Abstract
Background
Lipoxygenase (LOX) is a multifunctional enzyme that is primarily related to plant organ growth and development, biotic and abiotic stress responses, and production of flavor-associated metabolites. In higher plants, the LOX family encompasses several isozymes with varying expression patterns between tissues and developmental stages. These affect processes including seed germination, seed storage, seedling growth, fruit ripening, and leaf senescence. LOX family genes have multiple functions in response to hormones such as methyl jasmonate (MeJA) and salicylic acid.
Results
In this study, we identified 30 and 95 LOX homologs in Medicago truncatula and Medicago sativa, respectively. These genes were characterized with analyses of their basic physical and chemical properties, structures, chromosomal distributions, and phylogenetic relationships to understand structural variations and their physical locations. Phylogenetic analysis was conducted for members of the three LOX subfamilies (9-LOX, type I 13-LOX, and type II 13-LOX) in Arabidopsis thaliana, Glycine max, M. truncatula, and M. sativa. Analysis of predicted promoter elements revealed several relevant cis-acting elements in MtLOX and MsLOX genes, including abscisic acid (ABA) response elements (ABREs), MeJA response elements (CGTCA-motifs), and antioxidant response elements (AREs). Cis-element data combined with transcriptomic data demonstrated that LOX gene family members in these species were most likely related to abiotic stress responses, hormone responses, and plant development. Gene expression patterns were confirmed via quantitative reverse transcription PCR. Several MtLOX genes (namely MtLOX15, MtLOX16, MtLOX20, and MtLOX24) belonging to the type I 13-LOX subfamily and other LOX genes (MtLOX7, MtLOX11, MsLOX23, MsLOX87, MsLOX90, and MsLOX94) showed significantly different expression levels in the flower tissue, suggesting roles in reproductive growth. Type I 13-LOXs (MtLOX16, MtLOX20, MtLOX21, MtLOX24, MsLOX57, MsLOX84, MsLOX85, and MsLOX94) and type II 13-LOXs (MtLOX5, MtLOX6, MtLOX9, MtLOX10, MsLOX18, MsLOX23, and MsLOX30) were MeJA-inducible and were predicted to function in the jasmonic acid signaling pathway. Furthermore, exogenous MtLOX24 expression in Arabidopsis verified that MtLOX24 was involved in MeJA responses, which may be related to insect-induced abiotic stress.
Conclusions
We identified six and four LOX genes specifically expressed in the flowers of M. truncatula and M. sativa, respectively. Eight and seven LOX genes were induced by MeJA in M. truncatula and M. sativa, and the LOX genes identified were mainly distributed in the type I and type II 13-LOX subfamilies. MtLOX24 was up-regulated at 8 h after MeJA induction, and exogenous expression in Arabidopsis demonstrated that MtLOX24 promoted resistance to MeJA-induced stress. This study provides valuable new information regarding the evolutionary history and functions of LOX genes in the genus Medicago.
Similar content being viewed by others
Background
Lipoxygenase (LOX) is a dioxygenase-containing non-heme iron that catalyses unsaturated fatty acids with linoleic acid (LA), α-linolenic acid and arachidonic acid as substrates. It is present in both plants and animals, but its catalytic processes differ between kingdoms. In animals, LOX catalyzes the formation of neuroprotectin from arachidonic acid (20 carbon) and eicosapentaenoic acid [1]. Substrates in other animals include linolenic acid and docosahexaenoic acid (DHA). In plants, LOX catalyzes the dehydrogenation and oxygenation of unsaturated fatty acids such as linoleic acid, linolenic acid, and arachidonic acid to form hydroperoxides. Subsequent specialized metabolic reactions form oxygen-containing derivatives such as the plant hormone jasmonic acid (JA), reactive oxylipids with epoxides, conjugated carbonyls or aldehydes, and antibacterial and antifungal compounds, such as leaf aldehydes or diethylene ethers [2,3,4,5]. In addition, some polyunsaturated fatty acids can be catalyzed by α-dioxygenase to form α-hydroxy polyunsaturated fatty acids (PUFAs) or α-peroxy PUFAs [6, 7].
Since the 1930s, studies have shown that soybean flavor formation is related to enzymatic reactions of unsaturated fatty acids. The key enzyme in these oxidation reactions is LOX [8]. Later studies showed that LOX is widespread among fungi, bacteria, algae, and protozoa, and is more prevalent in plants than in animals [9]. To date, several LOX genes have been cloned in model plants such as Arabidopsis thaliana [10], rice, wheat, barley, maize, and tomato [11]. Furthermore, these genes show high activity levels in legumes.
Members of the LOX family can be classified based on their optimal pH, subcellular localization, and reaction location for specific substrates. Most researchers divide the LOX family into 9-LOXs and 13-LOXs based on the reaction sites of the catalytic substrates linoleic acid (LA, 18:2) and linolenic acid (LeA, 18:3) during oxidation; enzymatic PUFA oxidation can occur on carbon 9 (9-LOX) or carbon 13 (13-LOX) [12]. Furthermore, 13-LOX proteins can be further divided into two subgroups, type I and type II. Type I 13-LOX proteins have no transport peptides, show high sequence similarity (> 75%) with each other, and are usually localized to the cytoplasm. Type II 13-LOX proteins are localized to the chloroplast by an N-terminal transport peptide and show only moderate sequence similarity (> 35%) between subfamily members [13]. In animals, LOXs can be divided into the 5-LOX, 8-LOX, 9-LOX, 11-LOX, 12-LOX, and 15–1-LOX subfamilies based on the insertion position of molecular oxygen for oxygenation reactions [14]. Numerous studies have reported the presence of LOX family genes in plant species. To date, six LOXs have been reported in Arabidopsis [10], 23 in Chinese white pear (Pyrus bretschneideri) [15], 18 in melon (Cucumis melo) [16], 36 in soybean (Glycine max), 28 in Medicago truncatula, 10 in Cicer arietinum, five in Lotus japonicus [17], 14 in rice [18], 64 among four cotton species (Gossypium hirsutum, Gossypium barbadense, Gossypium arboretum, and Gossypium raimondii) [19], 41 among three banana species (Musa acuminata, Musa balbisiana, and Musa itinerans) [20], 14 in diploid woodland strawberry (Fragaria vesca) [21], and 20 in Artemisia annua [22]. Some researchers have shown that genes in the 9-LOX subfamily play important roles in seed, organ, and fruit development. They also function in defense responses against pathogens such as Phytophthora parasitica varocotianae (PPN) in tobacco [23], yellow branch mold in tomato [24] and Phytophthora infestans in potato [25]. Genes in the 13-LOX subfamily have key roles in plant development, senescence, and biotic stress responses (such as pathogen infection and insect herbivory) [17, 26, 27]. These genes also have essential roles in responses to hormones, including salicylic acid (SA), methyl jasmonate (MeJA), and abscisic acid (ABA) [21]. Most soybean LOX genes in the 13-LOX subfamilies are induced by parasitic nematodes and exhibit crosstalk with other signaling networks, such as the ethylene and SA pathways. They may also interact with members of other gene families, such as WRKYs [28]. In summary, genes in different LOX subfamilies within a species may have similar or opposing functions and show differential expression between tissues. Similarly, LOX genes in the same subfamily may also have distinct functions and spatiotemporal expression patterns.
LOX is known to contribute to biosynthesis of oxylipins and JA, both of which mediate the JA signaling pathway and play important roles in plant development and defense against biotic stressors [29]. Exogenous JA induces plant defense responses in a concentration-dependent manner. Low concentrations of MeJA can promote plant growth, induce JA synthesis, enhance expression of downstream defense-related genes, and alleviate stress-induced damage. However, high concentrations of MeJA and oxylipin inhibit seed or pollen germination [30, 31], which may be due to inhibitory effects of jasmonate on root meristem recombination, cell division, cell elongation, and cell precocity [31]. MeJA treatment can prolong A. thaliana flowering under short-day conditions [32]. MeJA-induced damage can also simulate insect defense responses [33]. This may be due to the influence of exogenous JA content, levels of volatile terpenes (e.g., α-terpene, α-pinene, β-phellandrene, β-caryophyllene, and myrcene) and specialized metabolites, such as flavonoids and phenolic compounds [34]. In Dunaliella salina, wheat, and tomato, exogenous MeJA treatment causes increases in β-carotene content and chlorophyll degradation, but reductions in the respiratory and photosynthetic rates [35]. MeJA treatment can increase peroxidase (POD) and superoxide dismutase (SOD) activity and enhance the plant capacity for scavenging active oxygen. Increased treatment time is associated with decreased SOD and POD activities, leading to active oxygen damage and enhanced membrane lipid peroxidation [36]. In simulated pest experiments with JA, SA has been shown to antagonize the JA signaling pathway [37, 38].
We here sought to characterize the LOX gene families in M. truncatula and M. sativa. Genome-wide analysis revealed 30 MtLOX and 95 MsLOX genes, which were comprehensively characterized via analyses of the gene structures, chromosomal distributions, phylogenetic relationships, promoter cis-elements, and expression patterns. We also determined the function of MtLOX24 during MeJA treatment based on exogenous expression in Arabidopsis. Our findings provide valuable insights regarding the complex network and functionality of LOX genes in legumes.
Results
Identification of LOX family members in M. truncatula and M. sativa and basic physicochemical properties analysis
We here identified 30 and 95 LOX genes in M. truncatula and M. sativa (cv. ‘Zhongmu No. 4’), respectively, and referred to them as MtLOX 1–30 and MsLOX 1–95, respectively (Additional file 1: Table S1; Additional file 2: Table S2). The lengths of the predicted products encoded by these genes ranged from 174 amino acids (aa) (MtLOX8) to 927 aa (MtLOX27) in M. truncatula, and from 248 aa (MsLOX65) to 2892 aa (MsLOX40) in M. sativa. The average length among all Medicago LOXs was 830 aa. The predicted MtLOX and MsLOX proteins varied in molecular weight from 20.56–104.81 kDa and from 28.29–324.36 kDa, respectively. The theoretical isoelectric points ranged from 5.3 (MtLOX15) to 8.87 (MtLOX14) in M. truncatula and from 5.12 (MsLOX77) to 7.99 (MsLOX1) in M. sativa. The average Grand Average of HYdropathicity (GRAVY) values of both the MtLOXs and the MsLOXs were negative, indicating that the proteins were primarily hydrophilic. Among the MtLOXs, eight were predicted to be localized to only the chloroplast, 17 to only the cytoplasm, and five to both the chloroplast and the cytoplasm. Among the MsLOX proteins, 30 were predicted to be localized to only the chloroplast, two to both the mitochondrion and the cytoplasm, 14 to both the chloroplast and the cytoplasm, and the remaining to the cytoplasm (Additional file 1: Table S1, Additional file 2: Table S2).
Chromosomal distribution of MtLOX and MsLOX genes
MtLOX genes were distributed on seven of the eight chromosomes in both M. truncatula and M. sativa (Additional file 3: Figure S1 A, and B). Chromosome (chr) 08 contained the largest number of MtLOX genes (17), followed by chr03 (five). There were only two MtLOX genes each on chr01, chr02, and chr04. Chr05 and chr07 contained only one MtLOX each, and there was no MtLOX gene on chr06. Chr08 also contained the largest number of LOX genes in M. sativa (42 genes, 44.21%), followed by chr03 (12 genes, 12.63%), chr04 (11 genes, 16.92%), chr01 (nine genes, 13.85%), and chr05 (seven genes, 10.77%). Many fewer genes were distributed on chr07 (four genes, 4.21%), chr02 (three genes, 3.16%), and chr06 (two genes, 2.11%). Several MsLOX genes were distributed on the four M. sativa homologous chromosomes, and four MsLOXs (namely MsLOX92, MsLOX93, MsLOX94, and MsLOX95) were identified in scaffolds.
Phylogenetic analysis of MtLOX and MsLOX genes
To understand the evolutionary relationships between LOX genes, a phylogenetic tree was constructed from LOX proteins encoded by M. truncatula, M. sativa, G. max, and the model plant Arabidopsis (Fig. 1). LOX family genes were categorized as 9-LOXs or 13-LOXs based on their classification in Arabidopsis, which depended on the reaction site of the catalytic substrates linoleic acid (LA, 18:2) and linolenic acid (LeA, 18:3) during oxidation [12]. Among them, all 9-LOXs amino acid sequences account for about a third of all amino acid sequences (Fig. 1, red colour), and 9-LOX differentiation occurs between the differentiation of type I 13-LOXs (Fig. 1, green colour) and type II 13-LOXs (Fig. 1, blue colour). Furthermore, 13-LOXs were divided into type I and type II based on their phylogenetic relationships in G. max. There were 11, 11, and eight 9-LOXs, type I 13-LOXs, and type II 13-LOXs, respectively, in M. truncatula and 30, 34, and 31 members of the same subfamilies, respectively, in M. sativa (Table 1, Additional file 4: Table S3, Additional file 5: Table S4).
Phylogenetic analysis of LOX proteins in Arabidopsis thaliana, Glycine max, Medicago truncatula, and Medicago sativa. The phylogenetic tree was generated from six AtLOXs, 40 GmLOXs, 30 MtLOXs, and 111 MsLOXs. The branch colors represent LOX subgroups. The color and shape at the end of each branch represents the species: Arabidopsis (blue square), soybean (purple circle), M. truncatula (yellow five-pointed star), or M. sativa (green triangle)
Structural and motif analyses of MtLOX and MsLOX genes
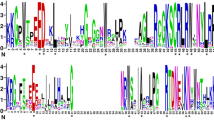
Evolutionary relationships between LOX family genes were next analyzed in M. truncatula and M. sativa based on predicted protein motifs (Figs. 2A, 3A). A total of 10 conserved motifs were identified in the LOXs. The distribution of conserved motifs was consistent with the relationships between genes depicted in the phylogenetic tree (Figs. 2B, 3B). Most MtLOXs contained 10 motifs, except four proteins: MtLOX1, MtLOX8, MtLOX27, and MtLOX29. MtLOX29 had lost motifs 9, 4, and 5, whereas MtLOX1 and MtLOX27 had gained additional copies of motifs 5 and 6, respectively. Three motifs were identified in MtLOX8. In M. sativa, all 9-LOXs contained 10 or fewer gene motifs. For example, MsLOX77 lacked motifs 3, 6, and 10, and MsLOX42 lacked motif 3. MsLOX45 lacked motif 3 and motif 1, and MsLOX68 had only two motifs. The type I 13-LOX proteins MsLOX62 and MsLOX54 had four motifs each, and MsLOX56 had seven motifs. The type II 13-LOXs MsLOX53, MsLOX58, MsLOX73, MsLOX86, and MsLOX4 had another nine motifs, MsLOX40 had another seven motifs, and MsLOX33 had three motifs. The conserved residues in MtLOXs and MsLOXs are shown in Additional file 6: Figure S2.
Gene structural analysis of Medicago truncatula LOXs. A Neighbor-joining phylogenetic tree showing relationships between MtLOX genes. B Motif analysis with the MEME suite yielded 10 motifs, each represented by a different color. C Conserved protein domain analysis with NCBI CDD predicted 12 conserved domains, each represented by a different color. D Exon–intron structural analysis. Yellow indicates exons, gray indicates introns, and green indicates untranslated regions (UTRs)
Gene structural analysis of Medicago sativa LOXs. A Neighbor-joining phylogenetic tree showing relationships between MsLOX genes. B Motif analysis with the MEME suite yielded 10 motifs, each represented by a different color. C Conserved protein domain analysis with NCBI CDD predicted 12 conserved domains, each represented by a different color. D Exon–intron structural analysis. Yellow indicates exons, gray indicates introns, and green indicates untranslated regions (UTRs)
Conserved structural domains were next investigated in the putative LOX proteins in M. truncatula and M. sativa (Figs. 2C, 3C; Additional file 7: Table S5; Additional file 8: Table S6). Most of the LOX family genes encoded PLAT (Pfam01477) and LOX (Pfam00305) domains. Members of the 9-LOX subfamily and the type I 13-LOX subfamily had similar conserved domains; most contained 13 domains, including PLN02337, LOX, PLN02264, PLN02305, PLAT_LH2, and PLAT. However, MtLOX29 and MtLOX8 lacked PLAT-related domains. Most type II 13-LOX subfamily proteins lacked one PLAT domain, except MtLOX14 and MsLOX38, which lacked two PLAT domains. MsLOX67, MsLOX76, MsLOX91, and MsLOX93 contained all 13 domains. MsLOX15, MsLOX17, MsLOX23, and MsLOX43 also had PRK06075 and complex1_49kDa superfamily (CL21493) domains. MsLOX11 had several additional domains: PRK06075, Complex1_49kDa superfamily, NuoD (COG0649), and NuoD (CL43223) SUPE domains.
Exon–intron structural analysis showed similar structures for most of the MtLOX genes, with eight or nine exons and seven or eight introns (Fig. 2D). However, MtLOX8, MtLOX11, and MtLOX29 contained only one or two exons each. The majority of MsLOX genes had between eight and 23 exons and seven or eight introns, although MsLOX68 contained only three exons (Fig. 3D). Most 9-LOX MsLOX genes contained eight or nine exons, similar to MtLOX genes. These results further indicated that structural evolution among members of the 13-LOX subfamily occurred later than that of 9-LOX subfamily members, and that the structures of 9-LOX genes were more highly conserved.
Cis-acting regulatory element analyses of MtLOX and MsLOX genes
To explore regulatory mechanisms associated with LOX gene expression, the 2-kb region upstream of the start codon for each MtLOX and MsLOX gene was analyzed to identify putative cis-acting regulatory elements. Most of these regulatory elements were related to plant development, phytohormone responses, and biotic and abiotic stress responses (Fig. 4). A large number of elements associated with hormone responses were present, specifically ABA response elements (ABREs) and MeJA response elements (CGTCA-motifs). In M. sativa, genes in the type II 13-LOX subfamily contained more ABREs than those in the other two subfamilies (Fig. 4A), but this was not the case in M. truncatula (Fig. 4B). MtLOX and MsLOX also contained stress-responsive cis-elements, including antioxidant response elements (AREs), stress response elements (STREs), and GT1 elements (GAAAAA). The promoters of these genes also contained several elements related to plant growth and development, including myeloblastosis (MBS) cis-elements, long terminal repeat (LTR) motifs, and wound-responsive elements (WUN-motifs); each gene contained three or fewer of these elements.
Promoter analysis of LOX genes in Medicago truncatula and Medicago sativa. A The number of cis-acting elements in the 2-kb promoter region upstream of the translation start site of each MsLOX gene. B The number of cis-acting elements in the 2-kb promoter region upstream of the translation start site of each MtLOX gene. C Distribution of specific types of cis-acting elements in MtLOX and MsLOX genes
MtLOX protein–protein interaction network analysis
A protein–protein interaction network was constructed for MtLOX proteins using the STRING database (Additional file 9: Figure S3 A). The network consisted of 24 edges (interactions) between 14 nodes (MtLOX proteins). Twelve MtLOXs were predicted to interact with MtLOX27 (AET01784) and with MtLOX14 (AET01336). STRING was further used to analyze a related network cluster (CL: 26,390), incorporating non-LOXs in the analysis; this produced a network consisting of 27 proteins (21 MtLOXs) with 133 interactions (Additional file 9: Figure S3 B). All MtLOXs were predicted to interact with AET05548, which was annotated as a pathogen-independent alpha dioxygenase. A 9- or 13-hydroperoxide lyase, hpl1 (AES89057), interacted with HPL3 (AES67192), MtLOX14 (AET01336), MtLOX27 (AET01784), and AES85033. HPL3, a 13-hydroperoxide lyase, interacted with MtLOX2 (AES62661), MtLOX5 (AES71573), MtLOX13 (AES82497), MtLOX14 (AET01336), and AES85033.
MtLOX expression profile analysis
Expression profiles for 26 MtLOX genes were downloaded from the M. truncatula Gene Expression Atlas (Additional file 10: Figure S4; Additional file 11: Table S7). Six MtLOXs (MtLOX10, MtLOX24, MtLOX25, MtLOX26, MtLOX27, and MtLOX30) were expressed three to six times higher in the leaves, petioles, stems, vegetative buds (Vegbuds), flowers, and pods than in the roots or seeds. MtLOX5, MtLOX9, and MtLOX16 were expressed more highly in the reproductive organs than in the vegetative organs, and MtLOX5 was most highly expressed in the flowers. In contrast, MtLOX19 was expressed at lower levels in the flowers and pods than in the petioles and stems. Seven MtLOX genes (MtLOX19, MtLOX20, MtLOX28, MtLOX22, MtLOX21, MtLOX12, and MtLOX15) had high expression levels at multiple nodule stages (Nod_4dpi and Nod_14dpi), RT_LCM_adjacent, RT_LCM_arbuscular, and RT_LCM_cortical. Only MtLOX3 and MtLOX4 were expressed specifically in the seeds, in which they were expressed five times higher than the other MtLOX genes.
MtLOX and MsLOX spatial expression patterns and exogenous MeJA responses
Quantitative reverse transcription (qRT)-PCR was used to confirm expression levels of MtLOX and MsLOX genes in the leaves, flowers, and stems of M. truncatula and M. sativa (Fig. 5). Several genes showed organ-specific high expression levels. For example, six MtLOX genes (MtLOX7, MtLOX11, MtLOX15, MtLOX16, MtLOX20, and MtLOX24) and four MsLOX genes (MsLOX23, MsLOX87, MsLOX90, and MsLOX94) were highly expressed in the flowers, suggesting roles in reproductive growth. Two MtLOX genes (MtLOX14 and MtLOX16) and two MsLOX genes (MsLOX87 and MsLOX94) were more highly expressed in the flowers and stems than in the leaves. Two MtLOX genes (MtLOX6 and MtLOX14) were highly expressed in the stem. MtLOX9, MtLOX21, MtLOX22, MsLOX30, and MsLOX85 were more highly expressed in the leaves than in the other two tissues. Other MtLOX genes showed similar expression patterns.
Expression patterns of LOX genes in different tissues of Medicago truncatula and Medicago sativa. Relative expression levels of MtLOX (A) and MsLOX (B) genes in the leaves, flowers, and stems as determined with qRT-PCR. Expression levels of MtLOX and MsLOX genes were normalized to levels of MtActin and MsActin, respectively. Hierarchical gene clustering was performed with the normalization method “standard score” on log2 transformed date. Colors indicates changes in expression compared to the leaf tissue, with red indicating an increase and blue indicating a decrease in expression
JA is a product of the fatty acid synthesis pathway that is involved in plant growth and development (Albechtová, 1994). To investigate the potential roles of LOX genes in response to stress induced by exogenous MeJA treatment (200 µM), the expression levels of 18 MtLOX genes and 13 MsLOX genes were determined in MeJA-treated plants via qRT-PCR (Fig. 6). Eight MtLOX genes (MtLOX6, MtLOX9, MtLOX11, MtLOX16, MtLOX20, MtLOX21, MtLOX22, and MtLOX24) and seven MsLOX genes (MsLOX18, MsLOX23, MsLOX30, MsLOX84, MsLOX85, MsLOX90, and MsLOX94) were up-regulated at 8 h of MeJA treatment; MtLOX16, MtLOX20, MtLOX21, MtLOX22, MsLOX18, MsLOX23, MsLOX84, and MsLOX85 were statistically significantly up-regulated. Three MtLOX genes (MtLOX2, MtLOX13, and MtLOX26) and three MsLOXs (MsLOX7, MsLOX14, and MsLOX87) were down-regulated after MeJA exposure.
Expression patterns of LOX genes in Medicago truncatula and Medicago sativa after exogenous methyl jasomate (MeJA) treatment. Relative expression levels of MtLOX (A) and MsLOX (B) genes at 0, 8, 12, and 48 h after treatment with 200 μM MeJA as determined with qRT-PCR. Expression levels of MtLOX and MsLOX genes were normalized to levels of MtActin and MsActin, respectively. Hierarchical gene clustering was performed with the normalization method “standard score” on log2 transformed data. Colors indicate changes in expression compared to the 0 h timepoint, with red indicating an increase and blue indicating a decrease in expression
Constitutive MtLOX24 expression in Arabidopsis increased tolerance to MeJA stress
MtLOX gene expression analysis in plants treated with exogenous MeJA (described above) showed that 44.4% of MtLOX genes were MeJA-inducible. Because LOXs are involved in JA synthesis, it is important to understand their responses to MeJA treatment. LOXs are also associated with abiotic stress, including insect pest stress (as simulated by MeJA treatment). In this study, we constructed 8 MtLOX24-overexpression (OE) lines in Arabidopsis and selected two lines (OE4, OE5) for further study. Gene expression level of the overexpression lines was confirmed using qRT-PCR (Additional file 13 Figure S5). In ½ × Murashige and Skoog (MS) medium, the Arabidopsis OE lines showed no differences in root length, fresh weight, or lateral root number compared to wild-type (WT) seedlings under normal growth conditions (Fig. 7). However, when 10 μM or 100 μM MeJA was added to the growth medium, the OE lines showed significantly higher fresh weights and lateral root growth than the WT seedlings (Fig. 7A-D). At 50 μM MeJA, the two OE lines had significantly more lateral roots than the WT, but differences in the root lengths and fresh weights were not significant (Fig. 7C-D). The root length-inhibiting effect of MeJA was not stronger in the OE lines than in the WT (Fig. 7B). In a separate experiment, MeJA was applied to three-week-old plants grown in soil; WT plants showed a higher degree of damage than the OE lines, with more yellow leaves (Fig. 8A), decreased chlorophyll content, increased H2O2 and increased relative conductivity levels (Fig. 8B-D).
Overexpression of MtLOX24 resulted in resistance to exogenous methyl jasmonate (MeJA) treatment of Arabidopsis seedlings. A Root growth among wild-type (WT) Arabidopsis seedlings and those overexpressing MtLOX24 (OE4 and OE5). Seven-d-old seedlings grown on ½ × Murashige and Skoog (MS) medium were transferred to ½ × MS medium supplemented with 0, 50, or 100 μM MeJA. Plants were assessed after two weeks. B Quantification of root length. C Fresh weight. D The number of lateral roots of Arabidopsis seedlings. Error bar represents the standard error calculated by three independent experiments
Overexpression of the Medicago truncatula gene LOX24 in Arabidopsis thaliana resulted in resistance to exogenous methyl jasmonate (MeJA) treatment among seedlings grown in soil. A Phenotypes of wild-type (WT) and MtLOX24-overexpression (OE4 and OE5) Arabidopsis plants grown in soil with or without MeJA treatment for three weeks. B Quantification of chlorophyll content. C H2O2 content. D Relative conductivity in Arabidopsis seedlings. Error bars represent the standard error calculated from three independent experiments
Discussion
The size of the LOX gene family varies greatly between species; there are 95 family members in M. sativa, 40 in G. max, 30 in M. truncatula, and only six in A. thaliana. These variations are largely consistent with differences in plant genome size: 2.74 Gb for alfalfa cv. ‘ZM-4’, 994 Mb for G. max, 450–500 Mb for M. truncatula, and -135 Mb for Arabidopsis [39,40,41]. Phylogenetic analysis has previously shown that the LOX family can be divided into two subfamilies, 9-LOX and 13-LOX. In M. truncatula and M. sativa, the 13-LOX subfamily can be further divided into two subfamilies: type I and type II 13-LOX. Evolutionary analysis here showed that GmLOX and MtLOX genes clustered together within each subfamily. This may be because GmLOX and MtLOX have a common ancestor [42]. Most MtLOX genes clustered together with three to five MsLOX genes each, indicating that numerous MsLOX genes may have arisen from gene duplication events after speciation. The type I 13-LOX cluster included the largest number of MsLOX genes. In this subfamily, only Glyma.15G026300 clustered together with the MtLOXs and MsLOXs, and the other seven GmLOX genes clustered together into a separate clade. This suggested that type I 13-LOX genes were significantly different in Medicago species compared to those in soybean. However, there were more 9-LOX and type II 13-LOX genes in G. max than in M. truncatula. This may be because the GmLOX family expanded as a result of ancient polyploidization events prior to speciation, leading to the emergence of soybean-specific gene duplicates [42].
As part of MtLOX and MsLOX characterization, we predicted subcellular localization. The 9-LOX and type I 13-LOX proteins in M. truncatula and M. sativa had no transport peptides, and most were therefore predicted to be localized to the cytoplasm. The exceptions were MtLOX21, MsLOX52, MsLOX53, MsLOX65, and MsLOX85, which were predicted to be localized only to the chloroplasts. These results were consistent with previous findings in rice [43] and banana [20]. Other proteins were predicted to be localized to both the cytoplasm and the chloroplasts; these included four MtLOXs (MtLOX2, MtLOX15, MtLOX17, and MtLOX18) and 13 MsLOXs (MsLOX5, MsLOX8, MsLOX12, MsLOX14, MsLOX51, MsLOX54, MsLOX56, MsLOX70, MsLOX73, MsLOX77, MsLOX82, MsLOX86, and MsLOX92). In addition to MsLOX4, MsLOX27, MsLOX31, and MsLOX40, all type II 13-LOX proteins in M. truncatula and M. sativa contained chloroplast transport peptides, and MtLOX6 and MsLOX32 were predicted to be localized to both the cytoplasm and the chloroplasts. This may have been due to sequences in these genes encoding chloroplast transport peptides, and the sequences have originated from different exons [44]. Type II 13-LOX genes localized to the chloroplasts may be involved in JA biosynthesis and resistance to insect herbivory, as previously reported for the tomato (Solanum lycopersicum) protein TomLoxD [45]. MsLOX27 and MsLOX31 were predicted to be localized to the mitochondria. LOXs in plant mitochondria may mediate the degradation of key membrane phospholipids and fatty acids [46].
Cis-acting regulatory elements are common among the promoters of stress-responsive genes. Identification and functional verification of such elements can aid in understanding transcriptional regulation of genes throughout the genome. Several phytohormone-responsive elements have been well characterized in plants. For example, the cis-element ABRE (PyACGTGGC) [47] was found in the promoter of the Arabidopsis dehydration-responsive gene RD29B, which is involved in transcriptional activation in response to ABA [48]. CGTCA-motifs are associated with MeJA responses, and TCA-elements are found in genes that participate in SA-related responses. Other cis-elements are associated with abiotic and biotic stress signaling. For example, the ARE element is found in antioxidant genes that can enhance environmental carcinogen detoxification [49]. In Neurospora crassa, the STRE motif is involved in regulation of a variety of cellular processes, including stress responses, carbohydrate metabolism restriction, and ethanol tolerance (through binding to Msn2p and Msn4p) [50, 51]. We here found numerous instances of the motifs mentioned above in MtLOX and MsLOX genes. Some genes, such as MtLOX5, MtLOX6, MtLOX8, MtLOX20, MtLOX23, MsLOX4, MsLOX15, MsLOX17, MsLOX43, and MsLOX94, contained multiple ABRE and CGTCA-motif elements. This suggested that these genes may play crucial roles in plant hormone regulation. The GT1-motif has often been studied as a light-responsive element; under light conditions, this motif is bound by the activation domain factor P, activating transcription [52]. The GT1-motif and the WUN-motif, the latter of which is associated with circadian rhythm, were found in the promoters of MtLOX1, MtLOX18, MtLOX24, MtLOX26, MsLOX55, MsLOX82, and MsLOX94, indicating that these genes may be light responsive. Few MBS and LTR motifs were found in MtLOXs and MsLOXs, likely because these elements are most often found in animal cell viruses, such as avian myeloblastosis virus [53] and avian leukosis virus [54].
LOX family genes show extensive spatiotemporal expression variation; these genes are involved in seed germination [55], seedling growth, and plant development and senescence [56]. Here, qRT-PCR showed that some MtLOX genes were highly expressed in the flowers and stems, suggesting that several MtLOX family genes played common roles in reproductive growth in M. truncatula. Six of the genes with high expression in the flowers, excluding MtLOX7 and MtLOX11, belonged to the type I 13-LOX subfamily, suggesting that type I 13-LOX genes play important roles in M. truncatula flower development. Other MtLOX genes, such as MtLOX11, also likely participate in flower and organ development. In contrast, in M. sativa, MsLOX genes from different subfamilies (e.g., MsLOX23, MsLOX87, MsLOX90, and MsLOX94) were highly expressed in the flowers. In Arabidopsis, the 13-LOX genes AtLOX3 and AtLOX4 have roles in plant vegetative growth and flower development [57]. In tea (Camellia sinensis), LOX1 is primarily expressed in the flowers, and is up-regulated during petal senescence and down-regulated during the bud opening period [56]. Analysis of previously published MtLOX expression data showed that they were highly expressed in the leaves, petioles, stems, VegBuds, flowers, and pods, but that some 9-LOXs and type I 13-LOXs (such as MtLOX12, MtLOX15, MtLOX19, MtLOX20, MtLOX21, MtLOX22, and MtLOX28) participated in root nodule synthesis and were induced by arbuscular mycorrhizal fungi. Similar results have also been reported in tomatoes. LOX activity is induced by inoculation with pathogens such as Funneliformis mosseae, Alternaria solani, and Cladosporium fulvum in tomato [58, 59], and C. fulvum tolerance is increased by TomloxD overexpression [24]. In Vicia faba [60], Phaseolus vulgaris [61], and G. max [62], the LOX genes VfLOX1, PvLOX5, and GmLOX1, respectively, show consistent expression levels during nodule development, suggesting that LOX genes are involved in the symbiotic nitrogen fixation process between rhizobia and legumes.
Lipoxygenase functions in both herbivore feeding and JA treatment via the octadecanoic acid pathway [45]. In Arabidopsis, potato, and tobacco (Nicotiana attenuata), silencing the 13-LOX gene LOX2 [63], a 13-LOX isoform (LOX-H3) [64], or LOX3 [65], respectively, decreases the tolerance of the plant to insect-induced stress. Analysis of LOX expression in response to MeJA exposure in both M. truncatula and M. sativa showed differences between LOX subfamilies in this study. Specifically, 9-LOX subfamily genes showed little change in expression levels after MeJA treatment. However, type I and II 13-LOX genes were strongly induced by MeJA in both species, suggesting that they may promote insect resistance by participating in the JA signaling pathway. Specifically, MtLOX5, MtLOX16, MtLOX20, MtLOX21, and MtLOX22 were up-regulated by 6–53 times at 8 h of MeJA treatment; MtLOX6, MtLOX9, MtLOX10, and MtLOX24 were induced by two to four times at the same timepoint. Similarly, MsLOX18, MsLOX23, MsLOX84, and MsLOX85 were induced by 4–47 times, whereas MsLOX30, MsLOX57, MsLOX90, and MsLOX94 were up-regulated by 1.5–3 times at 8 h after MeJA treatment. In our study, it was found that overexpression of the MtLOX24 in Arabidopsis could alleviate the damage caused by MeJA treatment, which may be due to the fact that MtLOX24, as a member of the 13-LOX gene subfamily, can catalyze α-LeA to produce 13-HPOT and increase the content of endogenous JA, which needs to be verified in subsequent studies.
Conclusion
In this study, a total of 30 MtLOX and 95 MsLOX genes were identified. LOX genes can be further divided into three subfamilies, 9-LOX, type I 13-LOX, and type II 13-LOX. Promoter element analysis predicted that LOXs were involved in abiotic stress response, hormone response and plant development. The expression profiles of MtLOX and MsLOX showed that the expression of 13-LOXs was different in different organs, and the expression of 13-LOXs was induced by MeJA. In addition, Arabidopsis overexpressing MtLOX24 showed resistance to exogenous MeJA and alleviated the oxidative damage of Arabidopsis leaves induced by MeJA, which may be involved in JA signaling pathways. In conclusion, this study provides important information on the potential function of alfalfa LOXs.
Methods
Plant materials, growth conditions, and stress treatments
M. truncatula R108 and M. sativa Zhongmu No. 4 were used in this study. Tetraploid alfalfa cultivar Zhongmu No. 4 was bred by the author Qingchuan Yang. Diploid M. truncatula ecotype R108 was provided by the Samuel Roberts Noble Foundation (U.S.A.) and preserved by our laboratory. The procedures of plant material collection complied with relevant institutional, national, and international guidelines and legislation. Sterilized seeds were germinated in petri dishes covered with double layer of filter paper. Five days after germination, seedlings were transferred to ½ × Hoagland solution for subsequent growth. For stress treatments, after growth for 3 weeks, MeJA was added to a final concentration of 200 µM. Samples were taken at 0, 8, 12, and 48 h after treatment. The stems, leaves, and flowers were collected for differential gene expression analysis. Plants were grown at 25/22 ℃ under a 16/8 h light/dark cycle with 60% relative humidity.
Genome-wide identification of LOX genes
LOX protein sequences for Arabidopsis, M. truncatula, and G. max were obtained from Phytozome (https://phytozome.jgi.doe.gov/pz/). The gene files for the ZM-4 alfalfa genome were downloaded from the following site: https://figshare.com/s/fb4ba8e0b871007a9e6c. The LOX domain (PF00305) was downloaded from Pfam (http://pfam.xfam.org/), and the Hidden Markov Model (HMM) Profile model was used to identify potential LOX family genes. A total of 95 putative LOX proteins were identified in the alfalfa ZM-4 genome that contained both PLAT (PF01477) and LOX (PF00305) domains, and these were selected as predicted MsLOX genes. ProtParam (https://web.expasy.org/protparam/) was used to analyze predicted MtLOX and MsLOX physical and chemical properties, including molecular weight, theoretical isoelectric point, and GRAVY values. Subcellular localization of each protein was predicted with Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/, accessed on 25 February 2021).
Analyses of phylogenetic relationships, gene structures, and conserved sequences
We used 171 full-length LOX gene sequences in the phylogenetic analysis, comprising six AtLOXs, 40 GmLOXs, 30 MtLOXs, and 95 MsLOXs. All sequences were aligned using the EMBL-EBI multiple sequence alignment tool (https://www.ebi.ac.uk/Tools/msa/). A phylogenetic tree was then constructed and visualized with the Interactive Tree Of Life (http://itol.embl.de/) using the default settings.
The LOX sequences from M. truncatula and M. sativa were aligned in MEGA5 [66] and a phylogenetic tree was constructed using the neighbor-joining (NJ) method with default parameters and 1000 bootstrap replicates. The conserved motifs of MtLOX and MsLOX proteins were identified with the MEME suite (http://meme-suite.org/tools/meme). Protein domain content was predicted with NCBI CDD (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). The intron–exon structures of MtLOXs and MsLOXs were determined using the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn). The final gene structures and conserved sequences were visualized with TBtools software [67].
Chromosome location analysis and protein–protein interaction network prediction
The general feature format (gff) files for MtLOX genes were downloaded from Ensembl plants (http://plants.ensembl.org/index.html) and the gff files for the alfalfa genome were downloaded from the following site: https://figshare.com/s/fb4ba8e0b871007a9e6c. M. truncatula and M. sativa genome length and gene mapping information were extracted with Tbtools [67]. A physical chromosomal location map was drawn with MapGene2Chrom web v2 (http://mg2c.iask.in/mg2c_v2.0/). The transcript identification numbers of MtLOX genes (Additional file 1: Table S1). The STRING database (https://cn.string-db.org/) was used to predict interactions among 14 MtLOXs using a protein–protein interaction (PPI) enrichment threshold of p < 1.0e-16.
Cis-acting regulatory element analysis
The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict cis-elements in the 2-kb promoter region upstream of the transcription start site of each MtLOX and MsLOX gene. The results were visualized with TBtools software [67].
Expression patterns of LOX genes in M. truncatula based on transcriptomic data
Expression data were obtained from the M. truncatula Gene Expression Atlas (https://medicago.toulouse.inrae.fr/MtExpress) for leaves, petioles, stems, VegBuds, flowers, pods, roots, seeds, nodules at multiple stages (Nod_4dpi and Nod_14dpi), RT_LCM_adjacent, RT_LCM_arbuscular, and RT_LCM_cortical. The sampling methods are described in detail by Benedito [68]. Dry mature seeds (DS) were collected from developing M. truncatula; the seeds were removed from the pods at 48-d intervals [69]. Transcripts of LCM-derived arbuscule-containing cells from mycorrhizal roots (RT_LCM_arbuscular), cortical cells of non-mycorrhizal roots (RT_LCM_cortical), and cortex cells adjacent to extracellular fungal hyphae (RT_LCM_adjacent) were obtained from Medicago (A17) genome arrays [70]. The expression data were log2 transformed [71] and visualized as heatmaps with TBtools.
Expression patterns of MtLOX and MsLOX genes based on qRT‑PCR
M. truncatula R108 and alfalfa ‘Zhongmu No. 4’ were treated with MeJA as described above. To analyze LOX expression patterns in response to this stress treatment, RNA was extracted from the stems, leaves, and flowers using the Eastep Total RNA Extraction Kit (Promega, Beijing, China). cDNA was generated using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara, Shiga, Japan). qRT-PCR was conducted on the CFX384 Touch Real-time PCR Detection System (Bio-Rad, Hercules, CA, USA). The MtActin and MsActin genes were used as the internal references for MtLOXs and MsLOXs, respectively, and expression levels were normalized using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The expression data were then log2 transformed [71] and visualized as heatmaps with TBtools. All primers used in this study were designed using NCBI Primer-Blast (nih.gov) and are shown in Additional file 12: Table S8.
Exogenous MtLOX24 expression in Arabidopsis
MtLOX24 (Medtr8g018690) was amplified from M. truncatula cDNA with the following oligonucleotide primers: 5′-GAAGTGAGCAAGCCACAAGC-3′ and 5′-ACATAGGAGGATGAGGGAAT-3′. The MtLOX24 coding sequence was inserted into the pCAMBIA3301 vector under the control of the CaMV 35S promoter. The resulting 35S::MtLOX24-GUS expression plasmid was transformed into Arabidopsis ecotype Columbia 0 using the floral dip method [72]. T3 homozygous plants were used in further experiments. Seeds were held at 4 ℃ in the dark for 2 d, then cultured on ½ × Murashige and Skoog (MS) medium in an incubator under a 16/8 h light/dark photoperiod at 22/18 ℃. After 7 d, seedlings were transferred to ½ × MS medium supplemented with MeJA (0, 50, or 100 µM) for 15 d. The fresh weight, root length, and lateral root number were then measured. Three-week-old transgenic Arabidopsis plants were also grown in soil treated with 0 or 200 µM MeJA solution every 3 d for a period of three weeks. For physiological index measurements, the chlorophyll content was measured according to Ergun [73], relative conductivity was mainly used to study the change trend of membrane permeability and were detected by method as described by Zhou B [74]. The concentration of H2O2 was determined according to the protocol of the H2O2 detection kit provided by the manufacturer (Nanjing JianchengInstitute of Bioengineering).
Statistical analysis
SPSS software was used for statistical analysis. The mean and standard deviation were calculated from three independent experiments with comparable results. All data collected were analysed using one-way ANOVA and Duncan's multiple mean comparison test was used to compare the control and treatment groups.
Availability of data and materials
The transcriptomic data were obtained from the M. truncatula Gene Expression Atlas (https://medicago.toulouse.inrae.fr/MtExpress).
References
Vang K, Ziboh VA. 15-lipoxygenase metabolites of gamma-linolenic acid/eicosapentaenoic acid suppress growth and arachidonic acid metabolism in human prostatic adenocarcinoma cells: possible implications of dietary fatty acids. Prostag Leukotr Ess. 2005;72(5):363–72.
Mack AJ, Peterman TK, Siedow JN. Lipoxygenase isozymes in higher plants: biochemical properties and physiological role. Isozymes. 1987;13:127–54.
Vick BA. Oxidative systems for modification of fatty acids: the lipoxygenase pathway. In: Biochemistry of Plants. Edited by Stumpf PK: Academic Press; 1987;9:53–90.
Hildebrand DF, Hamilton-Kemp TR, Legg CS, Bookjans G. Plant lipoxygenases: Occurrence, properties and possible functions. Curr Top Plant Biochem Physiol. 1988;7:201–19.
Siedow JN. Plant lipoxygenase:structure and funnction. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:145–88.
Hamberg M, Sanz A, Castresana C. alpha-oxidation of fatty acids in higher plants. Identification of a pathogen-inducible oxygenase (piox) as an alpha-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J Biol Chem. 1999;274(35):24503–13.
Saffert A, Hartmann-Schreier J, Schön A, Schreier P. A dual function alpha-dioxygenase-peroxidase and NAD(+) oxidoreductase active enzyme from germinating pea rationalizing alpha-oxidation of fatty acids in plants. Plant Physiol. 2000;123(4):1545–52.
Andre E, Hou KW. The presence of a lipoid oxidase in soybean. Glycine soya Lieb CR Acad Sci. 1932;194:645–7.
Forster C, North H, Afzal N, Domoney C, Hornostaj A, Robinson DS, Casey R. Molecular analysis of a null mutant for pea (Pisum sativum L.) seed lipoxygenase-2. Plant Mol Biol. 1999;39(6):1209–20.
Bannenberg G, Marta Martínez, Hamberg M, Castresana C. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids. 2009;44(2):85–95.
Mariutto M, Duby F, Adam A, Bureau C, Fauconnier ML, Ongena M, Thonart P, Dommes J. The elicitation of a systemic resistance by Pseudomonas putida BTP1 in tomato involves the stimulation of two lipoxygenase isoforms. BMC Plant Biol. 2011;11:29.
Liavonchanka A, Feussner I. Lipoxygenases: occurrence, functions, and catalysis. J Plant Physiol. 2006;163:348–57.
Feussner I, Wasternack C. The lipoxygenase pathway. Annu Rev Plant Biol. 2002;53(1):275–97.
Schneider C, Pratt DA, Porter NA, Brash AR. Control of oxygenation in lipoxygenase and cyclooxygen lase catalysis. ChemBiol. 2007;14:473–88.
Meng L, Li L, Dunwell JM, Xin Q, Xing L, Zhang S. Characterization of the lipoxygenase (LOX) gene family in the Chinese white pear (Pyrus bretschneideri) and comparison with other members of the Rosaceae. BMC Genomics. 2014;15(1):1–12.
Zhang C, Jin Y, Liu J, Tang Y, Cao S, Qi H. The phylogeny and expression profiles of the lipoxygenase (LOX) family genes in the melon (Cucumis melo L) genome. Sci Hortic. 2014;170:94–102.
Song H, Wang P, Li CS, Han SY, Wang XJ. Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci Rep. 2016;6(1):35245.
Umate P. Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal Behav. 2011;6(3):335–8.
Shaban M, Ahmed MM, Sun H, Ullah A, Zhu L. Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genomics. 2018;19(1):599.
Liu F, Li H, Wu J, Wang B, Tian N, Liu J, Sun X, Wu H, Huang Y, Lü P, Cheng C. Genome-wide identification and expression pattern analysis of lipoxygenase gene family in banana. Sci Rep. 2021;11(1):9948.
Li Z, Xie Q, Yan J, Chen J, Chen Q. Genome-wide identification and characterization of the abiotic-stress-responsive GRF gene family in diploid woodland strawberry (Fragaria vesca). Plants (Basel). 2021;10(9):1916.
Meng Y, Liang Y, Liao B, He W, Liu Q, Shen X, Xu J, Chen S. Genome-wide identification, characterization and expression analysis of lipoxygenase gene family in Artemisia annua L. Plants (Basel). 2022;11(5):655.
Rancé I, Fournier J, Esquerré-Tugayé MT. The incompatible interaction between Phytophthora parasitica var. nicotianae race 0 and tobacco is suppressed in transgenic plants expressing antisense lipoxygenase sequences. Proc Natl Acad Sci U S A. 1998;95(11):6554–9.
Hu TZ, Zeng H, Hu ZL, Qv XX, Chen GP. Overexpression of the tomato 13-lipoxygenase gene TomloxD increases generation of endogenous jasmonic acid and resistance to Cladosporium fulvum and high Temperature. Plant Mol Biol Rep. 2013;31(5):1141–9.
Kolomiets MV, Chen H, Gladon RJ, Braun EJ, Hannapel DJ. A leaf lipoxygenase of potato induced specifically by pathogen infection. Plant Physiol. 2000;124(3):1121–30.
Chauvin A. Caldelari, Wolfender J, Farmer EE. Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 2013;197(2):566–75.
Springer A, Kang C, Rustgi S, von Wettstein D, Reinbothe C, Pollmann S, Reinbothe S. Programmed chloroplast destruction during leaf senescence involves 13-lipoxygenase (13-LOX). Proc Natl Acad Sci U S A. 2016;113(12):3383–8.
Klink VP, Hosseini P, Matsye PD, Alkharouf NW, Matthews BF. Syncytium gene expression in Glycine max([PI 88788]) roots undergoing a resistant reaction to the parasitic nematode Heterodera glycines. Plant Physiol Biochem. 2010;48(2–3):176–93.
Nahar K, Kyndt T, De Vleesschauwer D, Höfte M, Gheysen G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011;157(1):305–16.
Maciejewska B, Kopcewicz J. Inhibitory effect of methyl jasmonate on flowering and elongation growth in Pharbitis nil. J Plant Growth Regul. 2003;21(3):216–23.
Tung P, Hooker TS, Tampe PA, Reid DM, Thorpe TA. Jasmonic acid: effects on growth and development of isolatedtomato roots cultured in vitro. Int J Plant Sci. 1996;157(6):713–21.
Albechtová JTP, Ullmann J. Methyl jasmonate inhibits growth and flowering in Chenopodium rubrum. Biol Plant. 1994;36(2):317–9.
Thaler JS, Stout MJ, Karban R, Duffey SS. Exogenous jasmonates simulate insect wounding in tomato plants (Lycopersicon esculentum) in the laboratory and field. J Chem Ecol. 1996;22(10):1767–81.
Lundborg L, Sampedro L, Borg-Karlson AK, Zas R. Effects of methyl jasmonate on the concentration of volatile terpenes in tissues of Maritime pine and Monterey pine and its relation to pine weevil feeding. Trees. 2019;33(1):53–62.
Liu L, Wei J, Zhang M, Zhang L, Li C, Wang Q. Ethylene independent induction of lycopene biosynthesis intomato fruits by jasmonates. J Exp Bot. 2012;63:5751–61.
Novacky PA. Use of dimethyl sulfoxide to detect hydroxyl radical during bacteria-Induced hypersensitive reaction. Plant Physiol. 1991;96(4):1157–60.
Kessmann H, Staub T, Hofmann C, Maetzke T, Herzog J, Ward E, Uknes S, Ryals J. Induction of systemic acquired disease resistance in plants by chemicals. Annu Rev Phytopathol. 1994;32:439–59.
Zhang L, Zhang F, Melotto M, Yao J, He SY. Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot. 2017;68(6):1371–85.
Long R, Zhang F, Zhang Z, Li M, Chen L, Wang X, Liu W, Zhang T, Yu LX, He F, Jiang X, Yang X, Yang C, Wang Z, Kang J, Yang Q. Genome Assembly of Alfalfa Cultivar Zhongmu-4 and Identification of SNPs Associated with Agronomic Traits. Genom Proteomics Bioinform. 2022;20(1):14–28.
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–83.
Branca A, Paape TD, Zhou P, Briskine R, Tiffin P. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc Natl Acad Sci U S A. 2011;108(42):E864–70.
Jin HS, Van K, Dong HK, Kim KD, Jang YE, Choi BS, Kim MY, Lee SH. The lipoxygenase gene family: a genomic fossil of shared polyploidy between Glycine max and Medicago truncatula. BMC Plant Biol. 2008;8:133.
Mizuno K, Iida T, Takano A, Yokoyama M, Fujimura T. A new 9-lipoxygenase cDNA from developing rice seeds. Plant Cell Physiol. 2003;44(11):1168–75.
Bruce BD. Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 2000;10(10):440–7.
Yan L, Zhai Q, Wei J, Li S, Wang B, Huang T, Du M, Sun J, Kang L, Li CB, Li C. Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 2013;9(12): e1003964.
Dupont J. Lipoxygenase-mediated cleavage of fatty acids in plant mitochondria. Physiol Plantarum. 1981;52(2):225–32.
Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803.
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A. 2000;97(21):11632–7.
Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–60.
Freitas FZ, Virgilio S, Cupertino FB, Kowbel DJ, Fioramonte M, Gozzo FC, Glass NL, Bertolini MC. The SEB-1 transcription factor binds to the STRE motif in neurospora crassa and regulates a variety of cellular processes including the stress response and reserve carbohydrate metabolism. G3 (Bethesda). 2016;6(5):1327–43.
Watanabe M, Tamura K, Magbanua JP, Takano K, Kitamoto K, Kitagaki H, Akao T, Shimoi H. Elevated expression of genes under the control of stress response element (STRE) and Msn2p in an ethanol-tolerance sake yeast Kyokai no. 11. J Biosci Bioeng. 2007;104(3):163–70.
Lam E, Chua NH. GT-1 binding site confers light responsive expression in transgenic tobacco. Science. 1990;248(4954):471–4.
Perbal B. Pathogenic potential of myeloblastosis-associated viruses. Infect Agents Dis. 1995;4(4):212–27.
Ruddell A, Linial ML, Groudine M. Tissue-specific lability and expression of avian leukosis virus long terminal repeat enhancer-binding proteins. Mol Cell Biol. 1989;9(12):5660–8.
Bailly C, Bogatek-Leszczynska R, Côme D, Corbineau F. Changes in activities of antioxidant enzymes and lipoxygenase during growth of sunflower seedlings from seeds of different vigour. Seed Scie Res. 2002;12(1):47–55.
Liu S, Han B. Differential expression pattern of an acidic 9/13-lipoxygenase in flower opening and senescence and in leaf response to phloem feeders in the tea plant. BMC Plant Biol. 2010;10(1):228.
Caldelari D, Wang G, Farmer EE, Dong XN. Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol Bio. 2011;75(1–2):25–33.
Song Y, Chen D, Lu K, Sun Z, Zeng R. Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front Plant Sci. 2015;6:786.
Peever TL, Higgins VJ. Electrolyte leakage, lipoxygenase, and lipid peroxidation induced in tomato leaf tissue by specific and nonspecific elicitors from Cladosporium fulvum. Plant Physiol. 1989;90(3):867–75.
Perlick AM, Albus U, Stavridis T, Fruhling M, Kuster H, Puhler A. The Vicia faba lipoxygenase gene VfLOX1 is expressed in the root nodule parenchyma. Mol Plant Microbe Interact. 1996;9(9):860–3.
Porta H, Rocha-Sosa M. A Phaseolus vulgaris lipoxygenase gene expressed in nodules and in Rhizobium tropici inoculated roots. Biochim Biophys Acta. 2000;1517(1):139–42.
Mohammadi M, Karr AL. Induced lipoxygenases in soybean root nodules. Plant Sci. 2003;164(4):471–9.
Bell E, Creelman RA, Mullet JE. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci U S A. 1995;92(19):8675–9.
Royo J, León J, Vancanneyt G, Albar JP, Rosahl S, Ortego F, Castañera P, Sánchez-Serrano JJ. Antisense-mediated depletion of a potato lipoxygenase reduces wound induction of proteinase inhibitors and increases weight gain of insect pests. Proc Natl Acad Sci U S A. 1999;96(3):1146–51.
Halitschke R, Baldwin IT. Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 2003;36(6):794–807.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202.
Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, Moreau S, Niebel A, Frickey T, Weiller G, He J, Dai X, Zhao PX, Tang Y, Udvardi MK. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55(3):504–13.
Verdier J, Lalanne D, Pelletier S, Torres-Jerez I, Righetti K, Bandyopadhyay K, Leprince O, Chatelain E, Vu BL, Gouzy J, Gamas P, Udvardi MK, Buitink J. A regulatory network-based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 2013;163(2):757–74.
Gaude N, Bortfeld S, Duensing N, Lohse M, Krajinski F. Arbuscule-containing and non-colonized cortical cells of mycorrhizal roots undergo extensive and specific reprogramming during arbuscular mycorrhizal development. Plant J. 2012;69(3):510–28.
Feng C, Wang H, Lu N, Chen T, He H, Lu Y, Tu XM. Log-transformation and its implications for data analysis. Shanghai Arch Psychiatry. 2014;26(2):105–9.
Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–43.
Ergun E, Demirata B, Gumus G. Simultaneous determination of chlorophyll a and chlorophyll b by synchronous fluorimetry. Anal Bioanal Chem. 2004;379(5–6):803–11.
Zhou B, Guo Z. Calcium is involved in the abscisic acid-induced ascorbate peroxidase, superoxide dismutase and chilling resistance in Stylosanthes guianensis. Biol Plant. 2009;53(1):63–8.
Acknowledgements
Not applicable.
Funding
This work was supported by Key Projects in Science and Technology of Inner Mongolia (2021ZD0031), China Agriculture Research System of MOF and MARA (CARS-34) and Agricultural Science and Technology Innovation Program (ASTIP-IAS14). The funding body played no role in the design of the study, the collection, analysis and interpretation of the data or the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
Qingchuan Yang and Ruicai Long planned and designed the research. Lei Xu and Xiaoxi Zhu performed the laboratory experiments. Lei Xu wrote the manuscript. Fengyan Yi, Yajiao Liu, and Bilig Sod analysed the data. Mingna Li, Lin Chen, Junmei Kang and Ruicai Long revised the manuscript. Ruicai Long supervised the research. Lei Xu and Xiaoxi Zhu contributed equally. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The plant material in this study does not involve endangered or protected species, and the collection and experimentation of plant material in this study complies with institutional, national and international guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Table S1.
Properties of LOX gene family in Medicago truncatula.
Additional file 2. Table S2.
Properties of LOX gene family in Medicago sativa.
Additional file 3. Figure S1.
Chromosomal locations of LOX genes in Medicago truncatula and Medicago sativa.
Additional file 4. Table S3.
Members of the LOX gene family in Medicago truncatula.
Additional file 5. Table S4.
Members of the LOX gene family in Medicago sativa.
Additional file 6. Figure S2.
Conserved residues in LOX family proteins.
Additional file 7. Table S5.
Conserved structural domains in Medicago truncatula LOXs.
Additional file 8. Table S6.
Conserved structural domains in Medicago sativa LOXs.
Additional file 9. Figure S3.
Protein-protein interaction network analysis.
Additional file 10. Figure S4.
Expression profiles of Medicago truncatula LOX genes in multiple plant organs and under several treatment conditions.
Additional file 11. Table S7.
Medicago truncatula LOX gene expression profiles. Data were downloaded from the M. truncatula Gene Expression Atlas.
Additional file 12. Table S8.
Primers used in this study.
Additional file 13. Figure S5.
The relative expression level of MtLOX from overexpressed Arabidopsis lines using qRT-PCR.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xu, L., Zhu, X., Yi, F. et al. A genome-wide study of the lipoxygenase gene families in Medicago truncatula and Medicago sativa reveals that MtLOX24 participates in the methyl jasmonate response. BMC Genomics 25, 195 (2024). https://doi.org/10.1186/s12864-024-10071-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10071-1