Abstract
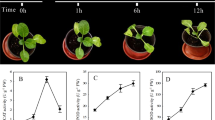
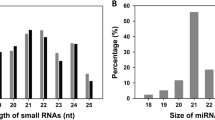
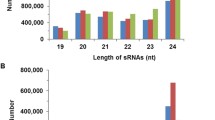
microRNAs (miRNAs) are a class of single-stranded endogenous non-coding RNAs that play critical roles in plant growth, development, and environmental stress responses. Temperature is one of the major physical parameters disturbing cellular homeostasis and causing leaf etiolation in plants. Previous studies have reported that several conserved and novel miRNAs were responsive to heat stress in plants. However, the characterization of miRNAs responsive to heat stress in radish remains poorly understood. To better understand miRNAs and their target genes under heat stress, two small RNA libraries were constructed from heat-treated (Heat24) and heat-untreated (CK) radish roots. Using Solexa system, totally, 26 known and 19 novel miRNAs were identified as differentially expressed under heat stress. Expression patterns of a set of heat-responsive miRNAs were validated by quantitative real-time PCR (qRT-PCR). Furthermore, 422 sliced targets for 25 known miRNAs were identified by degradome sequencing technology, and most of the identified targets are involved in multiple biological processes including transcriptional regulation and response to biotic and abiotic stresses. Moreover, some miRNAs and their corresponding targets, which are related to the accumulation of heat stress transcription factors and heat shock proteins, played important roles in thermo-tolerance in radish. These findings could enhance the understanding of molecular mechanisms underlying miRNAs and their targets in regulating plant responses to heat stress.







Similar content being viewed by others
References
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18:758–762
Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25:130–131
Barakat A, Sriram A, Park J, Zhebentyayeva T, Abbott DMaA (2012) Genome wide identification of chilling responsive microRNAs in Prunus persica. BMC Genomics 13:481
Bita CE, Zenoni S, Vriezen WH, Mariani C, Pezzotti M, Gerats T (2011) Temperature stress differentially modulates transcription in meiotic anthers of heat-tolerant and heat-sensitive tomato plants. BMC Genomics 12:384
Bowen J, Lay-Yee M, Plummer K, Ferguson I (2002) The heat shock response is involved in thermotolerance in suspension-cultured apple fruit cells. J Plant Physiol 159:599–606
Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22:268–280
Chen L, Ren Y, Zhang Y, Xu J, Sun F, Zhang Z, Wang Y (2012a) Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene 504:160–165
Chen L, Wang T, Zhao M, Tian Q, Zhang WH (2012b) Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 235:375–386
Chen L, Zhang Y, Ren Y, Xu J, Zhang Z, Wang Y (2012c) Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem Biophys Res Commun 417:892–896
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Jason S, Cumbie SAG, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of miRNA genes. PLoS One 2:e219. doi:10.1371/journal.pone.0000219
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15:2038–2043
German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De Paoli E, Lu C, Schroth G, Meyers BC, Green PJ (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26:941–946
Guan Q, Lu X, Zeng H, Zhang Y, Zhu J (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J 74:840–851
Hao DC, Yang L, Xiao PG, Liu M (2012) Identification of Taxus microRNAs and their targets with high-throughput sequencing and degradome analysis. Physiol Plant 146:388–403
Huang SQ, Peng J, Qiu CX, Yang ZM (2009) Heavy metal-regulated new microRNAs from rice. J Inorg Biochem 103:282–287
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kawashima CG, Yoshimoto N, Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi H, Dalmay T (2009) Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J 57:313–321
Li XP, Phippard A, Pasari J, Niyogi KK (2002) Structure–function analysis of photosystem II subunit S (PsbS) in vivo. Funct Plant Biol 29:1131–1139
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967
Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62:742–759
Li B, Qin Y, Duan H, Yin W, Xia X (2011) Genome-wide characterization of new and drought stress responsive microRNAs in Populus euphratica. J Exp Bot 62:3765–3779
Li H, Mao W, Liu W, Dai H, Liu Y, Ma Y, Zhang Z (2013) Deep sequencing discovery of novel and conserved microRNAs in wild type and a white-flesh mutant strawberry. Planta 238:695–713
Liu HT, Sun DY, Zhou RG (2005) Ca2+and AtCaM3 are involved in the expression of heat shock protein gene in Arabidopsis. Plant Cell Environ 28:1276–1284
Mahale BM, Fakrudin B, Ghosh S, Krishnaraj PU (2014) LNA mediated in situ hybridization of miR171 and miR397a in leaf and ambient root tis sues revealed expressional homogen eity in response to shoot heat shock in Arabidopsis thaliana. J Plant Biochem Biotechnol 23:93–103
McCormick KP, Willmann MR, Meyers BC (2011) Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence 2:2
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK (2008) Criteria for annotation of plant MicroRNAs. Plant Cell 20:3186–3190
Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat? Trends Biochem Sci 37:118–125
Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, Walther D, Scheible WR (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150:1541–1555
Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J (2010) Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J 62:960–976
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425
Ruiz-Ferrer V, Voinnet O (2009) Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60:485–510
Shamimuzzaman M, Vodkin L (2012) Identification of soybean seed developmental stage-specific and tissue-specific miRNA targets by degradome sequencing. BMC Genomics 13:310
Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I (2014) Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26:1792–1807
Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687
Wahid A, Gelani S, Ashraf M, Foolad M (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61:199–223
Wang L, He Q (2005) Chinese radish. Scientific and Technical Documents Publishing House, Beijing
Wang L, Yu X, Wang H, Lu YZ, de Ruiter M, Prins M, He YK (2011a) A novel class of heat-responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa). BMC Genomics 12:289
Wang T, Chen L, Zhao M, Tian Q, Zhang WH (2011b) Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12:367
Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol 10:123
Xu L, Wang Y, Xu Y, Wang L, Zhai L, Zhu X, Gong Y, Ye S, Liu L (2013a) Identification and characterization of novel and conserved microRNAs in radish (Raphanus sativus L.) using high-throughput sequencing. Plant Sci 201–202:108–114
Xu L, Wang Y, Zhai L, Xu Y, Wang L, Zhu X, Gong Y, Yu R, Limera C, Liu L (2013b) Genome-wide identification and characterization of cadmium-responsive microRNAs and their target genes in radish (Raphanus sativus L.) roots. J Exp Bot 64:4271–4287
Yu X, Wang H, Lu Y, de Ruiter M, Cariaso M, Prins M, van Tunen A, He Y (2012) Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot 63:1025–1038
Zeng QY, Yang CY, Ma QB, Li XP, Dong WW, Nian H (2012) Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol 12:182
Zhai L, Xu L, Wang Y, Huang DQ, Yu R, Limera C, Gong Y, Liu L (2014) Genome-wide identification of embryogenesis-associated microRNAs in radish (Raphanus sativus L.) by high-throughput sequencing. Plant Mol Biol Rep. doi:10.1007/s11105-014-0700-x
Zhang XN, Li X, Liu JH (2014) Identification of conserved and novel cold-responsive micrornas in trifoliate orange (Poncirus trifoliata (L.) Raf.) using high-throughput sequencing. Plant Mol Biol Rep 32:328–341
Zhou M, Luo H (2013) MicroRNA-mediated gene regulation: potential applications for plant genetic engineering. Plant Mol Biol 83:59–75
Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61:4157–4168
Zhou ZS, Song JB, Yang ZM (2012a) Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. J Exp Bot 63:4695–4613
Zhou ZS, Zeng HQ, Liu ZP, Yang ZM (2012b) Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ 35:86–99
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgments
This work was in part supported by grants from the NSFC (31171956, 31372064), Key Technology R & D Program of Jiangsu Province (BE2013429), JASTIF [CX (12)2006, CX (13)2007, (13)5082] and the PAPD.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, R., Xu, L., Zhu, X. et al. Transcriptome-Wide Characterization of Novel and Heat-Stress-Responsive microRNAs in Radish (Raphanus Sativus L.) Using Next-Generation Sequencing. Plant Mol Biol Rep 33, 867–880 (2015). https://doi.org/10.1007/s11105-014-0786-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-014-0786-1