Abstract
Background
Poplar anthracnose, which is one of the most important tree diseases, is primarily caused by Colletotrichum gloeosporioides, which has been detected in poplar plantations in China and is responsible for serious economic losses. The characteristics of 84K poplar that have made it one of the typical woody model plants used for investigating stress resistance include its rapid growth, simple reproduction, and adaptability.
Results
In this study, we found that the resistance of 84K poplar to anthracnose varied considerably depending on how the samples were inoculated of the two seedlings in each tissue culture bottle, one (84K-Cg) was inoculated for 6 days, whereas the 84K-DCg samples were another seedling inoculated at the 6th day and incubated for another 6 days under the same conditions. It was showed that the average anthracnose spot diameter on 84K-Cg and 84K-DCg leaves was 1.23 ± 0.0577 cm and 0.67 ± 0.1154 cm, respectively. Based on the transcriptome sequencing analysis, it was indicated that the upregulated phenylpropanoid biosynthesis-related genes in 84K poplar infected with C. gloeosporioides, including genes encoding PAL, C4H, 4CL, HCT, CCR, COMT, F5H, and CAD, are also involved in other KEGG pathways (i.e., flavonoid biosynthesis and phenylalanine metabolism). The expression levels of these genes were lowest in 84K-Cg and highest in 84K-DCg.
Conclusions
It was found that PAL-related genes may be crucial for the induced resistance of 84K poplar to anthracnose, which enriched in the phenylpropanoid biosynthesis. These results will provide the basis for future research conducted to verify the contribution of phenylpropanoid biosynthesis to induced resistance and explore plant immune resistance-related signals that may regulate plant defense capabilities, which may provide valuable insights relevant to the development of effective and environmentally friendly methods for controlling poplar anthracnose.
Similar content being viewed by others
Background
As one of the three important fast-growing tree species, poplar is crucial for the construction of protection forests, while also serving as a source of industrial timber. China contains the largest poplar plantation area, which now exceeds 8.5 million hectares [1]. With the expansion of the poplar planting area worldwide, poplar diseases have become a major concern in the forestry industry and among ecologists [2]. During growth, poplar trees are susceptible to multiple pathogens and pests. Some of these pathogens are responsible for anthracnose, which is the most severe poplar disease that causes extensive damage to the leaves. Previous studies on poplar anthracnose identified the causative pathogens as Colletotrichum species, including C. graminicola [3], C. gloeosporioides, and C. poplar [4], of which C. gloeosporioides is the main pathogen causing poplar anthracnose [3, 4]. Hence, C. gloeosporioides was analyzed in this study. The notable characteristics of the poplar hybrid (Populus alba × Populus glandulosa) designated as 84K include its rapid growth, relatively simple reproduction, and adaptability, which may help to explain why it is increasingly being used as one of the typical woody model plants for stress resistance research.
The interaction between plants and surrounding organisms, including microorganisms, is a common phenomenon. The long-term co-evolution of plants and microorganisms resulted in the development of adaptive mechanisms enabling plants to withstand microbial infections [5]. These mechanisms facilitate the recognition of microbial signaling molecules and the activation of specific physiological responses. Secondary metabolites are the ultimate manifestation of microbe-induced changes in gene and protein expression, with the types and contents of secondary metabolites reflecting the physiological and pathological status of plants [1].
There are two common mechanisms mediating plant disease resistance, namely rapid local reactions (e.g., hypersensitive response) and systemic acquired resistance [6]. Induced resistance involves several physiological and biochemical reactions that occur in plants after certain pathogen-related stimuli are perceived. Examples include increases in the contents of soluble carbohydrates and phenolic compounds, the production and accumulation of plant defense hormones, changes in various enzyme activities, and the production of pathogenesis-related proteins, ultimately leading to increased resistance to pathogens [1]. Phenylalanine ammonia lyase (PAL) is a key enzyme for the synthesis of phytoalexins, lignin, and phenols. When plants are induced, PAL activity increases significantly, thereby affecting phenylpropanoid biosynthesis [7].
Phenylpropanoid biosynthesis is an important secondary metabolic pathway in plants, with the metabolites produced by this pathway accounting for approximately 20% of the secondary metabolites in plants. The molecular basis of phenylpropanoid biosynthesis and the mechanism precisely regulating metabolite biosynthesis have been studied extensively in recent decades [8]. Structural genes and transcription factor genes in the phenylpropanoid biosynthesis pathway have been cloned and functionally verified in a variety of plants [1, 9]. However, the effects of the phenylpropanoid biosynthesis pathway on induced resistance in poplar have not been thoroughly characterized. Most of the existing studies on poplar anthracnose mainly focused on how C. gloeosporioides infects poplar, the interaction between the metabolic pathway and C. gloeosporioides, and the changes in resistance-related compounds in different poplar species. Unfortunately, the infection of poplar by C. gloeosporioides has rarely been explored from the perspective of the poplar tree [1, 10, 11].
In this study, we analyzed the induced resistance of 84K poplar to anthracnose by comparing the poplar samples under different conditions. The results of this study may provide new insights into the molecular basis of the induced resistance of 84K poplar to anthracnose.
Methods
Plant and fungal materials
Tissue-cultured 84K poplar seedlings and C. gloeosporioides strain CFCC 80,308 (Cg) were provided by the Forest Pathology Laboratory of Beijing Forestry University [12]. Two 84K poplar seedlings were cultivated in one tissue culture bottle at 25 ± 2°C with a 16-h light (2,000 lx):8-h dark cycle and 60–70% relative humidity. The seedlings were grown until they had six leaves and then they were inoculated with C. gloeosporioides (Figure S1).
After C. gloeosporioides was grown on potato dextrose agar medium for 7 days, sterile water was repeatedly added to cover the medium surface and then collected. The concentration of the resulting spore (conidia) solution was adjusted to 106/mL, after which 30 µL was used to inoculate the 84K poplar seedlings. The control 84K poplar seedlings were treated with 30 µL sterile water (i.e., 84K-No samples). Of the two seedlings in each tissue culture bottle, one (84K-Cg) treated with 30 µL spore (conidia) solution was inoculated for 6 days, whereas the 84K-DCg sample treated with 30 µL spore (conidia) solution was another seedling inoculated at the 6th day and incubated for another 6 days (the 12th day) under the same conditions. The follow-up experiments were conducted using the three 84K inoculated seedlings (i.e., 84K-No, 84K-Cg, and 84K-DCg samples) (Figure S1). Meanwhile, the seedling symptoms were based on the size of lesion on the 6th inoculation day. The experiment was completed using three biological replicates and five technical replicates.
Total RNA extraction
Total RNA was extracted from the 84K poplar samples (whole plants) using the RNAprep Pure Plant kit (Tiangen, Beijing, China) and then the RNA integrity was evaluated using the Agilent 2100 Bioanalyzer. For the transcriptome sequencing analysis, cDNA libraries were prepared using the Illumina Kit (Illumina, San Diego, CA, USA) and then sequenced on the Illumina NovaSeq 6000 platform. Trimmomatic v.0.33 (RWTH Aachen University, Aachen, Germany) was used to screen for quality and filter the original Illumina reads to remove low-quality reads. The Populus alba × Populus tremula genome was selected as the reference genome (https://db.cngb.org/search/project/CNP0000339/).
Assembly and quality evaluation
The transcriptome analysis was performed using three biological replicates. Trinity (2.0.6) was used to assemble clean reads (after removing PCR duplicates to increase the assembly efficiency) and then Tgicl (2.1) was used to cluster and eliminate redundant transcripts to obtain unigenes. Next, Tgicl was used to cluster and remove redundant high-quality unigenes from each sample to obtain non-scalable and non-redundant unigenes for the subsequent analysis. The integrity and quality of the assembled transcriptome were evaluated using conserved sequences from the Benchmarking Universal Single-Copy Orthologs database.
Gene expression analysis
Bowtie2 (2.2.5) was used to map the clean reads of each sample to the assembled transcriptome. According to the comparison results, RSEM (1.2.8) was used to quantify the mapped reads for each sample and determine the expression level of each gene in nine poplar samples. The gene expression levels were standardized according to fragments per kilobase of exon model per million mapped reads (FPKM) values. On the basis of a coefficient of variation threshold of ≤ 0.8, three biological replicates were screened for genes with highly correlated expression levels.
Functional characterization of unigenes
To functionally annotate all unigenes, BLASTX (Basic Local Alignment Search Tool) was used to compare the transcripts with sequences in public databases (E-value ≤ 10− 5 set as the threshold for significance). The following databases were analyzed: Nr (http://www.ncbi.nlm.nih.gov), Swiss-Prot (http://www.ncbi.nlm.nih.gov), COG (http://www.ncbi.nlm.nih.gov/COG), and KEGG (http://www.genome.jp/kegg). Additionally, on the basis of the Nr annotations, Blast2GO (2.5.0) was used to assign GO terms to all unigenes (https://www.biocloud.net).
Identification of differentially expressed genes
Pairwise comparisons of different treatments were performed and the differentially expressed genes (DEGs) were identified using DEseq2 and the following criteria:|log2(fold-change)| ≥ 1 and p ≤ 0.1. A hierarchical clustering analysis of the DEGs was performed using the pheatmap function in the R software. According to the GO annotation results, KEGG annotation results, and official classifications, the DEGs were functionally annotated and an enrichment analysis was conducted using the phyper function in R (4.0.3). An FDR correction was performed for the p values. Functions with a Q value ≤ 0.05 were considered significantly enriched.
Quantitative real-time PCR (qRT-PCR)
The expression of key DEGs was analyzed by qRT-PCR. Total RNA was reverse transcribed using the Hifair® II 1st Strand cDNA Synthesis SuperMix for qPCR (with gDNA digester plus) (Yeasen Biotechnology, Shanghai, China). The primer pairs (Table S1) for the candidate genes were designed using Primer Premier 6.0. The qRT-PCR analysis was performed using the CFX Connect Real-Time PCR instrument (Bio-Rad, USA) and the Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (Yeasen Biotech-nology). The 18 S gene served as an internal reference in this study. Relative gene expression was calculated using the 2−∆∆Ct method [13].
Phenylpropanoid and flavonoid extraction and analyses by HPLC-MS/MS
Samples from different treatments (84K-No, 84K-Cg, and 84K-DCg) were lyophilized at −40°C for 24 h and then stored in an ultra-low temperature freezer. Frozen samples were ground to a powder (30 Hz, 1 min) using a MM 400 grinder (Retsch, Haan, Germany). The ground material was stored at −20°C until analysis. The analysis methods of high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) were as described in Zhang et al. [1]. For the qualitative analysis of phenylpropanoid and flavonoid metabolites, the type and relative content of each compound was determined from HPLC chromatograms.
Data analysis
The generated data were statistically analyzed using SPSS 17.0 (SPSS Co., Ltd., Chicago, Illinois, USA) and Origin 8.0 (Origin Lab, Northampton, Massachusetts, USA). A one-way analysis of variance involving Duncan’s test was used to determine the significance (p < 0.05) of the differences between treatments.
Data deposition
All transcriptome data have been deposited in National Center for Biotechnology Information (NCBI) under accession codes of PRJNA 1,052,950.
Results
Disease symptoms of the 84K poplar seedlings under different conditions
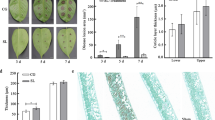
After 84K poplar seedlings (Fig. 1A) were inoculated with C. gloeosporioides, the leaf lesion area was examined for 6 consecutive days. The lesion size on the 84K-Cg leaf blades expanded rapidly (Fig. 1B and D). In contrast, the lesion area on the 84K-DCg leaf blades expanded slowly and the blackening of the lesion was less extensive (Fig. 1C and E). The average anthracnose spot diameter on the 84K-Cg and 84K-DCg leaves at 6 days post-inoculation was 1.23 ± 0.0577 cm and 0.67 ± 0.1154 cm, respectively; this difference was significant (Fig. 1F), which were measured at the 6th inoculation day.
Transcriptome sequencing and data assembly
We sequenced the transcriptomes of the 84K-No, 84K-Cg, and 84K-DCg samples to explore the changes in gene expression potentially related to the induced resistance of 84K poplar to anthracnose. The raw reads generated for the 84K-No, 84K-Cg, and 84K-DCg samples comprised 33,894,057 bp, 35,675,816 bp, and 35,226,466 bp, respectively (Table 1 and Fig. S2). For each sample, at least 5.92 Gb clean data were generated, with a quality score of Q30 for at least 94.29% of the data. The clean reads for each sample were mapped to the reference genome, with a mapping rate of 85.66–97.06%.
Differentially expressed genes identified by the transcriptome analysis
The sequenced transcriptomes were compared to identify the DEGs among the 84K-No, 84K-Cg, and 84K-DCg seedlings. A volcano map was produced to visualize the distribution of each group of DEGs. And we generated Venn diagrams to profile the DEG distribution among the 84K-No vs. 84K-Cg, 84K-No vs. 84K-DCg, and 84K-Cg vs. 84K-DCg. The comparison between 84K-No and 84K-Cg detected 2,042 DEGs (1,015 upregulated and 1,027 downregulated) (Fig. 2A and D). The comparison between 84K-No and 84K-DCg revealed 1,266 DEGs (969 upregulated and 297 downregulated) (Fig. 2B and D). The comparison between 84K-Cg and 84K-DCg identified 1,146 DEGs (599 upregulated and 547 downregulated) (Fig. 2C and D).
GO analysis of DEGs
The Gene Ontology (GO)-based functional annotations indicated that molecular-function-related GO terms were shown in the Table 2. The GO terms in the molecular function category were assigned to 10 upregulated DEGs (84K-No vs. 84K-Cg, 84K-No vs. 84K-DCg, and 84K-Cg vs. 84K-DCg) related to metabolic activities, including the following: phenylpropanoid biosynthesis; flavonoid biosynthesis; phenylalanine metabolism; phenylalanine, tyrosine, and tryptophan biosynthesis; tyrosine metabolism; isoquinoline alkaloid biosynthesis; and tropane, piperidine, and pyridine alkaloid biosynthesis (Table 2). In addition, 10 downregulated DEGs (84K-No vs. 84K-Cg, 84K-No vs. 84K-DCg, and 84K-Cg vs. 84K-DCg) included genes related to galactose metabolism, carbohydrate transport and metabolism, and ABC transporters (Table 2).
To analyze the associations between individual DEGs, we performed GO and KEGG enrichment analyses of the DEGs detected by the different comparisons. The GO enrichment analysis was performed to functionally characterize and classify all DEGs. The categories were divided into 56 subcategories. The top 10 categories in terms of the number of DEGs are presented in Fig. 3. In the biological process category, metabolic process (GO:0008152) was the most common term assigned to the DEGs. The GO analysis indicated that metabolic process was a significantly enriched GO term among the DEGs in all three comparisons (Fig. 3), of which the 84K-No vs. 84K-Cg comparison had the most DEGs (770) associated with metabolic processes, followed by the 84K-No vs. 84K-DCg comparison (437 DEGs) and the 84K-Cg vs. 84K-DCg comparison (393 DEGs).
KEGG analysis of DEGs
To further clarify the DEG functions, we conducted a KEGG enrichment analysis of all DEGs. A total of 117, 109, and 106 metabolic pathways were enriched among the DEGs detected by the 84K-No vs. 84K-Cg, 84K-No vs. 84K-DCg, and 84K-Cg vs. 84K-DCg comparisons, respectively. The 20 most enriched KEGG metabolic pathways (Fig. 4) included the following: MAPK signaling pathway–plant (ko04016), plant–pathogen interaction (ko04626), and phenylpropanoid biosynthesis (ko00940). Phenylalanine metabolism (ko00360) was not a significantly enriched KEGG pathway among the DEGs detected by the 84K-No vs. 84K-DCg comparison, but it was significantly enriched among the DEGs revealed by the 84K-No vs. 84K-Cg and 84K-Cg vs. 84K-DCg comparisons. Interestingly, the DEGs between the 84K-No and 84K-DCg samples were significantly associated with phenylpropanoid biosynthesis (ko00940). These results suggest that the 84K poplar response to a C. gloeosporioides infection may vary depending on the inoculation mode. Moreover, phenylpropanoid biosynthesis (ko00940) may be related to the induced resistance of 84K poplar to anthracnose.
Verification of the expression of the genes related to phenylpropanoid biosynthesis
Phenylpropanoid biosynthesis (ko00940) was an enriched KEGG pathway in all three groups, but not to the same extent. Thus, we analyzed the DEGs associated with this metabolic pathway in greater detail. The phenylpropanoid biosynthesis-related genes were generally more highly expressed in 84K-DCg than in 84K-No and 84K-Cg, with the lowest expression levels detected in 84K-Cg (Fig. 5). For example, the expression levels of a PAL-encoding gene (Pop_A06G085807) in 84K-No, 84K-Cg, and 84K-DCg were 276.43, 75.45, and 711.70, respectively. Accordingly, the expression level was more than 9-times higher in 84K-DCg than in 84K-Cg. Furthermore, in addition to being associated with phenylpropanoid biosynthesis (ko00940; Figure S3), this gene was also related to phenylalanine metabolism (ko00360; Figure S4). In contrast, the expression of the berberine bridge enzyme gene (Pop_A01G081808), which was also associated with phenylpropanoid biosynthesis (ko00940; Figure S3), was more similarly expressed between 84K-DCg and 84K-Cg (only 1.92-times higher in 84K-DCg than in 84K-Cg). The 16 phenylpropanoid biosynthesis-related DEGs were classified in eight gene families encoding various proteins, including PAL, cinnamate 4-hydroxylase (C4H), 4-coumaric acid-coenzyme A ligase (4CL), shikimate O-hydroxycinnamyl transferase (HCT), cinnamoyl coenzyme A reductase (CCR), caffeic acid 3-O-methyltransferase (COMT), ferulate-5-hydroxylase (F5H), and cinnamyl alcohol dehydrogenase (CAD). To verify the accuracy of the transcriptome data, we selected the phenylpropanoid biosynthesis-related DEGs in the 84K-No, 84K-Cg, and 84K-DCg samples for qRT-PCR, such as PAL, C4H, 4CL, HCT, CCR, and CAD (Figure S5). And the qRT-PCR analysis confirmed the RNA-seq data were reliable and accurate (Table S2). The enriched KEGG pathways assigned to these genes were phenylpropanoid biosynthesis (ko00940) and flavonoid biosynthesis (ko00941). For a more intuitive analysis of expression, we combined the gene expression levels and the processes associated with these two pathways (Fig. 5). Meanwhile, many phenylpropanoid and flavonoid metabolites downstream of these two pathways (ko00940 and ko00941), such as naringenin, rutin, apigenin, quercetin, catechin, and scutellarein, showed similar trends in their accumulation among different treatments, with highest concentration in the 84K-DCg samples (Table S3), which were consistent with the transcriptome results.
Discussion
The induced resistance of a plant host activated by a local infection involves a mechanism that mediates the production of a class of chemicals and signaling molecules that are transmitted through the phloem to the uninfected parts of the plant, thereby activating various defense responses [14]. In the current study on 84K poplar inoculated with C. gloeosporioides spores, the lesion area was smaller and the blackening of the lesion was less extensive on the 84K-DCg leaves than on the 84K-Cg leaves at 6 days post-inoculation. Additionally, the 84K-DCg leaf surface had substantially less mycelia than the 84K-Cg leaf surface. These differences may be related to the diversity in the expression levels of defense response-related genes between 84K-DCg and 84K-Cg. The reason for the decrease in the lesion area of the 84K-DCg leaves and how induced resistance is generated were unclear. Hence, we conducted a transcriptome analysis of the 84K-No, 84K-Cg, and 84K-DCg samples to elucidate the mechanism underlying the induced resistance of 84K poplar to anthracnose.
The transcriptome analysis revealed significant differences in gene expression levels in the 84K-No, 84K-Cg, and 84K-DCg seedlings, among which 84K-No and 84K-Cg had the most DEGs, with substantial changes in the expression of various genes, especially in 84K-DCg. For example, the expression levels of the genes encoding PAL, C4H, and 4CL were significantly upregulated. Both PAL and C4H regulate the synthesis of lignin monomers, while also contributing to other pathways downstream of the phenylpropanoid metabolic pathway. These findings suggest that in 84K poplar, phenylpropanoid metabolism may be induced in response to anthracnose. Specifically, metabolic pathways associated with multiple secondary compounds, including flavonoids and lignin, may be activated. According to the results of the GO and KEGG analyses, the identified DEGs were primarily involved in metabolic processes, especially phenylpropanoid metabolism. Phenylpropanoid biosynthesis, flavonoid biosynthesis, and isoflavone biosynthesis depend on the upregulated expression of genes encoding various enzymes, including PAL, C4H, and 4CL, which affect the accumulation of cinnamic acid, coumaric acid, shikimic acid, lignin, and caffeic acid [1]. Meanwhile, many phenylpropanoid and flavonoid metabolites downstream of ko00940 (Figure S3) and ko00941 (Figure S4) pathways, such as naringenin, rutin, apigenin, quercetin, catechin, and scutellarein, showed the highest concentration in the 84K-DCg samples (Table S3), which suggested that phenylpropanoid biosynthesis (ko00940) and flavonoid biosynthesis (ko00941) may be related to the induced resistance of 84K poplar to anthracnose.
Phenylpropanoid biosynthesis is one of the most important metabolic pathways in plants. In this pathway, phenylalanine derived from the shikimate pathway is involved in reactions catalyzed by PAL, C4H, and 4CL, with the resulting metabolites used as substrates [15, 16]. Because PAL is a key inducible enzyme linking the shikimate pathway to the phenylpropanoid biosynthesis pathway, its activity and concentration directly affect the abundance of downstream secondary metabolites (e.g., lignin, flavonoids, and isoflavones). Previous research confirmed that these compounds have important physiological roles influencing plant growth and development as well as resistance to biotic and abiotic stresses [17]. The PAL gene family has been identified in a variety of plants, although the number of PAL-encoding genes varies among plant species. Of the nine PAL genes in rice, PAL4 mediates broad-spectrum resistance to various pathogens [18]. The infection of tea plants by different pathogens leads to the significant upregulated expression of all six PAL genes [19]. In the present study, the phenylpropanoid and flavonoid metabolites (e.g., lignin, flavonoids, and isoflavones) were differentially expressed in the 84K-No and 84K-DCg seedlings (Table S3 and Fig. 5). Meanwhile, PAL activity and concentration directly affect the abundance of downstream phenylpropanoid and flavonoid metabolites in the 84K-DCg seedlings, such as naringenin, rutin, apigenin, quercetin, catechin, and scutellarein, which suggested that PAL-related genes may play an important role in the induced resistance of 84K poplar to anthracnose. On the other hand, some genes of 84K-Cg seedlings involved in phenylpropanoid biosynthesis such as PAL were down-regulated compared to 84K-No and 84K-DCg. It was reported that the appressorium of C. gloeosporioides may release effectors during infection to inhibit plant immune responses [20, 21], which may be the adaptation of C. gloeosporioides to host plants [12].
Compared with the corresponding expression levels in 84K-No and 84K-Cg, the genes involved in phenylpropanoid biosynthesis (ko00940) and flavonoid biosynthesis (ko00941) were more highly expressed in 84K-DCg, implying that phenylpropanoid metabolism was more active in 84K-DCg than in the other two samples. Our data also indicated that the genes encoding HCT and COMT were expressed at significantly higher levels in 84K-DCg than in 84K-No and 84K-Cg. Both HCT and COMT are considered to be the key factors controlling lignin production in the plant phenylpropanoid metabolic pathway [22]. The synthesis of lignin monomers involves a branch of the phenylpropanoid pathway. Specifically, three lignin monomers are oxidatively polymerized by laccase and peroxidase to generate high-molecular-weight lignin. In addition, lignin is tightly bound to polysaccharide molecules in the cell wall. Cross-linking forms a dense hydrophobic structure that increases the mechanical strength of plant cells and tissues, thereby acting as a natural physical barrier that inhibits the entry of pathogens [23]. There is currently considerable interest in the enzyme-catalyzed reactions in the lignin monomer synthesis pathway as well as the effects of lignin-related changes on plant immune responses, including those involving PAL, COMT, and CAD [24, 25]. In tobacco and Arabidopsis, the silencing or loss of the PAL gene reportedly decreases the overall immune response and increases the sensitivity to Pseudomonas syringae and tobacco mosaic virus [26, 27]. An earlier study demonstrated that Arabidopsis COMT mutants have enhanced mechanisms mediating responses to a variety of bacteria and fungi [28]. In the current study, the expression of genes encoding enzymes, such as PAL, COMT, HCT, and CAD, was significantly higher in 84K-DCg than in 84K-No and 84K-Cg, which may be related to the increase in the resistance of 84K-DCg to anthracnose. The improved immune response mechanism of 84K-DCg was likely critical for the observed decrease in the severity of the leaf lesions, which may be indicated phenylpropanoid biosynthesis mediated the induced resistance of 84K poplar and inproved the 84K-DCg treatments resistance to C. gloeosporioides.
The 84K-Cg and 84K-DCg samples were grown in the same tissue culture bottle, but they exhibited significantly different symptoms. Therefore, we hypothesized that there may be flavonoids or volatile gases in the air that directly inhibit pathogen infections or serve as a signal that stimulates an induced resistance-related mechanism. We will test this hypothesis as part of our future research. External biotic or abiotic stimuli can significantly induce the expression of plant genes related to phenylpropanoid metabolism [29], ultimately leading to a systemic immune response and the development of induced resistance.
Conclusions
In this study, we analyzed the DEGs among the differentially inoculated 84K-No, 84K-Cg, and 84K-DCg samples using transcriptomics techniques. We subsequently focused on the DEGs significantly associated with the phenylpropanoid biosynthesis pathway. Meanwhile, PAL activity and concentration directly affect the abundance of downstream phenylpropanoid and flavonoid metabolites in the 84K-DCg seedlings, such as naringenin, rutin, apigenin, quercetin, catechin, and scutellarein, which indicated that PAL-related genes enriched in the phenylpropanoid biosynthesis may be crucial for the induced resistance of 84K poplar to anthracnose. These results will provide the basis for future research conducted to verify the contribution of phenylpropanoid biosynthesis to induced resistance and explore plant immune resistance-related signals that may regulate plant defense capabilities.
Data availability
The datasets generated during the current study are available in National Center for Biotechnology Information (NCBI) repository, accession number: PRJNA 1052950, which will be released upon publication.
References
Zhang L, Bao H, Meng F, Ren Y, Tian C. Transcriptome and metabolome reveal the role of flavonoids in poplar resistance to poplar anthracnose. Ind Crops Prod. 2023;197:116537.
Jayawardena R, Hyde K, Damm U, Cai L, Liu M, Li X, Zhang W, Zhao W, Yan J. Notes on currently accepted species of Colletotrichum. Mycosphere. 2016;7:1192–260.
Dipak S, Chandra J, Singh A. Response of fungicides and antibiotics against anthracnose of poplar caused by Colletotrichum graminicola. Indian Forester. 1999;125:566–72.
Li Z, Liang YM, Tian CM. Characterization of the causal agent of poplar anthracnose occurring in the Beijing region. Mycotaxon. 2012;120:277–86.
Gong Q, Wang Y, He L et al. Molecular basis of methyl-salicylate-mediated plant airborne defence. Nature, 2023: 1–10.
Van Loon LC. Induced resistance in plants and the role of pathogenesis-related proteins. Eur J Plant Pathol. 1997;103:753–65.
Zeneli G, Krokene P, Christiansen E, et al. Methyl jasmonate treatment of mature Norway spruce (Picea abies) trees increases the accumulation of terpenoid resin components and protects against infection by Ceratocystis Polonica, a bark beetle-associated fungus. Tree Physiol. 2006;26:977–88.
Biała W, Jasiński M. The phenylpropanoid case–it is transport that matters. Front Plant Sci. 2018;9:1610.
Fraser CM, Chapple C. The phenylpropanoid pathway in Arabidopsis. Volume 9. The Arabidopsis Book/American Society of Plant Biologists; 2011.
Wang X, Lu D, Tian C. Mucin Msb2 cooperates with the transmembrane protein Sho1 in various plant surface signal sensing and pathogenic processes in the poplar anthracnose fungus Colletotrichum gloeosporioides. Mol Plant Pathol. 2021;22:1553–73.
Liu N, Meng F, Tian C. Transcriptional network in Colletotrichum gloeosporioides mutants lacking Msb2 or Msb2 and Sho1. J Fungi. 2022;8:207.
Meng F, Tian C. Gene family expansion during the adaptation of Colletotrichum gloeosporioides to woody plants. J Fungi. 2023;9:1185.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods. 2001;25:402–8.
Chen Z, Silva H, Klessig DF. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science. 1993;262:1883–6.
Zhang X, Liu C-J. Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol Plant. 2015;8:17–27.
Muro-Villanueva F, Mao X, Chapple C. Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr Opin Biotechnol. 2019;56:202–8.
Mauch-Mani B, Slusarenko AJ. Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora Parasitica. Plant Cell. 1996;8:203–12.
Tonnessen BW, Manosalva P, Lang JM, Baraoidan M, Bordeos A, Mauleon R, Oard J, Hulbert S, Leung H, Leach JE. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol Biol. 2015;87:273–86.
Wu Y, Wang W, Li Y, Dai X, Ma G, Xing D, Zhu M, Gao L, Xia T. Six phenylalanine ammonia-lyases from Camellia sinensis: evolution, expression, and kinetics. Plant Physiol Biochem. 2017;118:413–21.
Yang G, Yang J, Zhang Q, Wang W, Feng L, Zhao L, An B, Wang Q, He C, Luo H. The effector protein CgNLP1 of Colletotrichum gloeosporioides affects invasion and disrupts nuclear localization of necrosis-induced transcription factor HbMYB8-Like to suppress plant defense signaling. Front Microbiol. 2022;13:911479.
Zhao L, Liao Z, Feng L, An B, He C, Wang Q, Luo H. Colletotrichum gloeosporioides Cg2LysM contributed to virulence toward rubber tree through affecting invasive structure and inhibiting chitin-triggered plant immunity. Front Microbiol. 2023;14:1129101.
Irani S, Todd CD, Wei Y, Bonham-Smith PC. Changes in phenylpropanoid pathway gene expression in roots and leaves of susceptible and resistant Brassica napus lines in response to Plasmodiophora brassicae inoculation. Physiol Mol Plant Pathol. 2019;106:196–203.
Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–46.
Bhuiyan NH, Selvaraj G, Wei Y, King J. Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J Exp Bot. 2009;60:509–21.
Adams-Phillips L, Briggs AG, Bent AF. Disruption of poly (ADP-ribosyl) ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol. 2010;152:267–80.
Elkind Y, Edwards R, Mavandad M, Hedrick SA, Ribak O, Dixon RA, Lamb CJ. Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc Natl Acad Sci. 1990;87:9057–61.
Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou Y-H, Yu J-Q, Chen Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010;153:1526–38.
Quentin M, Allasia V, Pegard A, Allais F, Ducrot P-H, Favery B, Levis C, Martinet S, Masur C, Ponchet M. (2009). Imbalanced lignin biosynthesis promotes the sexual reproduction of homothallic oomycete pathogens. PLoS Pathog 5, e1000264.
Wong JH, Namasivayam P, Abdullah MP. The PAL2 promoter activities in relation to structural development and adaptation in Arabidopsis thaliana. Planta. 2012;235:267–77.
Funding
This work was supported by the National Natural Science Foundation of China (32201564) and the Beijing Forestry University Undergraduate Training Programs for Innovation and Entrepreneurship (X202210022019).
Author information
Authors and Affiliations
Contributions
FM conceived the project. FM, FX and LZ designed the experiments. FX, LZ and WG performed the experiments and analyzed the data. FX, LZ, WG and HF wrote the manuscript. FM and CT revised the manuscript. All authors discussed the manuscript, read, and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xing, F., Zhang, L., Ge, W. et al. Comparative transcriptome analysis reveals the importance of phenylpropanoid biosynthesis for the induced resistance of 84K poplar to anthracnose. BMC Genomics 25, 306 (2024). https://doi.org/10.1186/s12864-024-10209-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10209-1