Abstract
Pinellia ternata (Thunb.) Briet., a valuable herb native to China, is susceptible to the “sprout tumble” phenomenon because of high temperatures, resulting in a significant yield reduction. However, the molecular regulatory mechanisms underlying the response of P. ternata to heat stress are not well understood. In this study, we integrated transcriptome and miRNAome sequencing to identify heat-response genes, microRNAs (miRNAs), and key miRNA-target pairs in P. ternata that differed between heat-stress and room-temperature conditions. Transcriptome analysis revealed extensive reprogramming of 4,960 genes across various categories, predominantly associated with cellular and metabolic processes, responses to stimuli, biological regulation, cell parts, organelles, membranes, and catalytic and binding activities. miRNAome sequencing identified 1,597 known/conserved miRNAs that were differentially expressed between the two test conditions. According to the analysis, genes and miRNAs associated with the regulation of transcription, DNA template, transcription factor activity, and sequence-specific DNA binding pathways may play a major role in the resistance to heat stress in P. ternata. Integrated analysis of the transcriptome and miRNAome expression data revealed 41 high-confidence miRNA-mRNA pairs, forming 25 modules. MYB-like proteins and calcium-responsive transcription coactivators may play an integral role in heat-stress resistance in P. ternata. Additionally, the candidate genes and miRNAs were subjected to quantitative real-time polymerase chain reaction to validate their expression patterns. These results offer a foundation for future studies exploring the mechanisms and critical genes involved in heat-stress resistance in P. ternata.
Similar content being viewed by others
Introduction
Global warming has led to an increased incidence of extreme heat, which is a main factor affecting plant yield [1]. Besides the impact on the plant phenotype, heat stress disrupts cellular homeostasis, severely affecting their growth and development, leading to death in some cases [2]. Heat stress, particularly high-temperature stress, inhibits a wide range of physiological and biochemical responses in plants, including changes in water use, cell membrane stability, photosynthesis, and secondary metabolite and phytohormone levels [3]. Consequently, examining plant reactions to high temperatures will enhance our understanding of thermal adaptation at the molecular level and advance the identification of heat-resistant cultivars through genetic enhancement.
Plants possess distinct adaptive mechanisms that protect them from environmental stress [4]. These mechanisms are typically governed by intricate regulatory networks comprising multiple genes. Exploring gene interactions and genetic mechanisms is challenging because traditional research methods are often limited to the structure and function of genes. Transcriptome sequencing is a crucial approach for analyzing gene expression levels, identifying differentially expressed genes, exploring functional genes, and studying genetic evolution [5]. The technology has been exploited to study plant hormone signaling pathways, transcription factors (TFs), and protein kinases that respond to heat stress in a wide range of plants [6,7,8].
MicroRNAs (miRNAs) are produced from a single-stranded RNA precursors with a hairpin secondary structure and typically ranges from 20 to 24 nucleotides in size [9]. Plant miRNAs have a variety of biological functions and are involved in regulating plant growth and development, hormone signaling, and environmental stress responses [10]. They act as epigenetic regulators and negatively regulate gene expression at the post-transcriptional level, either by directly cleaving the target mRNA or by inhibiting the translation of the target gene by recognizing and binding to their mRNAs [11, 12]. High-throughput sequencing allows the easy and rapid identification of miRNAs in plants, which can be further combined with transcriptome and bioinformatics analyses to screen for miRNA target genes and enrich their regulatory networks.
Pinellia ternata (Thunb.) Briet. is a perennial herb of the Araceae family that has been traditionally used in Chinese medicine [13]. Its tuber is highly valued for medicinal uses and known for its antiemetic, antitussive, analgesic and sedative properties [14, 15]. The complex components of P. ternata, including alkaloids, organic acid, polysaccharose, proteins, and nucleosides, have been well-documented [16, 17]. Among these, alkaloids are recognized as the main active ingredients and believed to exert anticancer effects [18]. Currently, the market demand for P. ternata is increasing. However, owing to the changing climate and increased indiscriminate logging, natural resources of P. ternata have been significantly reduced [19]. Consequently, artificial cultivation has become the primary production method. High temperatures are one of the most influential factors in the average growth of P. ternata; Atmospheric temperature above 30 °C can cause “sprout tumble” in plants, where the above-ground parts rapidly wilt and die [20]. The “sprout tumble” in P. ternata is a response to heat stress rather than a necessary physiological process [21]. One small heat-shock protein (sHSP), two heat-shock proteins (HSPs), and one stearoyl-ACP-protein desaturase (SAD) were found in an SSH library of P. ternata under heat stress [22]. The overexpression of PtsHSP17.2 in tobacco increased the water retention and antioxidant capacity of transgenic plants, leading to a significant increase in heat tolerance [23]. In addition, overexpression of PtSAD in P. ternata reduced heat tolerance and increased the proportion of unsaturated fatty acids in transgenic plants [24]. Recent studies have shown that the PtNAC66 transcription factor enhances tolerance to heat stress in transgenic Arabidopsis by binding to and repressing the expression of the promoter regions of CYP707A3, MYB102, and NAC055, respectively [25]. Although progress has been made, our understanding of the molecular regulatory mechanisms underlying heat stress tolerance in P. ternata remains limited.
In this study, high-throughput sequencing was performed to investigate the changes in the transcriptome and miRNAs of P. ternata under heat stress. We screened for differentially expressed genes (DEGs) and identified miRNAs involved in the heat-stress response of P. ternata, providing a new theoretical basis for studying the molecular mechanisms and key resistance genes associated with “sprout tumble.”
Materials and methods
Plant material, growth conditions and treatments
The P. ternata variety used in the study was tested using tubers 1 cm in diameter from the Huaibei Normal University’s experimental field. The tubers were planted in pots and placed in a constant temperature-and-light incubator with a temperature of 25 ℃, light intensity of 4,000 lx, and a photoperiod of 16 h of light followed by 8 h of darkness. The conditions for the heat treatment of P. ternata have been previously described [24]. Seedlings that displayed consistent growth trends and reached a height of 15 cm were selected for the heat treatment experiment. The selected seedlings were subjected to a temperature of 40 ℃ for either 0–24 h while keeping all other conditions the same as those in the control group. Samples of leaves were collected, immediately frozen in liquid nitrogen, and stored at − 80 ℃ for further analysis. The experiments were performed three times, with three biological replicates per treatment.
DAB, NBT staining and biochemical index determination
Leaves of P. ternata seedlings were soaked in 1 mg/mL DAB and 0.5 mg/mL NBT staining buffers, vacuum-infiltrated for 20 min, stained in the dark at 28 ℃ for 8 h, and boiled in ethanol: lactic acid: glycerin (3:1:1) for 5 min before observation [26]. The specimens gathered before and after heat treatment were examined, with the concentration of chlorophyll in the foliage and stalks gauged using the Lichtenthaler method [27]. Malondialdehyde (MDA) content and peroxidase (POD) activity in the plant materials were assessed according to previously published methods [23].
Total RNA extraction, cDNA library construction and sequencing
Leaves from P. ternata seedlings were collected under normal and heat-treated (40 ℃ for 24 h) conditions at the three-leaf stage. TRIzol reagent (Sangon Biotech, Shanghai, China) was used to extract total RNA from the tissue samples. To ensure the accuracy of the experiment, the concentration and purity of RNA were determined using a NanoDropND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The integrity of total RNA was assessed using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Next, 1.0 µg RNA from each group was randomly fragmented. Equal amounts of RNA from each sample were sequenced on an Illumina HiSeq2500 sequencer (Illumina, San Diego, CA, USA) at the Sangon Biotech Company (Shanghai, China) to construct strand-specific RNA-seq libraries. For the RNA-seq analysis, three biological replicates were used for each condition.
Data processing and identification of DEGs
The initial output generated by the sequencing instrument was designated as raw reads. Raw reads obtained from each sample were subjected to a series of filtering steps. First, reads containing more than 10% of unknown nucleotides were removed. Additionally, reads with more than 40% low-quality bases were discarded. Finally, adapter sequences were eliminated from the reads using the Illumina adapter list. The remaining good-quality reads were subsequently subjected to de novo assembly using the Trinity software [28]. Transcript abundance reflects gene expression levels; the higher the transcript abundance, the higher the level of gene expression. To make the estimated gene expression levels comparable between different samples, the TPM algorithm in the Salmon software was used to measure the relative abundance of transcripts in each group of samples. DEGs in the samples were screened by DESeq; q-value < 0.05 and log2(fold change) > 1 were used as screening criteria to determine the significance of expression levels.
Gene annotation
To annotate the identified genes, alignments were performed based on the following databases: NCBI non-redundant protein sequences (NR) (http://ncbi.nlm.nih.gov/), NCBI nucleotide sequences (NT) (http://ncbi.nlm.nih.gov/), Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/cdd/), Swiss-Prot (http://www.uniprot.org/), TrEMBL (http://www.expasy.ch/sprot/), Gene Ontology (GO) (http://www.geneontology.org/), Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/), euKaryotic Ortholog Groups (KOG) (http://www.ncbi.nlm.nih.gov/KOG/), and Pfam (http://pfam.xfam.org/) using BLAST version 2.2.26 [29].
Construction of small RNA libraries and sequencing
Six samples (C: control, G: heat stress; each with three biological replicates) were harvested for small RNA library construction and sequencing. Total RNA was extracted from the samples using TRIzol reagent (Sangon Biotech, Shanghai, China). A total of 1.0 µg of RNA was utilized in the preparation of a small RNA library, following the guidelines provided by the TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA). Single-end sequencing, with a length of approximately 50 bp, was performed using an Illumina Hiseq2500 at Sangon Biotech Company (Shanghai, China).
Identification of miRNAs and prediction of their target genes
The raw data was processed using cutadapt software to eliminate the 3’-end connector (TGGAATTCTCGGGTGCCAAGGAACTC) and retain reads within the length range of 17–35 bp after connector removal. The reads were processed using the trimmomatic software to exclude bases with a quality score below 20 at both the 5’- and 3’-ends. Additionally, reads including four consecutive bases with an average quality value below 20 and reads shorter than 17 bases were filtered out. Raw data were filtered by removing joints and low-quality sequences to generate clean data. Clean reads were compared with the rRNA, sRNA, snRNA, and snoRNA annotation data from the RFAM database using BLASTN. The aligned reads were filtered according to the subsequent comparative criteria: gapopen = 0, evalue < 0.01, and mismatch ≤ 1. Any reads mapped to these databases were excluded from the analysis.
The miRNA levels in the heat-treated samples were analyzed for differential expression using the edgeR algorithm, with read counts serving as input data [30]. To remove miRNAs with low expression levels before performing differential analysis, the absolute value of log2(FoldChange) > 1 and p-value < 0.05 were set as screening criteria. psRNATarget (a plant small RNA target analysis server) (http://plantgrn.noble.org/psRNATarget/) (V2, 2017 release), based on the imperfect complementarity between miRNA sequences and their target genes, was used to predict miRNA target genes with default parameters and a maximum expectation value of four [31].
Quantitative RT-PCR analysis
To validate the transcriptome data, the SYBR green-based qPCR method was employed, and the Pt18SrRNA gene was used as an internal control for normalization [24, 32]. Primer sequences used for qRT-PCR are listed in Supplemental Table S1. miRNA expression was assessed using the Mir-XTM miRNA First-Strand Synthesis Kit (Clontech) for reverse transcription. Primers were specifically constructed for the six selected miRNAs, with the U6 snRNA serving as an internal control (Supplemental Table S1). qRT-PCR analyses were conducted three times, each containing three replicates for all genes. The 2−ΔΔCT method was used to calculate the relative expression level of each gene [33].
Results
Phenotypic analysis under heat stress
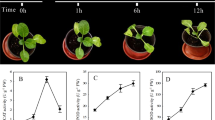
Wild-type P. ternata seedlings of uniform size at the three-leaf stage were subjected to stress treatment at 40 ℃ for 24 h and evaluated for their phenotypic response to heat stress. As shown in Fig. 1A, P. ternata seedlings subjected to heat stress showed no significant changes compared to control conditions. However, after 24 h of heat treatment, plants exhibited obvious deleterious phenotypes, such as curled and wilted leaves, shortened plant height, and growth inhibition, compared to the control plants. Reactive oxygen species (ROS) are produced in plants in response to stresses, such as heat stress. To further understand the cellular damage induced by ROS accumulation, MDA content, chlorophyll content, and POD activities in P. ternata seedlings were compared before and after heat treatment. The MDA content of the heat-treated P. ternata seedlings was two-fold higher than that of the control seedlings. Conversely, chlorophyll content and POD activity of the heat-treated plants were significantly lower than those of the control plants (Fig. 1B). This indicated that wild-type P. ternata seedlings exhibited a high degree of susceptibility to heat stress.
Physiological responses of Pinellia ternata seedlings before and after heat treatment. (A) P. ternata seedlings phenotypes in response to heat stress. Plants were grown in soil to the three-leaf stage and subjected to a 40 ℃ stress treatment for 24 h (left, scale bar = 4 cm). DAB and NBT staining from the same part of P. ternata seedlings before and after heat treatment (right, scale bar = 15 mm). (B) MDA and chlorophyll contents, and POD activities in P. ternata seedlings under control and heat treatment for 24 h. Except where noted, all data are presented as mean (n = 3) and standard deviation. Data were analyzed by Student’s t-test and one-way ANOVA. * and ** indicate significant differences between control and heat treatment plants at the P < 0.05 and P < 0.01 levels, respectively
Transcriptome sequencing in P. ternata under heat stress
To examine the gene expression patterns of P. ternata under heat stress, six libraries were constructed from two leaf samples, each with three biological replicates (C: control; G: heat stress). Following the removal of low-quality reads, each library’s total reads and total bases ranged from 42.34 million to 53.72 million and 6.10 billion to 7.69 billion, respectively. The Q30 and GC contents were consistently high across all libraries, with values ranging from 93.83 to 94.36% and 55.64–58.43%, respectively, indicating the exceptional quality of the transcriptome sequencing data (Supplemental Table S2). A comprehensive collection of 574,168 transcripts was obtained from cDNA libraries. These transcripts were subsequently filtered and assembled, resulting in the formation of 208,217 unigene clusters. Notably, these clusters exhibited an N50 value of 552 bp. A comprehensive summary of the transcriptome sequencing analysis of P. ternata is presented in Table 1.
All assembled unigene clusters were aligned against the GO, KEGG, Pfam, SwissProt, TrEMBL, CDD, KOG, NR, and NT databases using DIAMOND 23 with a threshold E-value of < 0.00001 [28]. The statistical results of the six authoritative databases are listed in Table 2.
Analysis of DEGs
Three comparisons of the two test conditions (C and G) were conducted to identify genes differentially regulated by heat stress in P. ternata. The cluster heat map (Supplemental Fig. S1A) and PCA plots (Supplemental Fig. S1B) demonstrated a high degree of reproducibility of the gene expression data across the three biological replicates of each sample. Notably, transcriptome expression levels exhibited contrasting patterns before and after subjecting P. ternata seedlings to heat treatment. As is evident from the scatter plot, the number of significantly upregulated transcripts (2,244) was lower than that of downregulated transcripts (2,716) among the 4,960 DEGs (Fig. 2A; Supplemental Table S3).
Transcriptome sequencing analysis of G and C. (A) scatter plot of DEGs in G vs. C. Significantly differentially expressed genes are represented by red dots (up-regulated) and green dots (down-regulated), whereas non-differentially expressed genes are represented by black dots. (B) Gene ontology (GO) classification of the DEGs in P. ternata seedlings under heat stress. The ordinate is the enriched GO term, and the abscissa is the percent and number of differentially expressed genes in this term. Different colors are used to distinguish biological processes, cellular components, and molecular functions. (C) Kyoto Encyclopedia of Genes and Genome (KEGG) enrichment of the DEGs in P. ternata under heat stress. The ordinate is the enriched KEGG term, and the abscissa is the percent and number of differentially expressed genes in this term. Different colors are used to distinguish biological processes, cellular components, and molecular functions
Functional annotation and enrichment analysis of the DEGs
GO enrichment analysis was performed to determine the functions of the identified DEGs, and 7,028 DEGs were annotated to 4,610 GO terms. Notably, we observed significant enrichment (p < 0.05) of 817 GO categories in P. ternata under heat stress (Supplemental Table S4). As depicted in Fig. 2B, the significantly enriched biological processes (BPs) encompassed cellular processes (GO:0009987), metabolic processes (GO:0008152), responses to stimuli (GO:0050896), and biological regulation (GO:0065007); the most significantly enriched cellular components (CCs) were cells (GO: 0005623), cell parts (GO:0044464), organelles (GO:0043226), and membranes (GO:0016020). In addition, catalytic activity (GO:0003824) and binding activity (GO:0005488) were significantly enriched in the molecular function (MF) categories.
To conduct a more comprehensive examination of the biological functions of the DEGs, pathway-based analysis was performed using KEGG pathway enrichment analysis. Carbohydrate metabolism, signal transduction, lipid metabolism, and energy metabolism were the most enriched pathways in P. ternata under heat stress (Fig. 2C). A total of 409 DEGs were assigned to 158 distinct pathways. Among these pathways, 78 were significantly enriched (p < 0.05) (Supplemental Table S4).
Several genes encoding TFs were differentially expressed
TF activity and protein binding (GO:0000988) were also important GO terms (Fig. 2B). In this study, 4,960 genes were identified, of which 38 encoded TFs belonging to 17 families. The TF families with the highest representation were No apical meristem, Arabidopsis transcription activation factor1/2, and Cup-shaped cotyledon2 (NAC), with a count of five. This was followed by basic helix-loop-helix (bHLH), APETALA2/Ethylene responsive factor, and WRKY TF, each with a count of four (Fig. 3). The vast majority of TFs showed opposite expression trends under control and heat treatment conditions, suggesting that the response to heat stress in P. ternata seedlings is regulated by a combination of factors.
Heatmap of the regulatory multiples of 38 transcription factors with different regulatory trends in the comparisons of control and heat stress treatments. Transcription factors of the same family are indicated by squares of the same color, and the number of genes is attached to the gene family name. The colored bars represent the 38 transcription factor values of control and heat treatment [log2(TPM + 1)]
Validation of RNA-Seq analysis by quantitative real-time PCR (qRT-PCR)
To validate the reliability of the heat stress-related gene expression data obtained from the RNA-Seq analysis of P. ternata seedlings, eight DEGs with different roles in the heat-stress response were selected from the control and heat-treated samples for qRT-PCR analysis (Supplemental Fig. S2 and Table S1). The trends in qRT-PCR and transcriptome sequencing results were consistent, further demonstrating the reliability of the transcriptome sequencing data.
MicroRNAome for heat stress responses to P. ternata
To comprehensively evaluate the influence of heat stress on the transcript profiles, six small RNA libraries were constructed. Following the removal of low-quality reads, each library’s total reads and total bases ranged from 1.50 million to 3.11 million and 0.36 billion to 0.72 billion, respectively. The Q30 and GC contents were consistently high across all libraries, with values ranging from 96.96 to 97.09% and 45.09–51.48%, respectively, indicating the exceptional quality of the miRNA sequencing data (Supplemental Table S5). PCA was conducted on three biological replicates of each sample, and the results demonstrated a strong connection (Supplemental Fig. S3), demonstrating high dependability of the replicates. The sequences of the clean reads obtained from the experiment were compared with those of miRNAs in plants such as Arabidopsis thaliana [34], Oryza sativa [35], and Medicago truncatula [36], sourced from from miRbase. A total of 1,597 known/conserved miRNAs were identified across all samples (Supplemental Table S6). Unaligned readings were analyzed to identify new miRNAs. Following the exclusion of small RNAs that did not satisfy the established criteria for plant miRNAs, finally a total of 92 upregulated and 231 downregulated differentially expressed miRNAs (DEMs) was obtained (Fig. 4A; Supplemental Table S7).
Analysis of differentially expressed miRNAs (DEMs) between G and C. (A) The number of DEMs in response to heat stress treatments in P. ternata seedlings. (B) Gene ontology (GO) enrichment analysis of stress-responsive DEMs under heat treatments. The ordinate is the enriched GO term, and the abscissa is the percent and number of differentially expressed genes in this term. Different colors are used to distinguish biological processes, cellular components, and molecular functions
Identification of the potential target genes
To gain a thorough understanding of the biological relevance of miRNAs and other short RNAs, the individual genes that they target must be identified. This information can be used to unravel the intricate regulatory network of miRNA-target interactions and their potential impact on various biological processes. We predicted 3,017 targets for 289 miRNAs that were differentially expressed during heat stress in P. ternata seedlings (Supplemental Table S8). Along with aly-miR4242 and bdi-miR7713-5p, 29 other miRNAs were predicted to interact with only one target gene. The remaining miRNAs exhibited a propensity to target several genes. The maximum number of targets was 117 for ppt-miR414, followed by 88 for osa-miR2102 and 87 for ppe-miR8123-3p. GO enrichment analysis was conducted to gain a deeper understanding of the potential biological functions associated with the target genes involved in heat-stress response (Fig. 4B; Supplemental Table S9). Among the enriched BPs, the most enriched GO terms were transcription, DNA-templated (GO:0006351) and regulation of transcription, DNA-templated (GO:0006355). In the cellular component group, the nucleus (GO:0005634) was the most overrepresented term. Among the enriched MFs, the most significant GO terms were DNA binding (GO:0003677) and transcription factor activity, sequence-specific DNA binding (GO:0003700). These observations suggest that the identified miRNAs may have a significant impact on DNA binding and transcriptional control within the nucleus.
Integrated analysis of miRNAome and transcriptome
As miRNAs negatively regulate the expression of their target mRNAs through target cleavage, their expression patterns are generally inversely correlated with those of their target. We collected miRNA-target gene pairs with negative regulatory interactions from the differential miRNA-differential target gene association analysis table and combined them with the transcriptome and miRNAome data (Table 3). For example, ata-miR2275a-5p was upregulated after heat stress, whereas its potential target gene (TRINITY_DN70714_c2_g1), an MYB family gene, was downregulated under heat stress conditions. Another potential target gene (TRINITY_DN70355_c4_g2) associated with calcium response was upregulated by heat stress, and its counterpart gma-miR172h-5p was downregulated. Moreover, our findings indicated that heat stress led to a decrease in the expression of ptc-miR156k, osa-miR166b-5p, and bra-miR172c-5p, whereas ghr-miR160 was upregulated following heat treatment. Further analysis indicated that 25 miRNAs were differentially expressed, corresponding to 41 distinct target genes, suggesting that a single miRNA has the potential to regulate multiple target genes.
We performed qRT-PCR on the six aforementioned miRNAs and their corresponding possible target genes to confirm the expected regulatory link between heat stress-associated miRNAs and their targets. The qRT-PCR results agreed with those obtained from Illumina sequencing. This concurrence provides additional support for the observed inverse expression patterns of the six miRNA-target pairs investigated in our study (Fig. 5). Expression profiling analyses of miRNA-mRNA interactions demonstrated a substantial level of confidence in the prediction of target genes. To further understand the regulatory mechanisms of the miRNAs, a molecular regulatory network diagram was constructed for these six miRNAs (Fig. 6). The results showed that all the miRNAs interacted closely with the genes. However, further testing of similar pairs is required to gain insight into the response of P. ternata seedlings to heat stress.
Validation of expression profile of miRNAs and their predicted target genes in P. ternata seedlings. Expression analysis of selected miRNAs (A) and one of their predicted targets genes (B) using qRT-PCR. Three biological replicates and two technical replicates were included in the study. Asterisks indicate significant differences between the control and the heat treatment sample (Student’s t-test, *P < 0.05, **P < 0.01)
Discussion
High temperatures, whether temporary or prolonged, can induce modifications in the morphological, physiological, and biochemical properties of plants, thereby affecting plant growth and development. This can lead to a significant decrease in yield, particularly in plant species vulnerable to high temperatures [24, 37]. P. ternata is a heat-sensitive plant, and heat is an important limiting factor in its production. The molecular mechanisms of the response to high temperatures in model plants have been well studied; however, the mechanisms of heat-stress response have been challenging to investigate in P. ternata because of its susceptibility to the “sprout tumble” phenomenon caused by high temperatures. However, the absence of a genome sequence for this species poses a challenge in unraveling the underlying molecular mechanisms. Omics-based approaches, including suppression subtractive hybridization [22], proteomics [38], and transcriptomics [39, 40], has been applied to P. ternata for the data mining of heat-responsive factors. However, none of the genes involved in heat-stress response have been functionally verified in P. ternata. Therefore, it is necessary to design an integrated multi-omics approach to elucidate the molecular regulatory network of P. ternata in response to heat stress. Our study combined transcriptome and miRNAome data to identify miRNAs and their target genes associated with heat stress.
The transcriptome sequencing results of P. ternata seedlings provided valuable insights into the molecular response to heat stress. The identification of a large number of transcripts (574,168) and their assembly into unigenes (208,217) highlighted the complexity of the transcriptome and the diversity of expressed genes in this species. The identification of 4,960 DEGs indicates substantial transcriptional changes in P. ternata seedlings under heat stress conditions. Interestingly, the number of downregulated genes (2,716) was higher than that of upregulated genes (2,244), suggesting that the plant response to heat stress may involve the repression of certain genes or pathways, as a probable strategy to conserve energy or redirect resources toward stress response mechanisms [41]. Functional annotation of the DEGs using GO and KEGG analyses provided insights into the biological processes and pathways potentially involved in plant responses to heat stress. The involvement of DEGs in the response to stimuli and signal transduction pathways suggests that these genes play crucial roles in sensing and responding to heat stress signals. The enrichment of DEGs in plant hormone signal transduction and the MAPK signaling pathway was particularly important because these pathways play key roles in plant stress responses [42]. Because of the complexity of this regulatory mechanism, their function in plant responses to heat stress is worth-exploring.
TFs play important roles in plant responses to heat stress. Members of the TF family such as NAC [43], bHLH [44], WRKY [45], MYB [46], AP2/ERF [47] and bZIP [48] are associated with heat stress. Our analysis revealed that the downregulated members (n = 14) of the TF families NAC, bHLH, WRKY, and bZIP surpassed the upregulated members (n = 7), supporting our earlier inference. NAC TFs have been confirmed to play an important role in the response to heat stress. In rice, SNAC3 regulates the heat-stress response by adjusting the redox homeostasis by controlling the expression of ROS-associated enzyme genes [43]. We also found that the NAC gene family had the largest representation among the TF genes screened. Of the five NAC genes, two were upregulated, while three were downregulated. Further comprehensive investigations are required to ascertain whether these genes are involved in distinct stress-signaling pathways. Heat-shock transcription factors (HSFs) play a pivotal role in the adaptation of plants to heat and other stress stimuli [49]. We identified two HSF genes that were significantly altered after heat treatment of P. ternata seedlings. Interestingly, the expression of these two genes was reversed, suggesting the presence of a multifaceted regulatory system in P. ternata in response to heat stress.
Small RNAs play an important role in the response to different stresses and regulation of gene expression during plant growth and development in plants [50]. Generally, a single miRNA can target several genes, and a single gene can be regulated by several miRNAs [51]. Through miRNAome analysis, 3,017 targets from 289 miRNA families were identified from miRNA libraries. Some miRNAs that have been previously known to respond to heat stress, such as miR156, miR160, miR166, and miR172, were also detected in our study. In Arabidopsis, plants overexpressing miR160 and arf deletion mutants have a significantly higher tolerance to high temperatures than wild-type plants [52]. In the present study, miR160 was upregulated after heat treatment, suggesting that it might have a function similar to that in other species. miR160, miR156, and miR172 also regulate plant tolerance to high temperatures by regulating the expression levels of target genes [53,54,55]. However, in the present study, these miRNAs were down-regulated during heat stress. One possible explanation for this phenomenon is the innate heat sensitivity of wild-type P. ternata, which may render it less capable of effectively enduring heat stress.
The integration of transcriptome and miRNAome expression datasets in response to heat stress in P. ternata led to the identification of crucial miRNA-mRNA modules. This approach provided a comprehensive understanding of the multiple regulatory networks involved in this biological process. To construct our regulatory network, we utilized only those miRNA-target pairs that showed inverse expression patterns in both our miRNAome and transcriptome datasets with high confidence. The analysis revealed that 25 miRNAs were differentially expressed, corresponding to 41 distinct target genes (Table 3). Among these, we identified an MYB-like and calcium-responsive gene that was significantly regulated by miRNAs. In Phaseolus vulgaris, the MYB family gene PvPHR1 is regulated by PvmiR399, and they play an important role in plant phosphorus deficiency signaling [56]. To date, no studies have provided evidence for the involvement of miRNAs in the regulation of MYB genes during heat stress. Therefore, in-depth studies are required to elucidate the underlying biological mechanisms. Calmodulin has a significant effect on the responses of plants towards heat stress [57]. In our study, we successfully identified a gene that is regulated by calcium and is a transcriptional co-activator that responds to calcium. However, the precise contribution of this gene to the heat-stress response of P. ternata seedlings via miRNA regulation remains to be investigated. Integrated analysis revealed the presence of several uncharacterized proteins. This observation could be attributed to the absence of comprehensive genetic data for P. ternata. However, this also implies the potential existence of additional stress-related genes among the target genes. Furthermore, we constructed a molecular regulatory network of these miRNAs to depict their intricate interactions with genes, thereby offering insights into the regulatory mechanisms underpinning P. ternata seedlings’ response to heat stress. Nevertheless, further experimental validation is necessary to fully elucidate miRNA-mediated regulatory pathways.
Conclusion
In summary, we studied small RNAs and their target genes in P. ternata using transcriptome and miRNAome profiles when exposed to heat stress. Although the complex miRNA-mediated regulatory networks require further study, our findings provide valuable information for the characterization of genes and miRNAs that respond to heat stress in P. ternata. Additionally, these results can be useful for other plant species, as they provide insights into the molecular mechanisms that govern plant responses to abiotic stress.
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) and GSE243965.
References
Shao J, Yuan T, Li Z, Li N, Liu H, Bai SH, Xia J, Lu M, Zhou X. Plant evolutionary history mainly explains the variance in biomass responses to climate warming at a global scale. New Phytol. 2019;222(3):1338–51.
Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol. 2005;16(2):123–32.
Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: an overview. Environ Exp Bot. 2007;61(3):199–223.
Zhu JK. Abiotic Stress Signaling and responses in plants. Cell. 2016;167(2):313–24.
Wang R, Xu S, Wang N, Xia B, Jiang Y, Wang R. Transcriptome Analysis of Secondary Metabolism Pathway, transcription factors, and transporters in response to Methyl Jasmonate in Lycoris aurea. Front Plant Sci 2017, 7.
Yang Y, Zhang C, Zhu D, He H, Wei Z, Yuan Q, Li X, Gao X, Zhang B, Gao H, et al. Identifying candidate genes and patterns of heat-stress response in rice using a genome-wide association study and transcriptome analyses. Crop J. 2022;10(6):1633–43.
Nahuel G-S, Dreni L, Lawas LMF, Galbiati M, Colombo L, Heuer S, Jagadish KSV, Kater MM. Genome-wide transcriptome analysis during Anthesis reveals New insights into the molecular basis of heat stress responses in tolerant and sensitive Rice varieties. Plant Cell Physiol 2016, 57(1).
Yang Z, Li W, Su X, Ge P, Zhou Y, Hao Y, Shu H, Gao C, Cheng S, Zhu G et al. Early response of Radish to heat stress by Strand-Specific Transcriptome and miRNA analysis. Int J Mol Sci 2019, 20(13).
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
Sattar S, Song Y, Anstead JA, Sunkar R, Thompson GA. Cucumis melo MicroRNA expression Profile during Aphid Herbivory in a resistant and susceptible Interaction. Mol Plant-Microbe Interact. 2012;25(6):839–48.
Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat Genet. 2006;38(Suppl):S31–6.
Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–87.
Moon BC, Kim WJ, Ji Y, Lee YM, Kang YM, Choi G. Molecular identification of the traditional herbal medicines, Arisaematis Rhizoma and Pinelliae Tuber, and common adulterants via universal DNA barcode sequences. Gen Mol Res 2016, 15(1).
Iwasa M, Iwasaki T, Ono T, Miyazawa M. Chemical composition and major odor-active compounds of essential oil from PINELLIA TUBER (dried rhizome of Pinellia ternata) as Crude Drug. J Oleo Sci. 2014;63(2):127–35.
Lin S, Nie B, Yao G, Yang H, Ye R, Yuan Z. Pinellia ternata (Thunb.) Makino Preparation promotes sleep by increasing REM sleep. Nat Prod Res. 2019;33(22):3326–9.
Gombodorj S, Yang M, Shang Z, Liu R, Li T, Yin G, Kong L. New phenalenone derivatives from Pinellia ternata tubers derived Aspergillus Sp. Fitoterapia. 2017;120:72–8.
Xu J, Dai C, Shan J, Xie T, Xie H, Wang M, Yang G. Determination of the effect of Pinellia ternata (Thunb.) Breit. On nervous system development by proteomics. J Ethnopharmacol. 2018;213:221–9.
Ji X, Huang B, Wang G, Zhang C. The ethnobotanical, phytochemical and pharmacological profile of the genus Pinellia. Fitoterapia. 2014;93:1–17.
Jie EY, Ryu YB, Choi SA, Ahn MS, Liu JR, Min SR, Kim SW. Mass propagation of microtubers from suspension cultures of Pinellia ternata cells and quantitative analysis of succinic acid in Pinellia tubers. Plant Biotechnol Rep. 2015;9(5):331–8.
Toshihiko EGUCHI, Hiroyuki TANAKA, Satoshi YOSHIDA, Ken MATSUOKA. Temperature effects on the yield and quality of the Medicinal Plant Pinellia ternata Breit. Environ Control Biol. 2019;57(3):83–5.
Juneidi S, Gao Z, Yin H, Makunga NP, Chen W, Hu S, Li X, Hu X. Breaking the summer dormancy of Pinellia ternata by introducing a heat tolerance receptor-like kinase ERECTA gene. Front Plant Sci 2020, 11.
Lu H, Xue T, Zhang A, Sheng W, Zhu Y, Chang L, Song Y, Xue J. Construction of an SSH Library of Pinellia ternata under heat stress, and expression analysis of four transcripts. Plant Mol Biol Rep. 2013;31(1):185–94.
Tian C, Zhang Z, Huang Y, Xu J, Liu Z, Xiang Z, Zhao F, Xue J, Xue T, Duan Y. Functional characterization of the Pinellia ternata cytoplasmic class II small heat shock protein gene PtsHSP17.2 via promoter analysis and overexpression in tobacco. Plant Physiol Biochem. 2022;177:1–9.
Zhang H, Zhang Z, Xiong Y, Shi J, Chen C, Pan Y, Xue T, Xue J, Duan Y. Stearic acid desaturase gene negatively regulates the thermotolerance of Pinellia ternata by modifying the saturated levels of fatty acids. Ind Crops Prod 2021, 166.
Bo C, Liu D, Yang J, Ji M, Li Z, Zhu Y, Duan Y, Xue J, Xue T. Comprehensive in silico characterization of NAC transcription factor family of Pinellia ternata and functional analysis of PtNAC66 under high-temperature tolerance in transgenic Arabidopsis thaliana. Plant Physiol Biochem. 2024;208:108539.
Chen K, Song M, Guo Y, Liu L, Xue H, Dai H, Zhang Z. MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol J. 2019;17(12):2341–55.
Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol 1987, 148 C:350–82.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–U130.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402.
Zhou X, Lindsay H, Robinson MD. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res 2014, 42(11).
Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–9.
Bo C, Su C, Teng J, Sheng W, Xue T, Zhu Y, Xue J. Transcriptome profiling reveals Differential Gene expression during the process of Microtuber formation in Pinellia ternata. Int J Mol Sci 2023, 24(14).
Livak KJ, Schmittgen TDL. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods. 2001;25(4):402–8.
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20(24):3407–4725.
Zeng H, Zhang X, Ding M, Zhu Y. Integrated analyses of miRNAome and transcriptome reveal zinc deficiency responses in rice seedlings. BMC Plant Biol 2019, 19(1).
Eyles RP, Williams PH, Ohms SJ, Weiller GF, Ogilvie HA, Djordjevic MA, Imin N. microRNA profiling of root tissues and root forming explant cultures in Medicago truncatula. Planta. 2013;238(1):91–105.
Matnur S, Jajoo A. Arbuscular mycorrhizal fungi protects maize plants from high temperature stress by regulating photosystem II heterogeneity. Ind Crops Prod 2020, 143.
Zhu Y, Zhu G, Guo Q, Zhu Z, Wang C, Liu Z. A comparative proteomic analysis of Pinellia ternata leaves exposed to heat stress. Int J Mol Sci. 2013;14(10):20614–34.
Ma G, Zhang M, Xu J, Zhou W, Cao L. Transcriptomic analysis of short-term heat stress response in Pinellia ternata provided novel insights into the improved thermotolerance by spermidine and melatonin. Ecotoxicol Environ Saf 2020, 202.
Yang J, Cui W, You Q, Liu M, Liu X, Zhao F, Zhu Y, Duan Y, Xue T, Xue J. Transcriptome Analysis Reveals Long Non-Coding RNAs Involved in Shade-Induced Growth Promotion in Pinellia ternata. FBL 2023, 28(9).
Zhang H, Sonnewald U. Differences and commonalities of plant responses to single and combined stresses. Plant J. 2017;90(5):839–55.
Danquah A, de Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv. 2014;32(1):40–52.
Fang Y, Liao K, Du H, Xu Y, Song H, Li X, Xiong L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J Exp Bot. 2015;66(21):6803–17.
Zuo Z-F, Lee H-Y, Kang H-G. Basic Helix-Loop-Helix transcription factors: regulators for Plant Growth Development and Abiotic stress responses. Int J Mol Sci 2023, 24(2).
Dang F, Wang Y, Yu L, Eulgem T, Lai Y, Liu Z, Wang X, Qiu A, Zhang T, Lin J, et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013;36(4):757–74.
Wu Z, Li T, Liu X, Yuan G, Hou H, Teng N. A novel R2R3-MYB transcription factor LlMYB305 from Lilium longiflorum plays a positive role in thermotolerance via activating heat-protective genes. Environ Exp Bot 2021, 184.
Xu Z, Chen M, Li L, Ma Y. Functions and application of the AP2/ERF Transcription Factor Family in Crop ImprovementF. J Integr Plant Biol. 2011;53(7):570–85.
Zhang X, Wollenweber B, Jiang D, Liu F, Zhao J. Water deficits and heat shock effects on photosynthesis of a transgenic Arabidopsis thaliana constitutively expressing ABP9, a bZIP transcription factor. J Exp Bot. 2008;59(4):839–48.
Scharf KD, Berberich T, Ebersberger I, Nover L. The plant heat stress transcription factor (hsf) family: structure, function and evolution. Biochim Biophys Acta. 2012;1819(2):104–19.
Li S, Castillo-Gonzalez C, Yu B, Zhang X. The functions of plant small RNAs in development and in stress responses. Plant J. 2017;90(4):654–70.
Shi R, Jiao W, Yun L, Zhang Z, Zhang X, Wang Q, Li Y, Mi F. Utilization of Transcriptome, small RNA, and Degradome sequencing to provide insights into Drought stress and Rewatering Treatment in Medicago ruthenica. Front Plant Sci 2021, 12.
Lin J, Kuo C, Yang I, Tsai W, Shen Y, Lin C, Liang Y, Li Y, Kuo Y, King Y et al. MicroRNA160 modulates Plant Development and Heat shock protein gene expression to Mediate Heat Tolerance in Arabidopsis. Front Plant Sci 2018, 9.
Matthews C, Arshad M, Hannoufa A. Alfalfa response to heat stress is modulated by microRNA156. Physiol Plant. 2019;165(4):830–42.
Ravichandran S, Ragupathy R, Edwards T, Domaratzki M, Cloutier S. MicroRNA-guided regulation of heat stress response in wheat. BMC Genomics 2019, 20.
Chen J, Pan A, He S, Su P, Yuan X, Zhu S, Liu Z. Different MicroRNA families involved in regulating high temperature stress response during cotton (Gossypium hirsutum L.) Anther Development. Int J Mol Sci 2020, 21(4).
VALDÉS-LÓPEZ O, ARENAS-HUERTERO C, RAMÍREZ M, SÁNCHEZ GIRARDL, VANCE CP F, LUIS REYES J, HERNÁNDEZ G. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 2008;31(12):1834–43.
Zhou R, Li B, Liu H, Daye. Progress in the participation of Ca2+–calmodulin in heat shock signal transduction. Prog Nat Sci. 2009;19(10):1201–8.
Funding
This work was supported by the National Natural Science Foundation of China (82274048 and 82373993), the Project of Natural Science Research of Universities in Anhui Province, China (KJ2021A0532 and 2023AH050352), Key Research and Development Program of Shandong Province, China (2019GNC106113) and Excellent Scientific Research and Innovation Team of University in Anhui Province (2022AH010029).
Author information
Authors and Affiliations
Contributions
T.X. and J.X. conceived this research. C.B. and M.L. performed most of the work. Q.Y., X.L., Y.Z. and Y.D. participated the experimental work. C.B. wrote the manuscript, D.W., T.X. and J.X. revised and edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bo, C., Liu, M., You, Q. et al. Integrated analysis of transcriptome and miRNAome reveals the heat stress response of Pinellia ternata seedlings. BMC Genomics 25, 398 (2024). https://doi.org/10.1186/s12864-024-10318-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10318-x