) were underexpressed (37 out of 38), whereas at the time of maximal neovascularization (P17), a large number of genes in the OIR retinas were overexpressed (855 out of 1370). Increased gene expression, though to a lesser extent, was still evident even in 28-day-old animals, age that corresponds to adolescence in a murine lifespan (107 out of 111).3
In the present study, we focused on analysis of biological processes and signaling pathways affected by the dysregulated genes in murine retinas with OIR at three time points: vaso-obliteration (P12), maximal neovascularization (P17), and after resolution of the retinal injury (P28), in an attempt to better understand the molecular mechanisms of oxygen-induced retinal damage and succeeding neovascularization regression.
Methods
Detailed methodology of the study was provided elsewhere.3 The Norwegian Animal Research Authority approved all experiments. Briefly, 7-day-old C57BL/6J mice litters (n = 60) with their mothers were exposed to 75% oxygen for 5 days (hyperoxia) and then returned to normoxic conditions for another 5 days, inducing relative hypoxia. Control animals (n = 60) were kept in room air (normoxia). One-third of the pups from both the experimental and control groups were sacrificed immediately after ending elevated oxygen exposure (P12), 5 days after their return to normoxic conditions (P17), and on the 28th day of life (P28). At each time point, the retinas were obtained and further processed for microarray analysis. Gene expression profiling was performed using SurePrint G3 Mouse Gene Expression 8 × 60 K Microarrays (Agilent, Santa Clara, USA), and the reverse transcription quantitative PCR (qPCR) method was used for validation of the results. Microarray data were analyzed by the GeneSpring GX software (Agilent, Santa Clara, USA). Raw microarray data were normalized, and subsequently, a moderated t test was applied to compare gene expression between the OIR and control group at each study time point. The results were corrected for multiple testing by the Benjamini–Hochberg procedure. A 0.05 significance level of the adjusted p values was used. In order to evaluate which predefined gene sets were significantly enriched between the study groups, the Database for Annotation, Visualization, and Integrated Discovery database (DAVID; version 6.8; https://david.ncifcrf.gov) annotation tools (gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis) were applied. Genes differentially expressed between the OIR and control group at each study time point, with p values adjusted for multiple testing to <0.05, were used as input for pathway enrichment analysis in DAVID.
Microarray gene expression data are available at GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession number: GSE130400.
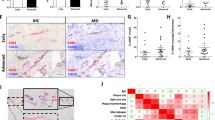
Eyes from randomly selected 36 pups (18 from the study group and 18 from the control group—6 animals per each harvesting time point) were used for the histologic verification of the model, according to the methodology provided by Connor et al.4 In brief, the eyes were enucleated and fixed with 4% paraformaldehyde solution (Boster Biological Technology, Pleasanton, CA). Then, under the dissecting microscope, the retinas were isolated, incubated in 0.5% Triton X-100 (Sigma, St. Louis, MO), and stained in the lectin solution (Alexa Fluor 594 – I21413, Molecular Probes). Then again, under a dissecting microscope, the retinas were mounted on a microscope slide and coverslipped with SlowFade medium (Thermo Fisher Scientific, Waltham, MA). Images were obtained using the Zeiss AxioCam MRc, Zeiss AxioObserver.Z1 microscope, and AxioVision 4.8 software (Fig. 1).
After further randomization, on P12, P17, and P28, 1/3 of the study and control animals were harvested, and retina tissue was obtained for histological and microarray analysis. a Comparison of the retinal flat mounts with retinal vessels stained with lectin from the OIR (H) and control (N) groups at different time points: P12, P17, and P28. b Volcano plots demonstrating alterations of specific genes in all analyzed time points, detected between control and OIR retinas colored by log2 fold change values. The vertical axis value—negative log of the p values, gray line—cut-off value for statistical significance (genes significantly differentially expressed between the study groups; corrected p value = 0.05), the horizontal axis—log2 change (FC fold change) between the two experimental conditions (OIR vs. normoxia). The number of underexpressed genes is shown in blue, whereas the number of overexpressed genes is shown in red.
Results
Identification of DEGs
A complete list of DEGs at every experimental time point with corrected p values and fold change values is provided in Supplementary Tables S1–S6.
GO term enrichment analysis
On P12, GO enrichment analysis of DEGs showed downregulation of biological processes involved in angiogenesis, cell adhesion, negative regulation of cell migration, and positive regulation of endothelial cell proliferation. On P17, the most downregulated biological process was transport, particularly regarding ions. Moreover, a significant overrepresentation of categories regarding relevant biological process, such as cell adhesion, angiogenesis, positive regulation of cell migration, response to hypoxia, and extracellular matrix organization, was observed in OIR retinas on P17. On the P28, immune system process, innate immune response, and response to lipopolysaccharide GO categories were upregulated in OIR retinas (Fig. 2). A complete list of the differentially enriched GO categories is provided in Supplementary Tables S8–S11.
Bar length represents the number of differentially expressed genes (DEGs) in OIR retinas compared with control retinas on P12, P17, and P28. Red bars indicate terms upregulated in OIR retinas; blue bars indicate terms downregulated in OIR retinas. On P17, the top five enriched terms in OIR retinas are shown. The color range is proportional to the log10 of the p value after Benjamini–Hochberg (BH) correction.
KEGG pathway analysis
To identify specific cellular pathways affected during the OIR course, we performed KEGG pathway analysis on the DEGs from the OIR vs. control mice retinas obtained on P12, P17, and P28. We did not find any significantly enriched pathways on P12. On P17, we identified a total of 45 and 19 significantly enriched pathways for upregulated and downregulated genes, respectively. The most significantly enriched pathways associated with neovascularization phase of OIR are presented in Table 1.
Genes of particular interest with altered expression involved in mitochondrial function—the oxidative phosphorylation (OXPHOS) signaling pathway—are presented in Fig. 3.
Figure 4 depicts correlation of selected genes of particular interest, with altered expression detected in OIR retinas at P17 involved in glycolysis, Krebs cycle, and OXPHOS.
On P28, only one KEGG pathway (osteoclast differentiation signaling pathway) was enriched with adequate statistical significance. A detailed list of all significantly enriched pathways is provided in Supplementary Table S7.
Identification of unique and jointly expressed genes
Comprehensive analysis of the genes differently expressed between the OIR and control groups at all experimental time points revealed a slight overlap (Fig. 5). Two genes (Ly6c1, Cxcl12) were continuously downregulated both on P12 and P17. Furthermore, 21 genes showed reversed expression during experimental time, from downregulation on P12 to upregulation on P17. These genes were associated with blood vessel formation and growth (PdgfB), immune response (Cd34, Cd248, Ly6c1, Cd93, Cxc112), or encoding adhesive molecules (Icam2). In case of four genes, we observed the inversion of the expression profile from downregulation on P12 to continued upregulation on P17 and P28 (Ctla2a, Tagln2, Plxnd1, Fn1).
We identified 77 genes significantly upregulated at the OIR group, both on P17 and P28. These were the ones known to be involved in apoptosis (Litaf, Dab2, Gadd45b, Lsp1, Ltbr, Rnf130, Tnfrsf1a), regulation of cell proliferation (Apobec1, Fgfr1, Ltbr, Sparc), cell differentiation (Elk, Dab2, Dclk1, Gadd45b, Pbxip1, Sema3b), and immune response (Ctss, H2-K1, Ltbr, Tnfrsf1a, Jak3, Adgre1, Csf1, Csf1r, Lilrb4a).
Discussion
The purpose of this study was to identify specific gene categories and signaling pathways altered over the course of murine OIR at three selected time points, such as the end of hyperoxia exposure (P12), maximal neovascularization (P17), and regression of neovascularization (P28).
Our comprehensive analysis of unique and jointly expressed genes between the OIR and control groups revealed that 5-day exposure to hyperoxia (determined at P12, vaso-obliteration phase) downregulated retinal genes involved in blood vessel formation, immune responses, and encoding adhesive molecules. Recovery from hyperoxia (determined at P17, maximal neovascularization phase) normalized the expression of those genes. Similarly, GO analysis of genes underexpressed at P12 revealed enrichment of GO terms associated with angiogenesis and positive regulation of endothelial cell proliferation, which might correspond to a loss of existing and delayed formation of new retinal vessels. Downregulation of the genes controlling cell adhesion might reflect the hyperoxia-driven damage of the cells and loss of their integrity. Our results are consistent with observations by Ishikawa et al., who identified the same categories of genes suppressed by hyperoxia in the OIR murine retinas.5
On P17, we found underexpression of the genes regulating the mitochondrial respiratory chain complex, including proteins involved in OXPHOS and ATP production. Most of these genes encode proteins involved in proton transport, therefore GO analysis revealed enrichment of transport and ion transport biological processes. Specifically, there were several underexpressed nuclear and mitochondrial genes encoding subunits of complex I (Ndufa1, Ndufa12, Ndufb3, Ndufb5, Ndufc2, Ndufs1, ND4). Ndufs1 and ND4 genes encoding the core subunits of the NADH dehydrogenase constitute a minimal protein cluster required for catalysis. We also detected underexpression of Uqcrc2, Uqcrfs1, Cox7c, and Cox5a genes encoding proteins essential in the complex III and IV assemblies. Moreover, we found diminished expression of the genes Atp5f1, Atp5g3, Atp5l, and Atp5k encoding these subunits of ATP synthase (complex V) that forms a proton channel. Simultaneous downregulation of numerous OXPHOS genes might disturb overall retinal ATP production. OXPHOS downregulation might be a part of hypoxia-induced factor-1-dependent adaptation to hypoxic conditions.6 However, we believe that the downregulation of OXPHOS genes at P17 was a maladaptive response to vaso-obliteration. By P17, neovascularization reached its peak. Since the metabolic rate is an important force behind angiogenesis, then non-oxidative ATP generation (glycolytic) must be initiated as OXPHOS becomes limited. Indeed, genes encoding enzymes important in other stages of cellular respiration showed significant alterations on P17. Namely, we observed overexpression of Pfkfb3 gene encoding a kinase that controls glycolysis and allows glucose shunt toward glycolysis. Overexpression of pyruvate dehydrogenase kinase (Pdk) gene might result in increased production of PDK enzyme, which acts to inactivate pyruvate dehydrogenase. In addition, genes (Pdhx, Pdp1) encoding subunits of pyruvate dehydrogenase complex, an enzyme located in the mitochondrial matrix and which converts pyruvate into acetyl-CoA linking glycolysis to Krebs cycle, were underexpressed. Consequently, pyruvate might be not converted into acetyl-CoA required in the mitochondrial energy production, instead it might be transformed into lactate. In addition, we observed overexpression of Ldha gene, product of which, lactate dehydrogenase, catalyzes the inter-conversion of pyruvate into lactate with concomitant inter-conversion of NADH and NAD+. Importantly, lactate directly or via its receptor GPR81 can trigger angiogenesis in OIR.7 We also detected underexpression of several genes encoding enzymes involved in Krebs cycle, such as Idh3a, Sucla2, Sdhaf4, and Mdh1. According to Sapieha et al., Krebs cycle metabolites, such as succinate, accumulate in the hypoxic retinas, where they act as potent mediators of vessel growth in addition to their feedback inhibition of some Krebs cycle enzymes.8
An interesting set of enriched pathways on P17 included those implicated in neurodegenerative disorders. In the complex pathomechanism of Alzheimer’s disease (AD), Parkinson’s disease (PD), and Huntington’s disease (HD), mitochondrial dysfunction plays an essential role.9,10,11 Namely, impaired (for example, by oligomeric intracellular β-amyloid in AD) mitochondrial respiratory chain complex activity results in ATP depletion, reduction of the mitochondrial membrane potential, increased ROS production, and consequently cell death. On the other hand, mitochondrial dysfunction itself may initiate abnormal production of the proteins involved in the pathogenesis of AD and PD.9 In our study, a majority of genes enriching signaling pathways associated with neurodegenerative diseases were the underexpressed and represent genes encoding proteins forming the respiratory chain and involved in OXPHOS. As the retina is a cerebral outgrowth, it is conceivable that pathological processes that occur in the developing retina also can happen in the brain. Since mitochondrial dysfunction was hypothesized to cause developmental origins of adult diseases, it would be interesting to verify whether the one observed after experimentally mimicking ROP development may trigger a long-lasting neurodegenerative process. Recently, Benterud et al. showed that perinatal oxygenation disturbances leading to oxidative stress might alter the level of β-amyloid in piglets.12
On P17, among the genes with increased expression, we identified several enriched pathways known to be involved in retinal hypoxia-driven angiogenesis, such as the phosphoinositide-3-kinase-Akt signaling pathway,13 focal adhesion signaling,14 and the extracellular matrix (ECM)–receptor interaction signaling pathway.15 Amebiasis signaling pathway shares several genes with ECM–receptor interaction signaling pathway. Therefore, we suppose that enrichment of this pathway points an extensive remodeling of retinal tissues occurring during the neovascularization OIR phase. Also, signaling pathways associated with cancers might be enriched owing to altered expression of genes related to angiogenesis and immunity.
Importantly, analysis of jointly and unique expressed genes revealed that genes involved in apoptosis, cell proliferation, cell differentiation, and immune response were constantly overexpressed both at P17 and P28 time points. As P28 was characterized by regression of OIR and normalization of retinal vasculature, it suggests that these genes are associated with regression from OIR and vascular normalization and that these processes are active from the neovascularization phase. In addition, signaling pathway analysis by pointing upregulation of osteoclast differentiation signaling pathway underlined the role of retinal microglia activation in the OIR regression.16 Retinal microglia are the resident ocular macrophages that play an important role in host defense against invading microorganisms, immunoregulation, and tissue repair.17 They are involved in phagocytosis of dying neurons.18 However, their prolonged activation could lead to neurodegeneration.19 We identified increased expression of Csf1 and Csf1r genes encoding colony-stimulating factor 1 (Csf1) and its receptor (Csf1r), respectively. Csfr1 signaling is essential in both microglia development as well as maintaining their viability.20 The prominent expression of the Tyrobp and Trem2 genes encoding cell-surface proteins (Dap12 and Trem2) involved in immune response were identified both on P17 (shown in Supplementary Materials) and P28. We assumed that they might reflect ongoing microglial activation in the OIR retinas. In addition, overexpression of Cyba gene, encoding the p22phox protein, one of the nicotinamide adenine dinucleotide phosphate oxidase components, might correspond to increased phagosome inflammatory response of the microglial cells.21 Our observations were in agreement with the findings of Fischer et al., who showed that activation of the retinal microglia correlated well with the retinal neovascularization in a mouse model of OIR.22
Limitations
We believe that the most significant limitation of our study is only partial characterization of OIR retinas, which was exclusively based on their genetic content and without any corresponding proteomic profiling. Moreover, we analyzed the entire retinas; thereby, we identified changes in gene expression and their assumed signaling contributions provided by all the retinal cells as a whole. Additional and more detailed studies are needed to evaluate fractional contribution from each specific component of the retina. For example, identification and cell sorting could be introduced to further support data regarding microglial activation. Also, our array results should be more widely verified by other methods to match the DEGs aligned to expected physiological changes at different time points. We believe that applicability of our findings might be further supported by a tighter time-dependent profile, such as qPCR analysis of gene expression of physiologically essential genes described above at P14 and P15 as well.
We are also aware of the fact that the mouse model of OIR, although mimics both ROP phases, deviates from the human situation in several aspects. Extremely low gestational age neonates (ELGANs) who are at the highest risk for ROP have incomplete retinal vascularization when exposed to hyperoxia, whereas 7-day-old mice pups when starting hyperoxia exposure have complete inner plexus vascularization. Moreover, the vast majority of ELGANs requiring supplemental oxygen with respiratory support frequently experience intermittent hypoxemic and hyperoxemic episodes caused by recurrent apneas due to immature development of the respiratory neuronal network.23,24 To minimize oxidative injury, high FiO2 is avoided. Therefore, constant exposure to 75% O2 for 5 days may be clinically irrelevant. The retinal genetic profile noted at P12 following constant stream of hyperoxia does not necessarily represent the retinal genetic profiles of ELGANs, who frequently experience intermittent hypoxemic and hyperoxemic episodes during oxygen therapy and who are at high risk for development of ROP. In addition, some pathological features of the mouse retinal vessels are different from human ROP. In mice, the central vessels, and not the peripheral vessels, are obliterated. In ROP, the peripheral vessels also fail to develop. Last but not least, in the mouse model, there is spontaneously resolution or regression of OIR with re-establishment of healthy-looking vessels by P25.25 ELGANs with severe ROP worsen without treatment and sometimes even with anti-ROP therapy.
Conclusions
To sum up, in this study, we showed that the OXPHOS signaling pathway is downregulated during neovascularization phase in a mouse model of OIR. In addition, our results point that hyperoxia exposure with subsequent relative hypoxia may have an impact on the altered expression of genes related to neurodegenerative conditions. Our study also demonstrated that genes involved in apoptosis, cell proliferation, cell differentiation, and immune responses are associated with regression from OIR and vascular normalization and that they are already overexpressed in the neovascularization phase.
References
Cavallaro, G. et al. The pathophysiology of retinopathy of prematurity: an update of previous and recent knowledge. Acta Ophthalmol. 92, 2–20 (2014).
Hellström, A., Smith, L. E. & Dammann, O. Retinopathy of prematurity. Lancet 382, 1445–1457 (2013).
Zasada, M. et al. Short- and long-term impact of hyperoxia on the blood and retinal cells’ transcriptome in a mouse model of oxygen-induced retinopathy. Pediatr. Res. https://doi.org/10.1038/s41390-019-0598-y (2019).
Connor, K. M. et al. Quantification of oxygen-induced retinopathy in the mouse: a model of vessel loss, vessel regrowth and pathological angiogenesis. Nat. Protoc. 4, 1565–1573 (2009).
Ishikawa, K. et al. Gene expression profile of hyperoxic and hypoxic retinas in a mouse model of oxygen-induced retinopathy. Invest. Ophthalmol. Vis. Sci. 51, 4307–4319 (2010).
Fuhrmann, D. C. & Brüne, B. Mitochondrial composition and function under the control of hypoxia. Redox Biol. 12, 208–215 (2017).
Madaan, A. et al. Müller cell-localized G-protein-coupled receptor 81 (hydroxycarboxylic acid receptor 1) regulates inner retinal vasculature via Norrin/Wnt pathways. Am. J. Pathol. 189, 1878–1896 (2019).
Sapieha P., et al. The succinate receptor GPR91 in neurons has a major role in retinal angiogenesis. Nat. Med. 14, 1067–1076 (2008).
Perez Ortiz, J. M. & Swerdlow, R. H. Mitochondrial dysfunction in Alzheimer’s disease: role in pathogenesis and novel therapeutic opportunities. Br. J. Pharmacol. 176, 3489–3507 (2019).
Park, J. S., Davis, R. L. & Sue, C. M. Mitochondrial dysfunction in Parkinson’s disease: new mechanistic insights and therapeutic perspectives. Curr. Neurol. Neurosci. Rep. 18, 21 (2018).
Carmo, C., Naia, L., Lopes, C. & Rego, A. C. Mitochondrial dysfunction in Huntington’s disease. Adv. Exp. Med. Biol. 1049, 59–83 (2018).
Benterud, T. et al. Perinatal asphyxia may influence the level of beta-amyloid (1-42) in cerebrospinal fluid: an experimental study on newborn pigs. PLoS ONE 10, e0140966 (2015).
Zhang, Z., Yao, L., Yang, J., Wang, Z. & Du, G. PI3K/Akt and HIF 1 signaling pathway in hypoxia ischemia (Review). Mol. Med. Rep. 18, 3547–3554 (2018).
Kornberg, L. J., Shaw, L. C., Spoerri, P. E., Caballero, S. & Grant, M. B. Focal adhesion kinase overexpression induces enhanced pathological retinal angiogenesis. Invest. Ophthalmol. Vis. Sci. 45, 4463–4469 (2004).
Khan, Z. A. & Chakrabarti, S. Cellular signaling and potential new treatment targets in diabetic retinopathy. Exp. Diabetes Res. 2007, 31867 (2007).
Weigelt, K. et al. Dap12 expression in activated microglia from retinoschisin-deficient retina and its PU.1-dependent promoter regulation. J. Leukoc. Biol. 82, 1564–1574 (2007).
Chen, L., Yang, P. & Kijlstra, A. Distribution, markers, and functions of retinal microglia. Ocul. Immunol. Inflamm. 10, 27–39 (2002).
Takahashi, K., Rochford, C. D. & Neumann, H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 201, 647–657 (2005).
Schuetz, E. & Thanos, S. Microglia-targeted pharmacotherapy in retinal neurodegenerative diseases. Curr. Drug Targets 5, 619–627 (2004).
Elmore, M. R. et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014).
Bodea, L. G. et al. Neurodegeneration by activation of the microglial complement-phagosome pathway. J. Neurosci. 34, 8546–8556 (2014).
Fischer, F., Martin, G. & Agostini, H. T. Activation of retinal microglia rather than microglial cell density correlates with retinal neovascularization in the mouse model of oxygen-induced retinopathy. J. Neuroinflammation 8, 120 (2011).
Di Fiore, J. M., Martin, R. J. & Gauda, E. B. Apnea of prematurity-perfect storm. Respir. Physiol. Neurobiol. 189, 213–222 (2013).
Martin, R. J., Di Fiore, J. M., Macfarlane, P. M. & Wilson, C. G. Physiologic basis for intermittent hypoxic episodes in preterm infants. Adv. Exp. Med. Biol. 758, 351–358 (2012).
Smith, L. E. et al. Oxygen-induced retinopathy in the mouse. Invest. Ophthalmol. Vis. Sci. 35, 101–111 (1994).
Acknowledgements
We thank Monika Atneossen-Asseng and Grethe Dyrhaug for technical assistance and Agate Noer and Wojciech Zasada for cooperation during procurement of the retina pictures. This work was supported by Polish-Norwegian Research Programme, operated by the National Centre for Research and Development under the Norwegian Financial mechanism 2009-2014 in the frame of Project Contract No. Pol-Nor/196065/54/2013.
Author information
Authors and Affiliations
Contributions
Substantial contributions to conception and design—M.Z., C.R., A.M.-T., A.G.W.R., O.D.S., M.B.-M., J.J.P., P.K.; acquisition of data—M.Z., A.M.-T., C.R., A.G.W.R., T.K., K.S., A.G.; analysis and interpretation of data—A.M.-T., M.Z.; drafting the article or revising it critically for important intellectual content—M.Z., A.M.-T., C.R., L.O.B., O.D.S., M.B.-M., J.J.P., P.K.; final approval of the version to be published—all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zasada, M., Madetko-Talowska, A., Revhaug, C. et al. Transcriptome analysis reveals dysregulation of genes involved in oxidative phosphorylation in a murine model of retinopathy of prematurity. Pediatr Res 88, 391–397 (2020). https://doi.org/10.1038/s41390-020-0793-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-020-0793-x
- Springer Nature America, Inc.
This article is cited by
-
Gene expression profile of the murine ischemic retina and its response to Aflibercept (VEGF-Trap)
Scientific Reports (2021)