Abstract
Background
Curcubita ficifolia Bouché (Cucurbitaceae) has high value as a food crop and medicinal plant, and also has horticultural value as rootstock for other melon species. China is home to many different cultivars, but the genetic diversity of these resources and the evolutionary relationships among them, as well as the differences between C. ficifolia and other Cucurbita species, remain unclear.
Results
We investigated the chloroplast (cp) genomes of 160 C. ficifolia individuals from 31 populations in Yunnan, a major C. ficifolia production area in China. We found that the cp genome of C. ficifolia is ~151 kb and contains 128 genes, of which 86 are protein coding genes, 34 encode tRNA, and eight encode rRNAs. We also identified 64 SSRs, mainly AT repeats. The cp genome was found to contain a total of 204 SNP and 57 indels, and a total of 21 haplotypes were found in the 160 study individuals. The reverse repeat (IR) region of C. ficifolia contained a few differences compared with this region in the six other Cucurbita species. Sequence difference analysis demonstrated that most of the variable regions were concentrated in the single copy (SC) region. Moreover, the sequences of the coding regions were found to be more similar among species than those of the non-coding regions. The phylogenies reconstructed from the cp genomes of 61 representative species of Cucurbitaceae reflected the currently accepted classification, in which C. ficifolia is sister to the other Cucurbita species, however, different interspecific relationships were found between Cucurbita species.
Conclusions
These results will be valuable in the classification of C. ficifolia genetic resources and will contribute to our understanding of evolutionary relationships within the genus Cucurbita.
Similar content being viewed by others
Introduction
The genus Cucurbita (Cucurbitaceae) is thought to have an American origin [1], and comprises 20–27 species [2], the majority of which are herbaceous. Five species are cultivated: C. argyrosperma, C. maxima, C. moschata, C. ficifolia and C. pepo; these are popular, economically important crops (gourds, squashes and pumpkins) and are widely cultivated in almost all regions having arable land. The genetic diversity and germplasm resources in four of these species (C. moschata, C. argyrosperma, C. maxima, and C. pepo) have been studied in some depth [3,4,5,6]. Furthermore, DNA barcoding [2], AFLP [7] and simple sequence repeats (SSRs) markers [8,9,10] have been developed for the study of genetic diversity and phylogenetic relationships in and between various Cucurbita species.
Cucubita ficifolia is a short-day plant. It is sensitive to temperature and is not heat-resistant. The plant is known as “black seed squash” in English, and is called “Black Seeded figleaf squash” in Chinese [11]. C. ficifolia originated in the Central-South American region [12], and is now grown world-wide in low-latitude/high-altitude regions. There are therefore no wild populations of C. ficifolia in China, although this species has a long history of cultivation in Yunnan, Sichuan, Guizhou and other higher altitude regions in the country [13]. Much of Yunnan Province has a climate suitable for the growth and cultivation of C. ficifolia, and this province has become the main production area in China. C. ficifolia is not as popular as a human food crop as other cultivated Cucurbita species, and has therefore received less research attention. However, C. ficifolia fruit yields are high, and the plants are strong, will grow on barren ground, and are resistant to cold, drought, and several diseases including Fusarium wilt. The species is therefore an important germplasm resource for the breeding of melon cultivars [7]. However, there have been only a few studies into the genetics of this species to date. Therefore, in order to effectively utilize and develop this resource, a better understanding of the genetic diversity of C. ficifolia, its differences from other Cucurbita species and the phylogenetic relationships within this group are required.
The chloroplast (cp) [14, 15] is important in various plant cell functions, including carbon fixation and photosynthesis as well as the stress response. The cp is semi-autonomous, having a semi-independent genome and encoding its own genetic system for the transcription translation and replication of DNA and RNA [16]. The structure of the cp genome is conserved. The double-stranded, circular DNA molecule has a quadripartite structure, comprising large (LSC) and small (SSC) single-copy regions usually separated by two inverted repeat (IR) regions [17,18,19]. The composition and order of the cp genes is highly conserved in most angiosperms [20], and the genome ranges between 120 and 160 kb in size [21]. Between 110 and 130 genes are usually present on the cp genome in flowering plants [22], and comprise genes related to photosynthesis, transcription/translation, and biosynthesis. Throughout the evolutionary history of the chloroplast, the cp genome has undergone certain major alterations, including the loss of specific introns, large-scale genomic rearrangements, and IR region contraction and expansion.
The cp genome is inherited from the maternal parent, is relatively small and simple in structure, with a low molecular weight and nucleotide substitution rate [23, 24]. The first cp genome to be assembled was that of Nicotiana tabacum [25], and with the rise of sequencing technology, the cp genomes of many plant species have been sequenced. Cp genomes, which are often used in species identification and analyses of genetic diversity [26,27,28], as well as phylogenetic, taxonomic and evolutionary studies [29] have even been called “DNA super barcodes” [23].
However, although the sequence and gene content of the plant cp genome are highly conserved [30], sequence variation occurs through loss or mutation of genes and pseudogenization [31]. These variants are valuable in species comparisons in the study of evolutionary relationships between taxa and plant taxonomy [32, 33].
We previously reported the cp genome from a single individual of C. ficifolia [34]. However, the genetic diversity in the cp genome in C. ficifolia resources from different regions are unknown. Compared with other cultivated melon species, C. ficifolia has received little research attention, and its systematic placement and relationships with other melons and gourds is unclear. In this study, we collected samples from 160 C. ficifolia landraces from Yunnan Province, China, and the cp genomes from these landraces were sequenced and assembled. In addition, we analyzed the GC content of the C. ficifolia cp genome, as well as the number of genes and repeat sequences, the codon usage bias and simple sequence repeats (SSR), and compared the IR region and gene differentiation in C. ficifolia with those in other Cucurbita species. The evolutionary relationships between C. ficifolia and other Cucurbitaceae species as inferred from their cp genomes is also discussed. These results will inform the wider utilization of C. ficifolia germplasm resources and further our knowledge of the evolution of this important family.
Materials and methods
Plant materials, DNA extraction and whole genome resequencing
We sampled a total of 160 C. ficifolia individuals from 31 different locations, and each location contained 1–15 individuals (Fig. 1 and Supplementary Table 1), Each sampled plant was at least 50 m apart. The C. ficifolia sampling locations were chosen to cover the main C. ficifolia growing areas in China. The materials were collected from artificial planting bases or from wild-growing plants, and official collection permits were not required because this species is not included on the Chinese List of National Key Protected Plants. The plant materials were formally identified by Yongjie Guo of the Kunming Institute of Botany, based on morphological characters. A voucher specimen of C. ficifolia has been deposited in the herbarium of the Kunming Institute of Botany, Chinese Academy of Sciences (KUN 1580438).
Collection locations and cp genome haplotype distribution of Cucurbita ficifolia germplasm resources. (A) Network map showed genetic analyses of 21 haplotypes. (B) Distribution of 21 haplotypes from 31 populations. Note: A1-A21 showed 21 haplotypes of C. ficifolia. The MAP is taken from CGIAR-CSI (https://srtm.csi.cgiar.org)
Fresh leaf tissue was stored at -80°C, and total DNA was extracted leaf samples using the CTAB method [35]. 1.0% Agarose gel electrophoresis (Omega Bio-Tek, Norcross, GA, United States) and a fluorometer (Qubit3.0, Thermo Fisher Scientific, Waltham, MA, United States) were used to quantify the DNA in each sample and assess its quality. DNA samples of sufficient quality were standardized to the same volume (10 µl) and quantity of DNA (200 µg). The Illumina NovaSeq6000 sequencing platform was used to randomly fragment the genomic DNA. Libraries were constructed from randomly fragmented genomic DNA (insert sizes ∼450 base-pairs (bp)), and 150 bp paired-end reads were generated. The raw sequencing data were filtered using fastp 0.21.0 with the parameters “fastp -q 10 -u 50 -y -g -Y 10 -e 20 -l 100 -b 150 -B 150”. Low-quality reads (50% or more of the bases with a quality score < 10) and poly-Ns (10% or more of the bases were Ns) were filtered out. Low-quality bases (Q ≤ 13) were removed, and adapters were removed from both ends. The clean data were used in the subsequent analyses.
Chloroplast genome assembly, gene annotation and sequence analysis
The clean reads were assembled using the GetOrganelle pipeline (https://github.com/Kinggerm/GetOrganelle). A reference genome C. moschata (Duch. ex Lam.) Duch. ex Poiret [36] was used to check the contigs, using BLAST (https://blast.ncbi.nlm.nih.gov/); the contigs were then aligned and oriented according to the reference genome. Annotation of the genome was automatic using the CpGAVAS pipeline [37] and Geneious 8.1 [38] was used to adjust the start/stop codons and intron/exon boundaries. The tRNA was identified using tRNAscan-SE v2.0 [39]. A physical map of the cp genome was generated using the online tool OGDraw v1.2 (http://ogdraw.mpimp-golm.mpg.de/) [40].
Analysis of the features of the chloroplast genome
Chloroplast genomes contain repetitive sequences, which are believed to be important in genome rearrangement and stabilization [41]. REPuter [42] was used to find forward tandem repeats, reverse repeats, complement repeats and palindromic repeats ≧ 16 bp in the cp genome of C. ficifolia, with a minimum alignment scored and maximum period size of 500. SSR markers in the genomes were identified using Phobos v3.3.12 [43] and SSRHunter [44], which use a recursive algorithm to identify dinucleotide and other multinucleotide repeats with lengths between two and six base pairs with at least four copies. Analysis of codon usage and calculation of relative synonymous codon usage (RSCU) were conducted using the MEGA v11 software [45].
Chloroplast genome genetic diversity analyses based on 160 individuals
For the identification of C. ficifolia varieties, we used MAFFT V7.471 (Kazutaka Katoh, Japan) which resulted in an alignment data matrix that could be used for DNAsp analysis [46]. Insertion/deletion polymorphisms (indels) and single nucleotide polymorphisms (SNPs) in the cp genome were then identified using DNAsp [47]. All indels found in the aligned sequences were included in the following analyses. DNAsp was also used to conduct a sliding window analysis [47], where the window length was set to 100 bp and the step size to 25 bp. Haplotype data files were generated in DNAsp and the haplotype diversity (Hd) was also calculated.
Structure of the C. ficifolia genome and comparison of the genome with others from the genus Cucurbita
Although the IRs are highly conserved in cp genomes, contraction and expansion at their borders are common in evolutionary time, and may significantly influence their boundaries with the LSC or SSC regions, as well as leading to size variations in different cp genomes [48,49,50] To compare the IR boundaries in several Cucurbita species, the cp genomes of seven Cucurbita species were downloaded from NCBI and compared with the C. ficifolia cp genome using IRscope10 [51].
Comparative analysis of different cp genomes is extremely important in genomics [52]. We used the online software mVISTA11 [53] using the Shuffle-LAGAN alignment model [54] to determine the differences between the cp genomes of the seven study Cucurbita species, with C. argyrosperma as a reference.
Phylogenetic reconstruction and population structure analysis
The cp genome sequences of 61 Cucurbitaceae species as well as an outgroup (Lavandula angustifolia, Lamiaceae) were downloaded from GenBank. MAFFT [46] was used to construct an alignment of the 61 downloaded sequences with the 21 C. ficifolia cp genome haplotype sequences from our study. To resolve the phylogenetic placement of C. ficifolia within the Cucurbitaceae, a maximum likelihood (ML) phylogenetic tree was reconstructed in MEGA v11 [45] using the cp genome sequences with the GTR + GAMMA substitution model and including a tree robustness assessment using 1000 replicates of rapid four bootstrap, the GTR + GAMMA model was chosen through “Find Best DNA model” in MEGA v11 [45].
Results
Characteristics of the C. Ficifolia chloroplast genome
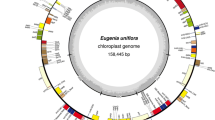
The collection localities of the 160 C. ficifolia study individuals are shown in Fig. 1. Resequencing these 160 individuals on an Illumina NovaSeq6000 sequencer generated 758.08 Gbp of clean data, with a total of 2.2 million 100 bp paired-end reads (332 Gb of sequence data), 93.63% of which had a Q value ≥ 30. The average rate of alignment of samples to the reference genome was 93.17%, the average depth of coverage was 10 × and the genome coverage was 66.11% (with at least one base coverage). The above resequencing data were then used to assemble and annotate the complete cp genome of C. ficifolia. We found that the C. ficifolia cp genome was circular and double-stranded, and that it ranged in size in our study individuals from 157,150 to 157,643 bp (Fig. 2). The genome comprised the LSC (87,730 − 88,210 bp), the SSC (18,136 − 18,144 bp), and IRa and IRb (25,638 − 25,597 bp). Throughout the genome, the GC content was 37.2% on average, with the IRa, IRb, SSC and LSC having 43.0, 43.0, 31.6 and 34.9% GC content, respectively (Table 1).
The C. ficifolia cp genome contained 128 genes in all individuals except one (FY_H2), where the ycf2 of FY_H2 was unannotated due to the presence of multiple termination codons. The atpA genes in individuals FY_H1, YB_H2 and YL_W1 were terminated prematurely and atpA was annotated as a pseudogene. The remaining individuals were all found to have 86 protein-coding genes (PCGs), 34 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes (Fig. 2). These genes were divided into three functional categories: photosynthesis (47 genes), self-replication (70), biosynthesis (6) and genes of unknown function (5) (Fig. 2; Table 2). In the IR regions, nineteen gene species were duplicated either completely or partially, including eight PCGs, (ndhB, ndhF, rpl2, rpl23, rps7, rps12, ycf2 and ycf15), seven genes encoding tRNAs (trnA-TGC, trnI-CAT, trnI-GAT, trnL-CAA, trnN-GTT, trnR-ACG and trnV-GAC), and the four genes encoding rRNAs (4.5 S, 5 S, 16 S and 23 S). The results of gene structure analysis suggested that the C. ficifolia genome included seven genes that contained introns, of which four were found in the LSC and one in the SSC (ndhA). Five genes contained a single intron (atpF, ropC1, ycf3, ndhA, ndhB) and two contained two introns (clpP and rpl2) (Table 3). The structural elements were almost identical between the 160 C. ficifolia varieties, suggesting that the structure of the cp genome is highly conserved in this species.
Chloroplast genome genetic diversity analyses based on 160 individuals
A total of 57 indels and 204 SNPs were found in the data matrix of our 160 individuals. Of the 204 SNPs, 149 were singleton variable sites and 55 were parsimony-informative sites (Fig. 3). A total of 21 haplotypes were resolved in the 160 sample C. ficifolia individuals and the haplotype diversity (Hd) was 0.598. Haplotype A1 was most widespread, appearing in 99 individuals from 28 populations, followed by A2, which appeared in 13 populations. 15 haplotypes occurred only once (Fig. 1). Populations JS and NH harbored five haplotypes and GJ had four. Most other populations had only one or two haplotypes. From the network analysis, the dominant haplotype A1 could form haplotypes A4, A5 and A12 through a single mutation, and A8 through two single mutations. The 21 haplotypes formed a network model, but not a linear model, meaning that they have a complex evolutionary relationship (Fig. 1). The sliding window analysis of 21 haplotypes showed that most variation occurred in four regions, especially around the position of 50,000 bp (Fig. 4A).
Types and distributions of repeat sequences and short sequence repeats (SSRs) in Cucurbita ficifolia chloroplast (cp) genomes. (A) Proportion of SSRs in C. ficifolia cp genomes. (B) Numbers of different types of repeat sequences in the C. ficifolia cp genomes. (C) Codon content for the 20 amino acids and stop codons in 86 protein-coding genes of C. ficifolia chloroplast genomes. (D) Sliding window analysis of 160 complete chloroplast (cp) genomes from Cucurbita ficifolia. The x-axis represents the midpoint of the window and the y-axis represents the nucleotide diversity (Pi) of each window. The window length is 600 bp with a 200-bp step size
SSRs, repeat sequences and codon usage bias in the C. Ficifolia chloroplast genome
Because A1 was the most widespread haplotype among the 160 C. ficifolia individuals, it was chosen for the following SSR, repeat and codon usage bias analyses. The cp genome of C. ficifolia haplotype A1 contained only 64 identified SSRs (Fig. 4A). There were 19 SSRs of type TA (4), and 9 of type AT (4), and most of the SSRs appeared only once. Dinucleotide, trinucleotide and tetranucleotide SSRs represented 99.4, 0.03, and 0.03% of the total SSRs, respectively (Fig. 4B). The LSC region contained the great majority of the SSRs (59.3%). Ten SSRs were found in the IR regions, and only five were found in the SSC. A/T repeats represented 70.3% of the C. ficifolia cp SSRs, indicating that there was an A/T nucleotide bias.
We then analyzed four types of repetitive sequences: forward, reverse, palindromic, and complement repeats. The C. ficifolia cp haplotype A1 contained 296 repeat sequences, with 106, 68, 96 and 26 forward, reverse, palindromic and complement repeats, respectively, which ranged in length from 16 to 200 bp, with most (accounting for 80.1% of the total) being 16–20 bp. A palindromic repeat in the LSC region was the longest at 166 bp. The locations of the repeats are given in Fig. 4C.
The protein-coding genes were then analyzed for codon usage. We found 45 codons with an RSCU > 1.0. The five most commonly used codons were UUU (4.24%), AAA (3.97%), AAU (3.69%), AUU (3.50%) and UAU (3.04%). The most common amino acids were Leu (L), Ser (S), Ile (I), all of which occurred > 4000 times. Conversely, the amino acids Met (M) and Trp (W) were used rarely, with fewer than 1000 occurrences (Fig. 4D). Codon preference analysis results showed that the 3’ ends of most codons, containing A or T, had RSCU values higher than 1, and that these codons were preferred.
IR expansion and contraction in the Cucurbita cp Genome
We then compared the IR boundaries characteristic of C. ficifolia cp genomes of haplotype A1 to the cp genomes of six other Cucurbita species (C. argyrosperma, C. maxima, C. moschata, C. okeechobeensis, C. pedatifolia, C. pepo). The complete cp genomes of these Cucurbita species ranged in length from 157,204 bp (C. maxima) to 158,614 bp (C. pedatifolia). All of the cp genomes included in this study had a structure typical of the angiosperms, being quadripartite and including a large and a small single-copy region, and two inverted repeat regions (Fig. 5; Table 4). We compared the genomic regions spanning the IR/LSC and IR/SSC junctions in our seven study species. The length of IR regions ranged from 25,555 in C. okeechobeenisis, which also had the smallest cp genome, to 26,582 bp in C. pedatifolia, which had the largest cp genome. Similarly, the LSC regions ranged in length from 87, 322 in C. pedatifolia, which also had the smallest cp genome, to 88, 387 bp in C. argyrosperma, which had the largest cp genome. There was no significant difference in the size of the SSC among these four species, and variation in the sizes of the IR and LSC regions appears to be the main reason for the differences in length seen in the different C. ficifolia cp genomes.
Comparison of border distance between adjacent genes and junctions of the LSC, SSC and two IR regions among the chloroplast genomes of seven Cucurbita species. Boxes above or below the main line indicate the adjacent border genes. The figure is not to scale with respect to sequence length, and only shows relative changes at or near the IR/SC borders
We found a few differences in the IR/LSC and IR/SSC junction regions among our study species. Five genes were present at the IR/LSC or IR/SSC boundaries: rps19, rpl2, ycf1, ndhF and trnH. We then analyzed the characteristics of the four boundaries IRa-SSC (JSA), IRa-LSC (JLA), IRb-LSC (JLB), and IRbSSC (JSB). We found that the JLB boundary lay in the intergenic region between rps19 and rpl2 in four Cucurbita cp genomes, but was located in the rpl2 gene in C. moschata and C. pepo, and in rps19 for C. pedatifolia. The JSB boundary was consistent throughout all the tested species and was located in the ndhF gene, and the JSA boundary was located in the ycf1 gene. The JLA boundary was located between the rpl2 and trnH genes (Fig. 5). The IR/LSC and IR/SSC junction regions are therefore relatively conserved between different Cucurbita species.
Comparative analysis of chloroplast genomes in Cucurbita species using mVISTA
Multiple alignments of the cp genomes from our seven study Cucurbita species were constructed in the mVISTA software, using C. argyrosperma as a reference (Fig. 6). Overall, the sequences of the cp genomes in Cucurbita species were highly conserved. Unsurprisingly, we found that the coding regions were more highly conserved than non-coding regions, and that the IR regions were less divergent than the LSC and SSC regions. Notably, intron-containing genes were found to have high levels of variability.
Comparison of four cp genomes using the mVISTA alignment program. The x-axis represents the coordinates in the cp genome. The y-axis indicates the average percent identity of sequence similarity in the aligned regions, ranging between 50% and 100%. Purple bars represent exons, blue bars represent untranslated regions (UTRs), pink bars represent noncoding sequences (CNS), gray bars represent mRNA, and white bars represent differences in genomics
The intergenetic spacers, including trnL-trnF, trnT-trnL, rpl32-trnL, rbcL-accD, trnS-trnR, rps12-trnV were the most highly divergent sequences in the seven cp genomes studied. The coding regions with the highest divergence were the accD, petD, ycf1 and ycf2 sequences. This is similar to the results obtained in previous studies [55,56,57], and suggest that these regions might evolve rapidly in Cucurbita, and could therefore be useful in the identification of Cucurbita species.
We used DNAsp to investigate nucleotide variability (π) and levels of sequence divergence within the aligned genome sequences from the seven study species. The nucleotide variability (π) was found to be 0.0034, showing that the genomes were relatively divergent despite the relatedness of the study species. 1,486 SNPs were found. The sliding window analysis of this genus showed that most variation occurred in the LSC and SSC regions, with the IR region being relatively conservative (Fig. 7). Our results suggest that the cp genome could informative for the reconstruction of species-level phylogenies this plant group, and that the LSC and SSC regions are a good choice when searching for loci for genetic diversity and phylogenetic analyses.
Phylogenetic analysis of 61 Cucurbitaceae species and 21 haplotypes of Cucurbita
To explore the evolutionary relationships among Cucurbitaceae species, the 21 cp genome haplotypes identified in C. ficifolia as well as the cp genome sequences of 61 other species in the Cucurbitaceae were aligned and used to reconstruct a phylogeny (Fig. 8). Lavandula angustifolia (Lamiaceae) was selected as an outgroup. ML trees were constructed using the whole cp genome. The different genera within the Cucurbitaceae can be distinguished in the phylogeny, meaning that the phylogeny reconstructed from cp genomic data was consistent with the traditional classification of this group. The Cucurbita species demonstrated a close genetic relationship and clustered together in a single branch. The 21 haplotypes identified from C. ficifolia also clustered together, reflecting the close evolutionary relationships among the different ecotypes of the same species. These results indicate that the whole cp genome is a reasonable choice for investigation of the evolutionary relationships within the Cucurbitaceae.
Discussion
Characteristics of the C. ficifolia chloroplast genome
The entire cp genome of C. ficifolia showed a conserved quadripartite structure. The length of the two reverse repeat regions was similar to those in most terrestrial plants. The IR region contained the rRNA genes, and had a lower GC content than that of the LSC and SSC regions. Overall, the cp genome had an AT content higher than the GC content, which reflects results reported from the chloroplast genomes from most higher plants [58]. A total of 7 genes in the C. ficifolia cp genome were found to contain introns. We then classified the cp genome of C. ficifolia using gene annotation, and divided the genes into three major categories according to function: genes for the photosynthetic system, genes for the genetic system and open reading frame and other genes. The results are basically consistent with those reported from other plants in Cucurbita [59, 60]. Codon preference analysis showed that the RSCU values of the 3’ ends of most codons containing A or T were higher than 1, and that these codons were preferred. We speculated that this might be due to the fact that the AT content of the whole C. ficifolia cp genome was enriched. Previous studies have shown that the second codon also has an AT bias [61, 62].
Repetitive sequences are widely found in cp genomes in higher plants and are an important source of genome variation [63]. The C. ficifolia cp genome contains 296 tandem repeats, suggesting that these repeats might lead to recombination or rearrangement of the cp genome during its evolution. Simple sequence repeats (SSR) are widely used as DNA markers [64]. We detected 67% of SSR markers in the C. ficifolia cp genome were found in the LSC and SSC regions, and a few in the IRs. This agrees with the results of many studies into the cp genome. We speculate that this number may be due to the repetitive nature of the IRs, which leads to sequence duplication and correction. In C. ficifolia, the cp genomic sequence has A/T base bias, and the SSR sequences mainly comprise poly-adenine (poly A) and poly-thymine (poly T) runs, which is consistent with our previous analysis of the C. ficifolia cp genome sequence. Chloroplast SSRs can be useful in phylogenetic analyses and species identification as well as in the study of species evolution and variation [65] and the SSRs detected in the C. ficifolia cp genome will therefore be important in future phylogenetic and population genetics studies in Cucurbita and the Cucurbitaceae.
Differences in the chloroplast genome and genetic diversity in C. ficifolia landraces
Several species from the genus Cucurbita are important as vegetables, and many different local cultivars and landraces have been developed [2]. Seed exchange has led to germplasm selection, and natural and artificial hybridization has contributed to genetic variation [66]. Collections of germplasm resources from different cultivars and landraces therefore represent a wide range of genetic diversity, which is of interest in the development of new cultivars with particular characteristics [67]. However, extensive genetic diversity studies are necessary before these germplasm resources can be effectively used. We sequenced the cp genomes of 160 individuals of C. ficifolia to investigate genetic diversity in this species. The haplotype diversity (Hd) was a little high as a domesticated specie. This could mean that C. ficifolia developed new genetic diversity in order to adapt to the local climate after its spread from the Central-South American region [12] and these new mutations represent important germplasm for the utilization of C. ficifolia. Network modeling of haplotypes showed complex genetic relationships within the C. ficifolia genetic resources. The variable geography and climate of southwest China may be the driving force behind this genetic variation, and anthropogenic intervention on the genetic structure of this species should also be considered.
Comparison of the chloroplast genomes of different Cucurbita species provides new insights into the phylogeny of this genus
We compared the cp genome sequences of seven species of Cucurbita. The genomes ranged in size from 157,204 bp (C. maxima) to 158,614 bp (C. pedatifolia), and all showed a conserved tetrad ring structure, consistent with cp genomes from other higher plants [68, 69]. We speculate that the different lengths of chloroplast genomic regions in Cucurbita is a result of shrinkage or expansion of the IR region compared to other Cucurbitaceae cp genomes [70,71,72]. IR regions shrinkage or expansion is relatively common [73, 74]. We found that SSC/IRa, SSC/IRb and LSC/IRa and had similar gene distribution patterns, and the ycf1 gene spanned the border between IRa and SSC. The rpl22 gene was located upstream of the LSC-IRa junction, and the rp12 gene overlapped the LSC-IRb region. This subtle span length can also be applied to species classification.
The maternally inherited cp genome evolves independently from the nuclear genome. It is also small and easy to isolate and sequence. These factors, as well as the moderate rate of base variation, means that the cp genome is often used as the basis for the study of phylogenetic relationships. The Cucurbitaceae family comprises about 1,000 species, and economically important plants in this family are widely cultivated in low latitudes with warm climates [75]. In order to reveal the phylogenetic relationships within the genus Cucurbita and their phylogenetic relationships with other species of Cucurbita, the cp genomes of 21 haplotypes of C. ficifolia, 6 species of Cucurbita and 56 further species from the Cucurbitaceae were selected as a data set with which to construct a phylogenetic tree. The 21 haplotypes of C. ficifolia formed a monophyletic group, and the six other species of Cucurbita clustered together. Schaefer et al. [76] investigated the history of the Cucurbitaceae using a multigene phylogeny for 114 species, and found that Cucurbita spp. have an apparent Central or South American origin, and that the split of the genus from its sister clade, Peponopsis, occurred about 16 (23 − 9) Myr ago. Chomicki et al. (2020) studied the phylogenetic distribution of cultivated Cucurbitaceae and made estimations of ancestral state on a phylogeny sampling 554 Cucurbitaceae species. The results suggested that the genus Cucurbita has a close relationship with Cucumis, Coccinia, Lagenaria and Citrullus. This close relationship is also indicated in our study.
In our study, the clade formed by C. ficifolia was sister to that comprising the rest of the Cucurbita species. This is consistent with the results from Kates et al. [77], Zheng et al. [78] and Sanjur et al. [79], who built phylogenies based on introns of single-copy nuclear genes, chloroplast and mitochondrial gene, respectively. However, the species-level topology of C. argyrosperma, C. maxima C. moschata, C. okeechobeensis and C. pepo in the phylogeny is obviously different when reconstructed different using different molecular markers (Fig. S1). Only one relationship among the six species in the clades remained consistent across all the previous molecular phylogenetic studies conducted to date: C. moschata is sister to C. argryosperma. However, using the whole cp genome, we found that C. moschata was sister to C. okeechobeensis. To date, only one of the three plant genomes at a time has been used to reconstruct the phylogenetic relationships in Cucurbita. The lack of congruence among these nuclear, mitochondrial or chloroplast-based phylogenetic trees might result from a lack of phylogenetically informative characters in one or more of the trees, or perhaps from reticulate evolution [80]. The phylogenetic reconstruction of Zheng et al. (2013), which was based on four chloroplast genes, also differed from the whole cp genome tree. This suggests that although many researchers use only a small number of gene loci to construct a phylogenetic tree, these few loci represent only a small amount of the information contained in the genome, which does not represent the evolutionary history of the whole genome. This should be considered when conducting phylogenetic analyses.
The cp genome has evolved independently of the nuclear genome. Its structure is conserved, although it contains many variable sites useful for analyzing the phylogenetic relationships between plants of the genus Cucurbita. However, the analysis of phylogenetic relationships using cp genome sequences has certain limitations when the study objects are species that undergo extensive interspecific or intergeneric hybridization, and the results may also be affected by introgression or incomplete lineage sorting. In order to make phylogenetic studies more instructive, more samples should be collected for analysis, and genetic analyses should contain not only chloroplast, mitochondrial and nuclear genetic information, but should also be combined with a knowledge of morphology, geography and domestication history. The analysis of the cp genomes of C. ficifolia and related species described here will provide basic theoretical data for further studies in this genus, and the phylogeny provides new insights into the phylogenetic taxonomic position of C. ficifolia within the Cucurbitaceae.
Data availability
The sequencing data generated in this study for the 160 samples have been submitted to the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA924019) under the BioProject accession PRJNA924019.
References
Wilson HD, Doebley J, Duvall M. Chloroplast DNA diversity among wild and cultivated members of Cucurbita (Cucurbitaceae). Theor Appl Genet. 1992;84:859–65. https://doi.org/10.1007/BF00227397.
Yoo E, Haile M, Ko HC, Choi YM, Cho GT, Woo HJ, Wang X, Sung P, Lee J, Lee J, et al. Development of SNP markers for Cucurbita species discrimination. Sci Hortic -Amsterdam. 2023;318. https://doi.org/10.1016/j.scienta.2023.112089.
Barrera-Redondo J, Ibarra-Laclette E, Vázquez-Lobo A, T.Gutiérrez-Guerrero Y, Vega GS, Piero D, Montes-Hernández S, Lira-Saade R, Eguiarte LE. The genome of Cucurbita argyrosperma (silver-seed gourd) reveals faster rates of protein- coding gene and long noncoding RNA turnover and neofunctionalization within Cucurbita (in Chinese). Mol Plant. 2019;12:506–20.
Zhu L, Zhu H, Li Y, Wang Y, Wu X, Li J, Zhang Z, Wang Y, Hu J, Yang S. Genome wide characterization, comparative and genetic diversity analysis of simple sequence repeats in Cucurbita species. Horticulturae. 2021;7:143. https://doi.org/10.3390/HORTICULTURAE7060143.
Ramjan M. Characterization of pumpkin (Cucurbita moschata Duch. Ex. Poir.) Germplasm through genetic variability, heritability and genetic advance. Electron J Plant Breed. 2021;12:91–6. https://doi.org/10.37992/2021.1201.014.
Wang W, Shi Y, Liu Y, Xiang C, Sun T, Zhang M, Shu Q, Qiu X, Bo K, Duan Y. Genetic relationships among Cucurbita pepo ornamental gourds based on EST-SSR markers. Czech J Genet Plant. 2021;57:125–39. https://doi.org/10.17221/27/2021-CJGPB.
Moya-Hernández A, Bosquez-Molina E, Serrato-Díaz A, Blancas-Flores G, Alarcón-Aguilar FJ. Analysis of genetic diversity of Cucurbita ficifolia Bouché from different regions of Mexico, using AFLP markers and study of its hypoglycemic effect in mice. S Afr J Bot. 2018;116:110–5. https://doi.org/10.1016/j.sajb.2018.02.409.
Formisano G, Roig C, Esteras C, Raffaella M, Fernando E. Genetic diversity of Spanish Cucurbita pepo landraces: an unexploited resource for summer squash breeding. Genet Resour Crop Evol. 2011;59:1169–84. https://doi.org/10.1007/s10722-011-9753-y.
Kaźmińska K, Sobieszek K, Targońska-Karasek Mg, Korzeniewska A, Niemirowicz-Szczytt K, Bartoszewski G. Genetic diversity assessment of a winter squash and pumpkin (Cucurbita maxima Duchesne) germplasm collection based on genomic Cucurbita -conserved SSR markers. Sci Hortic -Amsterdam. 2017;219:37–44. https://doi.org/10.1016/j.scienta.2017.02.035.
Kong Q, Chen J, Liu Y, Ma Y, Liu P, Wu S, Huang Y, Bie Z. Genetic diversity of Cucurbita rootstock germplasm as assessed using simple sequence repeat markers. Sci Hortic -Amsterdam. 2014;175:150–5. https://doi.org/10.1016/j.scienta.2014.06.009.
Lai Z. Cucurbita ficifolia, excellent germplasm resources (in Chinese). Yunnan Agricultural Sci Technol. 1991;4:40–2.
Pérez DM, Donadio LC, Carlos L. Caracterización De Frutos, semillas y plántulas de portainjertos de cítricos. Réanimation Urgences. 1999;2:259–66. https://doi.org/10.1016/S1164-6756(05)80470-0.
Kehua C. Cultivation techniques of Cucurbita ficifolia (in Chinese). Changjiang Vegetables. 1994;6:9–11.
Du Z, Lu K, Zhang K, He Y, Wang H, Chai G. The chloroplast genome of Amygdalus L. (Rosaceae) reveals the phylogenetic relationship and divergence time. BMC Genomics. 2021;22:645. https://doi.org/10.1186/s12864-021-07968-6.
Li C, Cai C, Tao Y, Sun Z, Jiang M, Chen L, Li J. Variation and evolution of the whole chloroplast genomes of Fragaria spp. (Rosaceae). Front Plant Sci. 2021;12:754209. https://doi.org/10.3389/fpls.2021.754209.
Favier A, Gans P, Boeri Erba E, Signor L, Muthukumar SS, Pfannschmidt T, Blanvillain R, Cobessi D. The plastid-encoded RNA polymerase-associated protein PAP9 is a superoxide dismutase with unusual structural features. Front Plant Sci. 2021;12:668897. https://doi.org/10.3389/fpls.2021.668897.
Song Y, Chen Y, Lv J, Xu J, Zhu S, Li M, Chen N. Development of chloroplast genomic resources for Oryza species discrimination. Front Plant Sci. 2017;8:1854. https://doi.org/10.3389/fpls.2017.01854.
Gao X, Zhang X, Meng H, Li J, Zhang D, Liu C. Comparative chloroplast genomes of Paris Sect. Marmorata: insights into repeat regions and evolutionary implications. BMC Genomics. 2018;19:878. https://doi.org/10.1186/s12864-018-5281-x.
Wang M, Wang X, Sun J, Wang Y, Ge Y, Dong W, Yuan Q, Huang L. Phylogenomic and evolutionary dynamics of inverted repeats across Angelica Plastomes. BMC Plant Biol. 2021;21:26. https://doi.org/10.1186/s12870-020-02801-w.
Chen J, Zang Y, Shang S, Liang S, Zhu M, Wang Y, Tang X. Comparative chloroplast genomes of Zosteraceae species provide adaptive evolution insights into seagrass. Front Plant Sci. 2021;12:741152. https://doi.org/10.3389/fpls.2021.741152.
Wang Y, Wang S, Liu Y, Yuan Q, Sun J, Guo L. Chloroplast genome variation and phylogenetic relationships of atractylodes species. BMC Genomics. 2021;22:103. https://doi.org/10.1186/s12864-021-07394-8.
Feng S, Zheng K, Jiao K, Cai Y, Chen C, Mao Y, Wang L, Zhan X, Ying Q, Wang H. Complete chloroplast genomes of four Physalis species (Solanaceae): lights into genome structure, comparative analysis, and phylogenetic relationships. BMC Plant Biol. 2020;20. https://doi.org/10.1186/s12870-020-02429-w.
Huang S, Ge X, Cano A, Salazar B, Deng Y. Comparative analysis of chloroplast genomes for five Dicliptera species (Acanthaceae): molecular structure, phylogenetic relationships, and adaptive evolution. PeerJ. 2020;8:e8450. https://doi.org/10.7717/peerj.8450.
Palmer JD, Stein DB. Conservation of chloroplast genome structure among vascular plants. Curr Genet. 1986;10:823–33. https://doi.org/10.1007/BF00418529.
Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–9. https://doi.org/10.1002/j.1460-2075.1986.tb04464.x.
Qiao J, Cai M, Yan G, Wang N, Li F, Chen B, Gao G, Xu K, Li J, Wu X. High-throughput multiplex cpDNA resequencing clarifies the genetic diversity and genetic relationships among Brassica napus, Brassica rapa and Brassica oleracea. Plant Biotechnol J. 2016;14:409–18. https://doi.org/10.1111/pbi.12395.
Jiao J, Yin Y. A strategy for developing high-resolution DNA barcodes for species discrimination of wood specimens using the complete chloroplast genome of three Pterocarpus species. Planta. 2019;250:95–104. https://doi.org/10.1007/s00425-019-03150-1.
Liu ZF, Ma H, Ci XQ, Li L, Li J. Can plastid genome sequencing be used for species identification in Lauraceae? Bot J Linn Soc. 2021;197:1–14. https://doi.org/10.1093/botlinnean/boab018.
Jheng C, Chen F, Lin TC, Chang JY. The comparative chloroplast genomic analysis of photosynthetic orchids and developing DNA markers to distinguish Phalaenopsis orchids. Plant Sci. 2012;190:62–73. https://doi.org/10.1016/j.plantsci.2012.04.001.
Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol. 2009;7:84. https://doi.org/10.1186/1741-7007-7-84.
Henriquez CL, Abdullah; Ahmed I, Carlsen MM, Mckain MR. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics. 2020;112:2349–60. https://doi.org/10.1016/j.ygeno.2020.01.006.
Song W, Chen Z, He L, Feng Q, Zhang H, Du G, Shi C, Wang S. Comparative chloroplast genome analysis of wax gourd (Benincasa hispida) with three Benincaseae species, revealing evolutionary dynamic patterns and phylogenetic implications. Genes. 2022;13:461. https://doi.org/10.3390/genes13030461.
Zhang YM, Han LJ, Yang CW, Yin ZL, Tian X, Qian ZG, Li GD. Comparative chloroplast genome analysis of medicinally important Veratrum (Melanthiaceae) in China: insights into genomic characterization and phylogenetic relationships. Plant Divers. 2022;44:13. https://doi.org/10.1016/j.pld.2021.05.004.
Zhang T, Xie J, Zhang J, Yang Z, Li X, He S. Analysis of Cucurbita ficifolia (Cucurbitaceae) chloroplast genome and its phylogenetic implications. Mitochondrial DNA Part B Resour. 2021;6:3033–5. https://doi.org/10.1080/23802359.2021.1959440.
Sue Porebski LG, Bernard B. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 1997;15:8–15. https://doi.org/10.1007/BF02772108.
Sun H, Wu S, Zhang G, Jiao C, Guo S, Ren Y, Zhang J, Zhang H, Gong G, Jia Z, et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol Plant. 2017;10:1293–306. https://doi.org/10.1016/j.molp.2017.09.003.
Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 2012;13:715. https://doi.org/10.1186/1471-2164-13-715.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. https://doi.org/10.1093/bioinformatics/bts199.
Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–7. https://doi.org/10.1093/nar/gkw413.
Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–74. https://doi.org/10.1007/s00294-007-0161-y.
Milligan BG, Hampton JN, Palmer JD. Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol Biol Evol. 1989;6:355–68. https://doi.org/10.1007/BF02270728.
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;4633–42. https://doi.org/10.1093/nar/29.22.4633.
Leese F, Mayer C, Held C. Isolation of microsatellites from unknown genomes using known genomes as enrichment templates. Limnol Oceanogr Methods. 2008;6:412–26. https://doi.org/10.4319/lom.2008.6.412.
Li Q, Wan JM. SSRHunter: development of a local searching software for SSR sites (in Chinese). Hereditas. 2005;27:808.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. https://doi.org/10.1093/molbev/mst197.
Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30. https://doi.org/10.1093/molbev/mst010.
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R, DnaSP. DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–7. https://doi.org/10.1093/bioinformatics/btg359.
Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 2007;8:174. https://doi.org/10.1186/1471-2164-8-174.
Xia Z, Wang YZ, Smith JF. Familial placement and relations of Rehmannia and Triaenophora (Scrophulariaceae s.l.) inferred from five gene regions. Am J Bot. 2009;96:519–30. https://doi.org/10.3732/ajb.0800195.
Yao X, Tang P, Li Z, Li D, Liu Y, Huang H. The first complete chloroplast genome sequences in Actinidiaceae: genome structure and comparative analysis. PLoS ONE. 2015;10:e0129347. https://doi.org/10.1371/journal.pone.0129347.
Amiryousefi A, Hyvönen J, Poczai P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–1. https://doi.org/10.1093/bioinformatics/bty220.
Huang Z, Xu J, Xiao S, Liao B, Gao Y, Zhai C, Qiu X, Xu W, Chen S. Comparative optical genome analysis of two pangolin species: Manis pentadactyla and Manis javanica. Gigaence. 2016;5:1–5. https://doi.org/10.1093/gigascience/giw001.
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046. https://doi.org/10.1093/bioinformatics/16.11.1046.
Brudno M, Malde S, Poliakov A, Do CB, Couronne O, Dubchak I, Batzoglou S. Glocal alignment: finding rearrangements during alignment. Bioinformatics. 2003;19:54–62. https://doi.org/10.1093/bioinformatics/btg1005.
He S, Yang Y, Li Z, Wang X, Guo Y, Wu H. Comparative analysis of four Zantedeschia chloroplast genomes: expansion and contraction of the IR region, phylogenetic analyses and SSR genetic diversity assessment. PeerJ. 2020;8:e9132. https://doi.org/10.7717/peerj.9132.
Park I, Kim WJ, Yeo SM, Choi G, Kang YM, Piao R, Moon BC. The complete chloroplast genome sequences of Fritillaria Ussuriensis Maxim. And Fritillaria Cirrhosa D. Don, and comparative analysis with other Fritillaria species. Molecules. 2017;22:982. https://doi.org/10.3390/molecules22060982.
Shen XF, Wu ML, Liao BS, Liu ZX, Bai R, Xiao SM, Li XW, Zhang BL, Xu J, Chen SL. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 2017;22:1330. https://doi.org/10.3390/molecules22081330.
Massouh A, Schubert J, Yaneva-Roder L, Ulbricht-Jones ES, Zupok A, Johnson MTJ, Wright S, Pellizzer T, Sobanski J, Bock R. Spontaneous chloroplast mutants mostly occur by replication slippage and show a biased pattern in the plastome of Oenothera. Plant Cell. 2016;28:911–29. https://doi.org/10.1105/tpc.15.00879.
Li B, Lin F, Huang P, Guo W, Zheng Y. Complete chloroplast genome sequence of Decaisnea insignis: genome organization, genomic resources and comparative analysis. Sci Rep -UK. 2017;7:10073. https://doi.org/10.1038/s41598-017-10409-8.
Chen C, Zheng Y, Liu S, Zhong Y, Xu M. The complete chloroplast genome of Cinnamomum camphora and its comparison with related Lauraceae species. PeerJ. 2017;5:e3820. https://doi.org/10.7717/peerj.3820.
Asaf S, Waqas M, Khan AL, Khan MA, Kang S-M, Imran QM, Shahzad R, Bilal S, Yun BW, Lee IJ. The complete chloroplast genome of wild rice (Oryza minuta) and its comparison to related species. Front Plant Sci. 2017;8:304. https://doi.org/10.3389/fpls.2017.00304.
Yin D, Wang Y, Zhang X, Ma X, He X, Zhang J. Development of chloroplast genome resources for peanut (Arachis hypogaea L.) and other species of Arachis. Sci Rep -UK. 2017;7:11649. https://doi.org/10.1038/s41598-017-12026-x.
Hu J, Gui S, Zhu Z, Wang X, Ke W, Ding Y, Min Xiang J. Genome-wide identification of SSR and SNP markers based on whole-genome re-sequencing of a Thailand wild sacred lotus (Nelumbo nucifera). PLoS ONE. 2015;10:e0143765. https://doi.org/10.1371/journal.pone.0143765.
Ebert D, Peakall R. Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol Ecol Resour. 2010;9:673–90. https://doi.org/10.1111/j.1755-0998.2008.02319.x.
Curci PL, Paola DD, Danzi D, Vendramin GG, Sonnante G. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PLoS ONE. 2015;10:e0120589. https://doi.org/10.1371/journal.pone.0120589.
Gomes RS, Machado Júnior RdA, Chagas CF, de Oliveira RR, Delazari RL, da Silva FT. Brazilian germplasm of winter squash (Cucurbita moschata D.) displays vast genetic variability, allowing identification of promising genotypes for agro-morphological traits. PLoS ONE. 2020;15:e0230546. https://doi.org/10.1371/journal.pone.0230546.
Rao NK. Plant genetic resources: advancing conservation and use through biotechnology. Afr J Biotechnol. 2004;3:136–45. https://doi.org/10.5897/AJB2004.000-2025.
Whitten WM, Neubig KM, Williams NH. Generic and subtribal relationships in Neotropical Cymbidieae (Orchidaceae) based on matK/ycf1 plastid data. Lankesteriana. 2013;13. https://doi.org/10.15517/lank.v13i3.14425.
Du YP, Bi Y, Yang FP, Zhang MF, Chen XQ, Xue J, Zhang XH. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Sci Rep. 2017;7:5751. https://doi.org/10.1038/s41598-017-06210-2.
Rousseau-Gueutin M, Bellot S, Martin GE, Boutte J, Chelaifa H, Lima O, Michon-Coudouel S, Naquin D, Salmon A, Ainouche K. The chloroplast genome of the hexaploid Spartina maritima (Poaceae, Chloridoideae): Comparative analyses and molecular dating. Proc. Natl. Acad. Sci. U.S.A 2015, 93, 5–16, https://doi.org/10.1016/j.ympev.2015.06.013.
Hao ZD, Cheng TL, Zheng RH, Zhou HB, YW; MP. The complete chloroplast genome sequence of a relict conifer glyptostrobus pensilis: comparative analysis and insights into dynamics of chloroplast genome rearrangement in Cupressophytes and Pinaceae. PLoS ONE. 2016;11:e0161809. https://doi.org/10.1371/journal.pone.0161809.
Zheng W, Chen J, Hao Z, Shi J. Comparative analysis of the chloroplast genomic information of Cunninghamia lanceolata (Lamb.) Hook with sibling species from the genera Cryptomeria D. Don, Taiwania Hayata, and Calocedrus Kurz. Int J Mol Sci. 2016;17(1084). https://doi.org/10.3390/ijms17071084.
Zhu A, Guo W, Gupta S, Fan W, Mower JP. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 2016;209:1747–56. https://doi.org/10.1016/j.ympev.2017.03.002.
He J, Yao M, Lyu RD, Lin LL, Cheng J. Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci Rep. 2019;9:15285. https://doi.org/10.1038/s41598-019-51601-2.
Wang J, Sun P, Li Y, Liu Y, Yang N, Yu J, Ma X, Sun S, Xia R, Liu X, et al. An overlooked paleotetra ploidization in Cucurbitaceae. Mol Biol Evol. 2018;35:16–26. https://doi.org/10.1093/molbev/msx242.
Schaefer H, Heibl C, Renner SS. Gourds afloat: a dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. P Roy Soc B -Biol Sci. 2008;276:843–51. https://doi.org/10.1098/rspb.2008.1447.
Kates HR, Soltis PS, Soltis DE. Evolutionary and domestication history of Cucurbita (pumpkin and squash) species inferred from 44 nuclear loci. Mol Phylogenet Evol. 2017;111:98–109. https://doi.org/10.1016/j.ympev.2017.03.002.
Zheng Y-H, Alverson AJ, Wang Q-F, Palmer JD. Chloroplast phylogeny of Cucurbita: evolution of the domesticated and wild species. J Syst Evol. 2013;51:326–34. https://doi.org/10.1111/jse.12006.
Sanjur OI, Piperno DR, Andres TC, Wessel-Beaver L. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: implications for crop plant evolution and areas of origin. Proc Natl Acad Sci U S A. 2002;99:535–40. https://doi.org/10.1073/pnas.012577299.
Knowles LL, Klimov PB. Estimating phylogenetic relationships despite discordant gene trees across loci: the species tree of a diverse species group of feather mites (Acari: Proctophyllodidae). Parasitology. 2011;138:1750–9. https://doi.org/10.1017/S003118201100031X.
Acknowledgements
We would like to thank Yongjie Guo, Mingjian Feng and Yan Zhao for help with the collection of samples.
Funding
The project was supported by the National Natural Science Foundation of China (Project number: 31500459), the Key Program of Agriculture-Related Special Funds (202301BD070001-027), the young talents of Yunnan Xingdian (XDYC-QNRC-2022-0233), the Yunnan Province Major Science and Technology Project (202402AE090012) and Expert Workstation of Zhangxiaolan project (202205AF150021).
Author information
Authors and Affiliations
Contributions
ZY and XL developed the research concepts. SH and BX directed most of the experimental and analytical work and wrote the manuscript. JX and HW collected the leaf material and participated in the experimental work. SC and GL directed analytical work, ZY and SH acquired the funding. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The experiments did not involve endangered or protected species. The collection of plant data was carried out with the permission of the relevant institutions, and complied with national or international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
He, S., Xu, B., Chen, S. et al. Sequence characteristics, genetic diversity and phylogenetic analysis of the Cucurbita ficifolia (Cucurbitaceae) chloroplasts genome. BMC Genomics 25, 384 (2024). https://doi.org/10.1186/s12864-024-10278-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10278-2