Abstract
Background
Grain yield is a complex and polygenic trait influenced by the photosynthetic source-sink relationship in wheat. The top three leaves, especially the flag leaf, are considered the major sources of photo-assimilates accumulated in the grain. Determination of significant genomic regions and candidate genes affecting flag leaf size can be used in breeding for grain yield improvement.
Results
With the final purpose of understanding key genomic regions for flag leaf size, a meta-analysis of 521 initial quantitative trait loci (QTLs) from 31 independent QTL mapping studies over the past decades was performed, where 333 loci eventually were refined into 64 meta-QTLs (MQTLs). The average confidence interval (CI) of these MQTLs was 5.28 times less than that of the initial QTLs. Thirty-three MQTLs overlapped the marker trait associations (MTAs) previously reported in genome-wide association studies (GWAS) for flag leaf traits in wheat. A total of 2262 candidate genes for flag leaf size, which were involved in the peroxisome, basal transcription factor, and tyrosine metabolism pathways were identified in MQTL regions by the in silico transcriptome assessment. Of these, the expression analysis of the available genes revealed that 134 genes with > 2 transcripts per million (TPM) were highly and specifically expressed in the leaf. These candidate genes could be critical to affect flag leaf size in wheat.
Conclusions
The findings will make further insight into the genetic determinants of flag leaf size and provide some reliable MQTLs and putative candidate genes for the genetic improvement of flag leaf size in wheat.
Similar content being viewed by others
Background
Wheat (Triticum aestivum L.) is one of the most important cereal crops worldwide and provides about one-fifth of the calories consumed in the global diet [1]. There is a major concern that the risk of a global food crisis is increasing, due to the ever-increasing population, extreme climate changes, and reduction in arable land [2]. It is also estimated that an additional 1 billion tons of grain per year will need to be grown by 2050 to meet food demands [3]. To address this issue, wheat breeders are emphasizing trait-based breeding using genotype complementation with elite agronomic traits to accelerate grain yield improvement [4, 5]. Further improvement in wheat genetic gain can likely be achieved by the breeding for key yield-related agronomic and physiological traits [6].
Grain yield is a complex and polygenic trait that is influenced by the photosynthetic source-sink relationship that determines changes in carbohydrate synthesis, accumulation, and distribution, especially in mature leaves [7,8,9,10,11]. Recent studies have shown that delaying leaf senescence in plants can contribute to maintain source-sink relationships, resulting in higher grain yields [12,13,14]. In wheat, the top three leaves, especially the flag leaf, are considered the major sources of photo-assimilates that accumulate in the grain [11, 15]. The flag leaf contributes about 45 to 58% of total photosynthetic activity [16] and over 40% of assimilates during grain filling [17]. The orientation and size of flag leaves are important in plant breeding, because they affect plant canopy morphology and photosynthetic efficiency [18]. The size of the flag leaf, consisting of leaf length, width, and area, is an extremely important factor that determines leaf structure and yield potential [17, 19]. Therefore, it is of high priority to understand the genetic mechanisms underlying flag leaf traits in wheat.
Flag leaf size is a typical quantitative trait, controlled by polygenes and highly influenced by environmental factors [20,21,22]. Several efforts have been made to explain the genetic mechanisms underlying flag leaf size by two strategies of quantitative trait loci (QTL) mapping [23,24,25] and genome-wide association studies (GWAS) [10, 26,27,28] in wheat. However, the significance of these QTL mapping results is strongly influenced by the experimental conditions, the type and size of mapping populations, density of genetic markers, and statistical methods used [29, 30]. In this context, these identified QTLs are often not robust enough to be used directedly in wheat breeding for marker-assisted selection (MAS) [31]. Therefore, the discovery of major and robust QTLs and closely associated markers with high potential for molecular breeding remains a challenge [32].
As another method for integrating QTL information, the meta-QTL (MQTL) analysis provides an effective strategy for identifying major genomic regions governing traits, regardless of the genetic backgrounds and environments [33]. This method has been used to identify consensus regions by examining QTL data from independent studies for their effect and consistency across different genetic backgrounds and environments, and to refine and confirm QTL positions on a consensus map by using mathematical models [34]. In wheat, MQTL analysis has also been successfully used to identify consensus QTL regions for yield-related traits [35,36,37,38], drought and heat tolerance [39,40,41,42], disease resistance [43,44,45,46], grain quality traits [31, 47,48,49], root-related traits [50,51,52], and so on. Likewise, MQTL has also been widely used for the different quantitative traits in different species such as rice (Oryza sativa L.) [53,54,55], maize (Zea mays L.) [56,57,58], barley (Hordeum vulgare L.) [59], and cotton (Gossypium hirsutum L.) [60]. MQTL analysis examined relevant QTL studies and refined the confidence intervals (CIs) of QTLs or QTL clusters to identify more reliable QTLs [38]. With the release of the high-quality genome sequence of Chinese Spring [61], there is an unprecedented likelihood of using these public resources to uncover the molecular mechanisms that influence important wheat agronomic traits at the genetic level [62]. In the same way, numerous transcriptomic data of wheat are available on a user-friendly platform [63, 64]. For instance, MQTL analysis combined with transcriptome assessment for important quality traits in wheat was performed [47], which led to the identification of 110 MQTLs, and finally 44 candidate genes with high probability of association with quality traits. Saini et al.(2022) [65] identified 141 MQTLs out of 2852 major MQTLs for yield and related traits and further predicted 1202 candidate genes within major MQTL regions and 50 homologs of associated genes for yield from other cereals. Eighty-six MQTLs for yield-related traits were also identified from 381 original QTLs under different environmental conditions, of which 18 genes or gene clusters associated with these MQTLs were validated in this study [36].
The aim of the present study was to conduct a meta-analysis for flag leaf size from independent QTL mapping studies published in the last decades and to deepen the genetic architecture underlying flag leaf size by discovering putative genes and incorporating transcriptomic studies. The results will provide further insight into the genetic determinants for flag leaf size, and some reliable QTLs and putative candidate genes will be suggested to be employed for the genetic improvement of these traits in wheat.
Results
Quantitative trait loci controlling flag leaf size in wheat
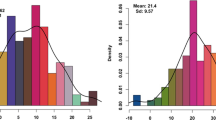
A total of 31 studies published between 2008 and 2020, involving 34 recombinant inbred line (RIL) populations, three double haploid (DH) populations, and two backcross (BC) populations, were thoroughly reviewed to compile information on available QTLs (Table 1). As a result, 521 initial QTLs associated with flag leaf size were collected and distributed among all 21 wheat chromosomes. Of the earlier reported 521 initial QTLs, 38.39% (200) were distributed to subgenome A, 39.54% (206) to subgenome B, and only 22.07% (115) to subgenome D (Fig. 1a). Only 333 QTLs were successfully projected onto the consensus map (Fig. 1a and additional file 3), whereas the associated markers of the remaining 188 QTLs were absent from the consensus map or had a low phenotypic variation explained (PVE) value or large CI. The projected 333 QTLs were identified on all 21 chromosomes except 2D, 3A, and 5D. The highest number of projected QTLs was on subgenome B with 164 QTLs, whereas the lowest number of projected QTLs was on subgenome D with 41 QTLs (Fig. 1a). Correspondingly, the 95% CI varied from 0.04 cM to 55.14 cM, with approximately 63.53% (331) of the collected initial QTLs having a CI of less than 10 cM and 84.26% (439) having a CI of less than 20 cM (Fig. 1b). The PVE values for individual QTLs ranged from 1.05 to 54.38% with a mean of 10.23%. Only 39.92% (208) of the initial QTLs had PVE values greater than 10% (Fig. 1c).
Projection of initial QTLs and identification of meta-QTL for flag leaf size
A total of 64 MQTLs were generated from the 333 projected QTLs based on the criteria of the lowest model value and at least two overlapping initial QTLs (Fig. 2, Table S1). The 95% CI of the identified MQTLs, ranging from 0.02 to 18.05 cM with an average of 1.74 cM, was 5.28-fold reduction than that of the initial QTLs (Fig. 3). This indicated that these MQTLs were mapped more accurately. In addition, there were significant differences in the average CIs of the MQTLs between different chromosomes. For example, the average CI of MQTLs on chromosomes 1A, 3B, 6A and 1D decreased 102.28-, 30.16-, 23.38- and 22.14-fold, respectively (Fig. 3). Based on the comparison of the flanking marker sequences, the physical locations of all 64 MQTLs ranged from 1.64 Mb (MQTL 2B.6) to 206.35 Mb (MQTL 5A.2) (Table S1). These MQTLs, were then selected for further identifying putative candidate genes. Of the MQTLs identified, 13 were core MQTLs, such as MQTL2A.1, MQTL 2A.2, MQTL 2A.3, MQTL 2A.5, MQTL 2B.4, MQTL 2B.6, MQTL 2B.8, MQTL 3B.4, MQTL 3B.5, MQTL 5A.4, MQTL 5B.3, MQTL 6B.6, and MQTL 7A.5, met the previously established criteria for the search for candidate genes in the databases available. Moreover, the physical distances of the core MQTLs ranged from 1.64 to 18.77 Mb, while the genetic distance ranged from 0.02 to 0.9 cM (Table 2).
MQTLs matching MTAs from previous genome-wide association studies
The physical positions of the MQTLs identified in this study and the marker trait associations (MTAs) from 11 previous studies were used for comparison to further determine the accuracy of MQTL for flag leaf size (Fig. 4, Table S2). Accordingly, 51.56% (33/64) of the identified MQTLs were co-located with 77 SNP peak positions early reported in GWAS for leaf size in wheat. This indicated that only half of the MQTL regions could be validated by the MTAs. The number of MTAs colocalized for each MQTL also varied from one to seven in 11 GWAS studies. Each of these 33 MQTLs was colocalized with at least one MTA. Of these, MQTL-2A.1, MQTL-4B.5 and MQTL 4A.1 were colocalized with 7, 6 and 5 MTAs, respectively.
Putative genes and in silico gene expression analysis
In this study, three approaches were developed to identify putative candidate genes associated with leaf size in wheat. An exhaustive search for known rice genes associated with flag leaf traits resulted in a collection of 97 candidate genes (Table S3) that were used to identify their corresponding homologs in wheat. Only 41 genes of these were identified within 22 MQTL regions (Table S4). These genes were reported to affect leaf-related traits of rice through a variety of proteins/products, such as auxin response factor, zinc finger protein, probable transcription factor, cyclin-dependent kinase inhibitor, growth regulating factor, and so on. The number of putative genes within each MQTL varied from one to five, with an average of 1.86 genes per MQTL. We identified 2262 genes within MQTL regions, including 41 genes with corresponding homologs for the leaf traits in rice (Table S4), and 2221 putative genes after eliminating duplicate genes in overlapping MQTLs (Table S5). Most putative genes (278 genes) were identified within the confidence region of MQTL-5B.3, whereas only one gene was found on chromosome 6A. These genes with similar function included 183 putative genes for F-box-like domain proteins, 78 for protein kinases, 48 for BTB/POZ domain-containing proteins, 33 for leucine-rich repeat domain proteins, 25 for glycosyltransferase family proteins, and 21 for cytochrome P450 proteins, etc. (Fig. S1).
Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to determine the functional classification of the identified genes. KEGG enrichment analysis revealed that these putative genes were highly involved in the peroxisome, basal transcription factor, and tyrosine metabolism pathways, with the greatest number of putative genes in the peroxisome pathway (Fig. 5). GO analysis revealed a range of GO terms, of which some of the most important and abundant GO terms included those involved in all three categories. The most enriched GO terms related to biological processes involved metabolic processes (621 genes) and cellular processes (506 genes). The most enriched GO terms related to molecular functions involved binding (955 genes) and catalytic activities (634 genes). Regarding cellular components, genes were mainly enriched in the cell (314 genes) and in cell parts (309 genes) (Fig. 6).
Some critical genes controlling leaf size that emerged from enrichment analysis of GO and KEGG pathways were considered as selected genes for further in silico expression analysis.
The expression analysis of the available genes revealed that 134 genes with > 2 transcripts per million (TPM) were highly and specifically expressed in the leaf (Fig. 7, Table S6). Based on their expression, these genes could be divided into three classes (Fig. 7). In class I, the genes showed high expression in leaf during seedling and tillering stage, while in class II, the genes showed high expression in leaf at 14-day development stage and finally in class III, the genes showed high expression in leaf at three-leaf stage. Despite in the same tissues, the expression of the genes (e.g., TraesCS2A02G072400, TraesCS4B02G36700, TraesCS6B02G063400 and so on) varied in different growth stages (Fig. 7). Consequently, the expression analysis of 134 putative candidate genes at different developmental stages allowed us to realize their potential roles in seedling leaf size, which were hypothesized to influence leaf size at the adult plant stage.
Discussion
Identification of key MQTL regions through meta-analysis
Extensive studies on QTL mapping of yield and other important agronomical traits in wheat have been conducted in recent decades. Nevertheless, most QTLs identified in these studies are each associated with a long CI and low PVE. This made these QTLs less useful for the marker-assisted breeding. In contrast, MQTLs with a narrow CI and a relatively high PVE are more compelling in proving useful for molecular breeding [65]. It was also found that the results of the MQTL analysis were significantly and positively correlated with the quality of the QTL mapping results [92]. In addition, new QTLs are regularly added to the databases as molecular genetics and QTL mapping methods continue to evolve. Therefore, it is very important to keep up with this pace to integrate new QTLs into more stable and reliable MQTLs [47]. In the present study, 521 initial QTLs were collected from 31 studies between 2008 and 2020 to identify genomic regions associated with flag leaf size in wheat (Table 1). Compared with subgenomes A and B, the subgenome D had a lower number of QTLs, which was consistent with previous MQTL analyses for grain yield and other yield-related traits [32, 36, 65, 92]. Only about 24.4% of the QTLs were mapped on subgenome D, while about 75.6% were found on subgenomes A and B (Fig. 1a). One possible reason for this phenomenon could be that the subgenome D has a low level of DNA polymorphism. Compared to the diploid progenitor species Aeegilops tauschii, very low genetic diversity has been observed for the subgenome D of wheat [93]. Meanwhile, there has also been a limited gene flow from Aegilops tauschii to cultivated wheat [94].
For the 64 MQTLs identified in the present study, the CI of the identified MQTLs with an average of 1.74 cM reduced 5.28-fold compared with the mean value of the corresponding initial QTLs (Fig. 3). In a similar study, the discovery of 13 MQTLs with an average CI of 13.6 Mb for the initial QTLs and 6.01 Mb for the MQTLs was found to be 2.26-fold reduction than that of the initial QTLs for drought tolerance in bread wheat [32]. Moreover, the definitive physical position of the 64 MQTLs in the present study was obtained by the publication of the wheat genome reference sequence of Chinese Spring, where the physical position of the identified 64 MQTLs varied from 1.64 Mb (MQTL 2B.6) to 206.35 Mb (MQTL 5A.2). Interestingly, 48 of the identified MQTLs contained a 95% genetic CI below 2 cM (Table S1).
It is widely accepted that optimizing flag leaf morphology, including the leaf length, width and area are important determinants to increase yield in wheat [82, 84, 95, 96]. Generally, leaf size is controlled by two main dimensions: leaf length and width which are sensitive to environmental factors [97, 98]. Previous studies have detected significant and positive correlation between FLW and FLA, also found that FLW was more crucial than FLL in determining FLA [73, 84]. Of the 64 MQTLs identified in the current study, 45 MQTLs were detected associated with FLA, of which 32 MQTLs correlated with FLW and 35 MQTLs related to FLL. It seemed that there was much the same of their contribution on FLA. Moreover, there were 22 MQTLs all associated with FLL, FLW and FLA. The comparison of initial QTLs PVE of 22 MQTLs found that 14 MQTLs possessed higher PVE with FLW than FLL. Similar to early studies, this result further demonstrated that FLW as the major contributor had more influences on FLA. Also, Li et al. (2018) reported that FLW can be used to select lines with large KN which is one of the main components of grain yield [25]. Therefore, individuals with wider flag leaves should be selected to increase FLA and also increase yield potential in wheat breeding programs.
It has been demonstrated that several QTL intervals for flag leaf traits were mapped to the same or similar chromosomal regions for yield-related traits in the previous studies. For example, Ma et al. (2020) found that the interval of QFll.sicau-2D.3, QFlw.sicau-2D and QFla.sicau-2D were closely related to QTL for spikelet number per spike, plant height (PH), anthesis date, thousand-grain weight (TGW), spike length (SL) and kernel number per spike (KN) [84]. Liu et al. (2018) confirmed that QFLA-4B.1 and QFLA-4B.2, were detected close to marker Xbarc20, which was also found to co-localize with QTL for PH, SL, spike number per plant, KN, GW, and TGW [18]. In addition, 34 MQTLs identified in the current study had their physical positions almost coincidence with those physical positions of MQTLs reported in three recent studies for yield-related traits [38] (Table S1). Given that MQTL for flag leaf traits that showed consistent relationships with yield-related traits, these MQTLs with a higher level of confidence may be described as pleiotropic regions which can effectively improve breeding efficiency for multiple traits.
There were 13 core MQTLs were selected basing on the preferred criteria of at least two overlapping initial QTLs with a physical distance < 20.0 Mb and a genetic distance < 1.0 cM [44] (Table 2), a higher level of confidence for further analysis and for identification of candidate genes. These 13 core MQTLs showed a smaller genetic CI (0.28 cM) compared with the initial QTLs (9.18 cM) with 32.79-fold reduction. Of these, MQTL-2A.1, MQTL-2A.3, MQTL-2B.4, MQTL-5A.4 and MQTL-5B.3 were validated by the MTAs. As for the 134 genes obtained via transcriptome and functional annotation, 39 genes were identified within the regions of 13 core MQTLs. It was worth mentioning that nearly half of (18/39) genes available from five core MQTL regions verified by the MTAs. Some of the significant features of these 13 core MQTLs detected in this study were described as follows: (i) They showed stability under different environments: MQTL-1A.1 consisted of nine initial QTLs for flag leaf length, width, and area with an average PVE of 6.54% from six different populations [22, 28], suggesting that MQTL-1A.1 exhibits strong stability for the flag leaf size trait. Apart from the above core MQTLs, the other core MQTLs also showed high stability under different environments. (ii) There were multiple core MQTLs accounting for the same traits. Except for MQTL-2A.1, 12 of the 13 core MQTLs were all based on the initial QTLs for flag leaf length. Likewise, MQTL-2A.1, MQTL-2A.2, MQTL-2A.3, MQTL-2B.4, MQTL-2B.6, MQTL-3B.5, MQTL-5A.4, MQTL-5B.3, and MQTL-7A.5 were composed of the initial QTLs for the flag leaf area trait (Table 2). These core MQTLs, not based on only one trait, were apparently more robust than initial QTLs. (iii) The core MQTLs showed pleiotropic effect. All 13 core MQTLs were responsible for controlling more than two traits. Of these, MQTL-5A.4 derived from nine initial QTLs, followed by MQTL-5B.3 and MQTL-3B.5 derived from seven initial QTLs for control of multiple traits (Table 2), suggesting that these MQTLs may represent a complex genomic region for control of more than one trait.
Potential candidate genes associated with leaf size in meta-QTL regions
To support the location of the MQTLs identified in this study, an extensive literature search was conducted to identify known genes within MQTL regions. For example, Siddiqui et al. (2021) identified two candidate genes TraesCS4B02G293600 and TraesCS4B02G293700 on wheat chromosome 4 B[99]. They were strongly expressed in leaves and stems as well as under drought stress conditions, suggesting that the two genes are involved in photosynthetic pathways and drought tolerance mechanisms. The two genes were also located in the MQTL-4B.4 region identified in the present study. Liu et al. (2018) found that the gene Ppd-D1 regulated leaf growth by controlling photoperiod, and its physical location was also found in the MQTL-5B.2 region in the current study [100]. When comparing the MQTLs identified in this study with the wheat gene TaCHLI-7A, encoding the protein CHLI involved in the biosynthesis of chlorophyll in common wheat [101], the gene was also located in the MQTL-7A.4 region. In addition, Muhammad et al. (2021) predicted 18 candidate genes for flag leaf length in wheat [27], where six of the predicted genes TraesCS5A01G487600, TraesCS5A01G487700, TraesCS5A01G487800, TraesCS5A01G487900, TraesCS5A01G488000, TraesCS5A01G488100, TraesCS5A01G488200 were located with the MQTL-5A.5 region in this study.
Another important finding in the present study was that 2262 putative genes related to flag leaf size were identified within the MQTL regions and showed the spatiotemporal and specific expression pattern (Table S4, Table S5). These candidate genes mainly encode the F-box-like domain proteins, protein kinases and BTB/POZ domain-containing proteins. The F-box protein FBX92 played a role in regulating leaf growth in Arabidopsis, where FBX92 regulates the rate of cell division [102]. PINOID kinase may actually function by influencing auxin accumulation and distribution leading to influence auxin metabolism and signaling indirectly, and finally suggest a role in leaf development as well in Arabidopsis [103]. The candidate genes associated with wheat leaf size within the MQTL regions were identified through the analysis of transcriptome data. Given the close evolutionary relationship between Gramineae species genomes [104], candidate genes with unknown functions in the wheat genome were evaluated in the MQTL regions of our study based on their orthologous genes in the rice genome [48]. For example, eight wheat homologs for rice genes, TraesCS1D02G393900, TraesCS1D02G394000, TraesCS1D02G394100, TraesCS4B02G064000, TraesCS5B02G508800, TraesCS7A02G059000, TraesCS7B02G484200, and TraesCS7D02G409700 were predicted in the early study [92], which were overlapped with six MQTLs identified in the present study. Of these, the first three genes were all present on MQTL-1D. 1, which was formed from five initial QTLs from three different populations. The last five genes were located on the MQTL-4B.5, MQTL-5B.3, MQTL-7A.1, MQTL-7B.1, and MQTL-7D.2, respectively. The involvement of these eight homologous genes in the different biological processes was associated with leaf size and chlorophyll content in rice, suggesting that these genes may be involved in the regulation of leaf size in wheat.
In addition, we annotated 2262 genes using GO or KEGG analysis (Figs. 5 and 6). KEGG and GO pathway enrichment analysis revealed that these putative genes were highly involved in the peroxisome, basal transcription factor, tyrosine metabolism, photosynthesis and plant hormone signal transduction pathways. Peroxisome get involved in the photorespiration and the synthesis of phytohormones, which are important for signaling pathways, including jasmonic acid, auxin, and salicylic acid [105, 106]. The CFL2 regulated by transcription factor Roc5, encoding a cytochrome P450 protein, is involved in the regulation of flag leaf shape by influencing epidermis and cell wall development [107]. Tocochromanols and plastoquinone produced in the tyrosine biosynthetic pathways are essential metabolites produced in all plants and other photosynthetic organisms [108]. Plastoquinone is required for photosynthesis as an electron carrier, an enzyme involved in carotenoid biosynthesis and is a cofactor of phytoene desaturase. Tocopherols have unexpected roles in photo-assimilate transport [109, 110]. Herein, a total of 134 putative genes with TPM > 2 in the robust and stable MQTL regions were listed based on significant gene expression in the leaf that may potentially affect leaf size in wheat (Table S6). For example, TraesCS4A02G149900, TraesCS4B02G165100, and TraesCS4D02G157200, encoding an ATP-dependent proteolytic subunit of Clp protease, were specifically expressed at the leaf seedling stage, while their homologous NAL9 genes caused reduced cell number in the lateral direction due to a significant reduction in the total number of vascular bundles in rice [111]. TraesCS4B02G341600, encoding a cytochrome P450 family protein, was also specifically expressed at the leaf seedling stage. Its homologous gene sd37 in rice was confirmed to encode the putative cytochrome P450 protein CYP96B4, and the sd37 transgenic leaves were smaller than those of the wild type, reflecting a decrease in cell number in the mutant [112]. TraesCS7D02G350500, encoding β-ketoacyl-CoA synthase, was strongly expressed at the third leaf stage. In rice, its homologous gene WSL1 showed a pleiotropic phenotype, including reduced growth and shortened leaves [113]. Although the relationship between these genes and leaf size in wheat has not been reported, their homologous genes have been shown to be involved in the regulation of leaf size in rice. In addition, 17 genes with TPM > 2 enriched in the peroxisome, photosynthesis, basal transcription factors and plant hormone signal transduction pathway. This suggests that these 134 putative candidate genes may have potential effects on leaf size regulation in wheat.
Conclusions
In this study, we deciphered key genomic regions controlling flag leaf size in wheat by integrating MQTL analysis and in silico transcriptome assessment. The 333 initial QTLs were successfully projected on the reference genetic map and refined into 64 MQTLs. Of these, 13 core MQTLs showed the mean CI was 32.79-fold reduction than initial QTLs and five core MQTLs were validated by the MTAs, suggesting as potential loci in MAS for flag leaf size in wheat. The 2262 putative candidate genes were mined within the MQTL regions by the genomic sequence comparison, where 134 candidate genes with more than 2 TPM were highly and specifically expressed in the leaf by in silico gene expression analysis. This suggested that, if these key MQTL regions and candidate genes will be further validated through biological experiment strategies, they have great application potential in molecular genetic improvement of flag leaf size in wheat.
Methods
Bibliographic collection of QTLs for flag leaf size and construction of reference map for QTL projection
For QTLs controlling for flag leaf size, a comprehensive bibliographic collection was performed using PubMed (http://www.ncbi.nlm.nih.gov/pubmed), Google Scholar (https://scholar.google.com/) and China National Knowledge Infrastructure (https://www.cnki.net/). QTL projection were conducted if all required information were available. For each study, QTL information collected included: (i) QTL name; (ii) three flag leaf related traits, including flag leaf length (FLL), flag leaf width (FLW) and flag leaf area (FLA); (iii) closely related flanking markers; (iv) position of the peak and associated 95% CI; (v) type and size of lines in the mapping population; (vi) LOD score; and (vii) the phenotypic explained variation (PVE) or R2 values of the QTLs. In cases where the log of odds ratio (LOD) and R2 values were missing for some QTLs detected in previous studies, they were assumed to be 3 and 10, respectively [44, 53]. When the peak position was missing, the midpoint between the two flanking markers was treated as the position [92]. In addition, for the initial QTLs that were missing flanking markers and CIs, the CIs were recalculated according to population type and size using the following standard formula: (i) F2 and backcross population, CI = 530/ (N× R2); (ii) recombinant inbred line (RIL) population, CI = 163/ (N× R2); and (iii) doubled haploid population, CI = 287/ (N× R2). Here, 530, 163, and 287 are the population-specific constants obtained from different simulations [114, 115], N is the size of the mapping population used for QTL analysis, and R2 is the phenotypic variation explained by QTL [32].The main markers used to generate genetic linkage maps in QTL mapping studies include Simple Sequence Repeat (SSR), Diversity Arrays Technology (DArT), and Single Nucleotide Polymorphism (SNP) markers [36]. The genetic reference map obtained from two dense genetic maps [51, 116] was integrated as a high-density reference map [35]. This map contained 14,548 markers, including SSR, DArT, SNP and other types of markers, with a total length of 4813.72 cM, ranging from 155.6 cM to 350.11 cM in the 21 linkage groups. The map was used as reference map for projection of individual QTLs identified in independent populations [48].
QTL projection and meta-QTL analysis
The initial QTLs data, the associated individual genetic maps from previous independent studies, and the reference genetic map were used as input files to create a consensus map and further perform the MQTL analysis [92]. BioMercator v4.2 software was used for projection [117, 118], the initial QTLs and the information of each QTL, for instance, CI, peak position, LOD score and and R2 were projected onto a reference map [117]. QTLs were discarded when they could not be projected onto the consensus map and those mapped to positions outside the consensus map [32].
After projection, MQTL analysis was performed on each chromosome using BioMercator v4.2 software [117, 118] via the Veyrieras two-step algorithm [118, 119]. Two different approaches were used based on the number of initial QTLs on each chromosome. In the first approach, the meta-analysis proposed by Goffinet and Gerber (2000) [119] was applied when the number of initial QTLs on a chromosome was less than 10. Based on this approach, the best MQTL model with the lowest AIC values for QTL integration and identification of consensus MQTL positions in BioMercator v4.2 software was selected. On the other hand, if the number of QTLs in a chromosome was at least 10, the second method proposed by Veyrieras [120] was used. According to this approach, meta-analyses were performed for individual chromosomes using a two-stage approach available in the software. In the first step, the collected QTLs on individual chromosomes are clustered using default parameters. The number of potential MQTLs per chromosome is then estimated based on the following five selection criteria, including Akaike information criterion (AIC), corrected Akaike information criterion (AICc), Akaike information criterion 3 (AIC3), Bayesian information criterion (BIC), and approximate weight of evidence (AWE). A QTL model that had the lowest values of the selection criteria was considered the best optimal model for the next step of meta-analysis. In the second step, the 95% CI and the positions of each MQTL were determined according to the optimal model selected in the previous step. The QTLs were integrated so that the peak position of the initial QTLs was in the MQTL CI [34], whereas the MQTLs without the minimum AIC values were to be discarded.
Identification of putative genes in MQTL regions
The putative genes are located within the regions identified basing on the positions of the flanking markers of the MQTL (or the marker closest to the flanking markers) [32]. After identifying the MQTLs, the next step was to find the flanking markers within the target MQTLs based on the 95% CI and the positions of each MQTL. The physical positions of each MQTL were obtained based on the positions of flanking markers which were searched in the Triticeae Multi-omics Center (http://wheatomics.sdau.edu.cn) annotated by IWGSC_v1.1_HC_gene. When the physical positions of the flanking markers were not found, the GrainGenes database (https://wheat.pw.usda.gov/GG3) or the DArT database (https://www.diversityarrays.com) were used to obtain their sequences. The sequences information was then aligned to the wheat reference genome in the Triticeae Multi-omics Center (http://wheatomics.sdau.edu.cn), using the BLASTN program to find the physical position of flanking markers [92].
MQTLs are considered potential genomic regions that are likely harbor putative genes for the traits [121]. Based on other meta-analyses published in recent years, three methods have been used to identify putative genes within MQTL regions [44, 92]. (i) In the first method, the strategy of orthologous comparison between wheat and rice was used to identify the major putative genes in the MQTL regions. For this purpose, the China Rice Data Center (https://www.ricedata.cn/gene/) was manually used to identify the genes for flag leaf associated traits in rice. In addition, the homologous genes of wheat were retrieved from the Triticease-Gene Tribe (http://wheat.cau.edu.cn/TGT/) based on the IWGSC RefSeq v1.1. (ii) To further refine the MQTL, those with at least two overlapping initial QTLs with a physical distance < 20.0 Mb and a genetic distance < 1.0 cM, referred to as core MQTLs, were selected from the second approach. (iii) The peak physical positions of the remaining MQTLs were calculated using 1-Mb region on each side of the MQTLs for mining relevant genes within the MQTL regions. The peak physical position of the MQTLs was calculated according to the method proposed by Saini [65]. Both the original and estimated range of physical positions were then entered into the search toolbox of the “Gene” in the WheatGmap database [122] to obtain details of gene models (locus ID information and functional descriptions) corresponding to MQTL regions.
Verification of MQTLs by GWAS and known wheat genes within MQTLs
To further validate the accuracy of the discovered MQTL regions, available genome-wide association studies for flag leaf size were reviewed to search for MTAs that could be compared with the MQTLs identified in this study. Considering the relatively large linkage disequilibrium decay in wheat (approximately 5 Mb), the MTAs obtained from GWAS near MQTLs in the 5 Mb physical region were considered to be related to MQTLs [92]. The physical positions of known genes associated with flag leaf size within MQTLs were obtained from the Triticeae Multi-omics Center (http://wheatomics.sdau.edu.cn). Subsequently, the physical positions of these genes were compared with the genomic regions of the MQTLs to identify genes that might correspond to individual MQTLs [65].
Expression of candidate genes within MQTL regions
Gene expression analysis examines how genes are transcribed to produce functional products such as RNA or proteins [47]. The GENEDENOVO cloud platform (https://www.omicshare.com) was used to perform the GO and KEGG analysis. For transcriptional expression analysis, the Expression Visualization and Integration Platform (expVIP, http://www.wheat-expression.com) with expression data from 18 tissues throughout the wheat growth period [64, 123] was used in this study. Only candidate genes showing at least 2 TPM expression were considered [124]. The expression characteristics of candidate genes were displayed by the heat map of TPM using the TBtools software [125]. In this study, the tissues and their corresponding stages were leaves at seedling, 14-day, three-leaf, and tillering stages; roots at seedling, 14-day, and three-leaf stages; stems at 1-cm spike, two-node, and anthesis stages.
Availability of data and materials
The relevant data and additional information are available in the supplementary files.
Abbreviations
- AIC:
-
Akaike information criterion
- AICc:
-
Corrected Akaike information criterion (AICc)
- AIC3:
-
Akaike information criterion 3
- AWE:
-
Approximate weight of evidence
- BC:
-
Backcross populations
- BIC:
-
Bayesian information criterion
- CI:
-
Confidence interval
- DArT:
-
Diversity arrays technology
- DH:
-
Double haploid populations
- expVIP:
-
Expression visualization and integration platform
- FLA:
-
Flag leaf area
- FLL:
-
Flag leaf length
- FLW:
-
Flag leaf width
- GO:
-
Gene ontology
- GWAS:
-
Genome-wide association study
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- KN:
-
Kernel number per spike
- LOD:
-
The log of odds ratio
- MAS:
-
Marker-assisted selection
- MQTL:
-
Meta-QTL
- MTAs:
-
Marker trait associations
- PH:
-
Plant height
- PVE:
-
The explained phenotypic variation
- QTL:
-
Quantitative trait loci
- RIL:
-
Recombinant inbred line populations
- R 2 :
-
The proportion of phenotype variation value
- SL:
-
Spike length
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeat
- TGW:
-
Thousand-grain weight
- TPM:
-
Transcripts per million
References
Cao S, Hanif M, Xia X, He Z. Genetic architecture underpinning yield component traits in wheat. Theor Appl Genet. 2020;133(6):1811–23.
Curtis T, Halford NG. Food security: the challenge of increasing wheat yield and the importance of not compromising food safety. Ann Appl Biol. 2014;164(3):354–72.
Hatfield JL, Beres BL. Yield gaps in wheat: path to enhancing productivity. Front Plant Sci. 2019;10:1603.
Bustos DV, Hasan AK, Reynolds MP, Calderini DF. Combining high grain number and weight through a DH-population to improve grain yield potential of wheat in high-yielding environments. Field Crop Res. 2013;145:106–15.
Liu H, Searle IR, Mather DE, Able AJ, Able JA. Morphological, physiological and yield responses of durum wheat to pre-anthesis water-deficit stress are genotype-dependent. Crop Pasture Sci. 2015;66(10):1024–38.
Tshikunde NM, Mashilo J, Shimelis H, Odindo A. Agronomic and physiological traits, and associated quantitative trait loci (QTL) affecting yield response in wheat (Triticum aestivum L.): A review. Front. Plant Sci. 2019;10:1428.
Attri H, Dey T, Singh B, Kour A. Genetic estimation of grain yield and its attributes in three wheat (Triticum aestivum L.) crosses using six parameter model. J Genet. 2021;100(2):1–9.
Guan P, Lu L, Jia L, Kabir MR, Zhang J, Lan T, et al. Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). front. Plant Sci. 2018;9:529.
Rigatti A, de Pelegrin AJ, Meier C, Lunkes A, Klein LA, Da Silva AF, et al. Combination capacity and association among traits of grain yield in wheat (Triticum aestivum L.): A review. J Agric Sci. 2018;10(5):179–87.
Gambín BL, Borrás L, Otegui ME. Kernel weight dependence upon plant growth at different grain-filling stages in maize and sorghum. Aust J Agric Res. 2008;59(3):280–90.
Khaliq I, Irshad A, Ahsan M. Awns and flag leaf contribution towards grain yield in spring wheat (Triticum aestivum L.). Cereal Res Commun. 2008;36(1):65–76.
Borrill P, Fahy B, Smith AM, Uauy C. Wheat grain filling is limited by grain filling capacity rather than the duration of flag leaf photosynthesis: A case study using NAM RNAi plants. PLoS One. 2015;10(8):e0134947.
Liang X, Liu Y, Chen J, Adams C. Late-season photosynthetic rate and senescence were associated with grain yield in winter wheat of diverse origins. J Agron Crop Sci. 2018;204(1):1–12.
Zhao D, Derkx AP, Liu DC, Buchner P, Hawkesford MJ. Overexpression of a NAC transcription factor delays leaf senescence and increases grain nitrogen concentration in wheat. Plant Biol. 2015;17(4):904–13.
Li Z, Pinson S, Stansel JW, Paterson AH. Genetic dissection of the source-sink relationship affecting fecundity and yield in rice (shape Oryza sativa L.). Mol Breed. 1998;4(5):419–26.
Ba Q, Zhang L, Chen S, Li G, Wang W. Effects of foliar application of magnesium sulfate on photosynthetic characteristics, dry matter accumulation and its translocation, and carbohydrate metabolism in grain during wheat grain filling. Cereal Res Commun. 2020;48(2):157–63.
Sharma SN, Sain RS, Sharma RK. The genetic control of flag leaf length in normal and late sown durum wheat. J Agric Sci. 2003;141(3-4):323–31.
Liu K, Xu H, Liu G, Guan P, Zhou X, Peng H, et al. QTL mapping of flag leaf-related traits in wheat (Triticum aestivum L.). Theor Appl Genet. 2018;131(4):839–49.
Guitman MR, Arnozis PA, Barneix AJ. Effect of source-sink relations and nitrogen nutrition on senescence and N remobilization in the flag leaf of wheat. Physiol Plant. 1991;82(2):278–84.
Coleman R, Gill G, Rebetzke G. Identification of quantitative trait loci for traits conferring weed competitiveness in wheat (Triticum aestivum L.). Aust J Agric Res. 2001;52(12):1235–46.
Sohei K, Yoshimichi F, Satoshi M, Tadashi S, Mitsuru O, Khush GS. Quantitative trait loci affecting flag leaf development in rice (Oryza sativa L.). Breed Sci. 2003;53(3):255–62.
Zhao C, Bao Y, Wang X, Yu H, Ding A, Guan C, et al. QTL for flag leaf size and their influence on yield-related traits in wheat. Euphytica. 2018;214(11):1–15.
Farokhzadeh S, Fakheri BA, Nezhad NM, Tahmasebi S, Mirsoleimani A. Mapping QTLs of flag leaf morphological and physiological traits related to aluminum tolerance in wheat (Triticum aestivum L.). Physiol Mol Biol Plants. 2019;25(4):975–90.
Jia H, Wan H, Yang S, Zhang Z, Kong Z, Xue S, et al. Genetic dissection of yield-related traits in a recombinant inbred line population created using a key breeding parent in China’s wheat breeding. Theor Appl Genet. 2013;126(8):2123–39.
Li F, Wen W, He Z, Liu J, Jin H, Cao S, et al. Genome-wide linkage mapping of yield-related traits in three Chinese bread wheat populations using high-density SNP markers. Theor Appl Genet. 2018;131(9):1903–24.
Li F, Wen W, Liu J, Zhang Y, Cao S, He Z, et al. Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol. 2019;19(1):1–19.
Muhammad A, Li J, Hu W, Yu J, Khan SU, Khan MHU, et al. Uncovering genomic regions controlling plant architectural traits in hexaploid wheat using different GWAS models. Sci Rep-UK. 2021;11(1):1–14.
Yan X, Zhao L, Ren Y, Zhang N, Dong Z, Chen F. Identification of genetic loci and a candidate gene related to flag leaf traits in common wheat by genome-wide association study and linkage mapping. Mol Breed. 2020;40(6):1–15.
Amo A, Soriano JM. Unravelling consensus genomic regions conferring leaf rust resistance in wheat via meta-QTL analysis. Plant Genome. 2022;15(1):e20185.
Swamy BP, Vikram P, Dixit S, Ahmed HU, Kumar A. Meta-analysis of grain yield QTL identified during agricultural drought in grasses showed consensus. BMC Genomics. 2011;12(1):319.
Singh R, Saripalli G, Gautam T, Kumar A, Jan I, Batra R, et al. Meta-QTLs, ortho-MetaQTLs and candidate genes for grain Fe and Zn contents in wheat (Triticum aestivum L.). Physiol Mol Biol Plants. 2022;28(3):637–50.
Kumar A, Saripalli G, Jan I, Kumar K, Sharma PK, Balyan HS, et al. Meta-QTL analysis and identification of candidate genes for drought tolerance in bread wheat (Triticum aestivum L.). Physiol Mol Biol Plants. 2020;26(8):1713–25.
Soriano JM, Colasuonno P, Marcotuli I, Gadaleta A. Meta-QTL analysis and identification of candidate genes for quality, abiotic and biotic stress in durum wheat. Sci Rep-UK. 2021;11(1):1–15.
Daryani P, Darzi Ramandi H, Dezhsetan S, Mirdar Mansuri R, Hosseini Salekdeh G, Shobbar Z-S. Pinpointing genomic regions associated with root system architecture in rice through an integrative meta-analysis approach. Theor Appl Genet. 2022;135(1):81–106.
Bilgrami SS, Ramandi HD, Shariati V, Razavi K, Tavakol E, Fakheri BA, et al. Detection of genomic regions associated with tiller number in Iranian bread wheat under different water regimes using genome-wide association study. Sci Rep-UK. 2020;10(1):1–17.
Liu H, Mullan D, Zhang C, Zhao S, Li X, Zhang A, et al. Major genomic regions responsible for wheat yield and its components as revealed by meta-QTL and genotype-phenotype association analyses. Planta. 2020;252(4):65.
Shamuyarira KW, Shimelis H, Mathew I, Zengeni R, Chaplot V. A meta-analysis of combining ability effects in wheat for agronomic traits and drought adaptation: implications for optimizing biomass allocation. Crop Sci. 2022;62(1):139–56.
Miao Y, Jing F, Ma J, Liu Y, Zhang P, Chen T, et al. Major genomic regions for wheat grain weight as revealed by QTL linkage mapping and Meta-analysis. Front Plant Sci. 2022;13:802310.
Acuña-Galindo MA, Mason RE, Subramanian NK, Hays DB. Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Sci. 2015;55(2):477–92.
Zhang J, Zhang S, Cheng M, Jiang H, Zhang X, Peng C, et al. Effect of drought on agronomic traits of rice and wheat: A Meta-analysis. Int J Environ Res Public Health. 2018;15(5):839.
Kumar S, Singh VP, Saini DK, Sharma H, Saripalli G, Kumar S, et al. Meta-QTLs, ortho-MQTLs, and candidate genes for thermotolerance in wheat (Triticum aestivum L.). Mol Breed. 2021;41(11):69.
Tanin MJ, Saini DK, Sandhu KS, Pal N, Gudi S, Chaudhary J, et al. Consensus genomic regions associated with multiple abiotic stress tolerance in wheat and implications for wheat breeding. Sci Rep-UK. 2022;12(1):13680.
Soriano JM, Royo C. Dissecting the genetic architecture of leaf rust resistance in wheat by QTL Meta-analysis. Phytopathology. 2015;105(12):1585–93.
Venske E, Dos Santos RS, Farias D, d R, Rother V, Da Maia L C, Pegoraro C, Costa de Oliveira A. Meta-analysis of the QTLome of Fusarium head blight resistance in bread wheat: refining the current puzzle. Front. Plant Sci. 2019;10:727.
Saini DK, Chahal A, Pal N, Srivastava P, Gupta PK. Meta-analysis reveals consensus genomic regions associated with multiple disease resistance in wheat (Triticum aestivum L.). Mol Breed. 2022;42(3):11.
Pal N, Jan I, Saini DK, Kumar K, Kumar A, Sharma PK, et al. Meta-QTLs for multiple disease resistance involving three rusts in common wheat (Triticum aestivum L.). Theor Appl Genet. 2022;135(7):2385–405.
Gudi S, Saini DK, Singh G, Halladakeri P, Kumar P, Shamshad M, et al. Unravelling consensus genomic regions associated with quality traits in wheat using meta-analysis of quantitative trait loci. Planta. 2022;255(6):115.
Shariatipour N, Heidari B, Tahmasebi A, Richards C. Comparative genomic analysis of quantitative trait loci associated with micronutrient contents, grain quality, and agronomic traits in wheat (Triticum aestivum L.). Frontiers. Plant Sci. 2021;12:2142.
Saini P, Sheikh I, Saini DK, Mir RR, Dhaliwal HS, Tyagi V. Consensus genomic regions associated with grain protein content in hexaploid and tetraploid wheat. Front Genet. 2022;13:1021180.
Darzi-Ramandi H, Shariati JV, Tavakol E, Najafi-Zarini H, Bilgrami SS, Razavi K. Detection of consensus genomic regions associated with root architecture of bread wheat on groups 2 and 3 chromosomes using QTL meta-analysis. Aust J Crop Sci. 2017;11(7):777–85.
Soriano JM, Alvaro F. Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci Rep-UK. 2019;9(1):1–14.
Saini DK, Chopra Y, Pal N, Chahal A, Srivastava P, Gupta PK. Meta-QTLs, ortho-MQTLs and candidate genes for nitrogen use efficiency and root system architecture in bread wheat (Triticum aestivum L.). Physiol Mol Biol Plants. 2021;27(10):2245–67.
Khahani B, Tavakol E, Shariati V, Fornara F. Genome wide screening and comparative genome analysis for Meta-QTLs, ortho-MQTLs and candidate genes controlling yield and yield-related traits in rice. BMC Genomics. 2020;21(1):1–24.
Li L, Peng Y, Tang S, Yu D, Tian M, Guo F, et al. Mapping QTL for leaf pigment content at dynamic development stage and analyzing Meta-QTL in rice. Euphytica. 2021;217(5):1–23.
Selamat N, Nadarajah KK. Meta-analysis of quantitative traits loci (QTL) identified in drought response in rice (Oryza sativa L.). Plants. 2021;10(4):716.
Guo J, Chen L, Li Y, Shi Y, Song Y, Zhang D, et al. Meta-QTL analysis and identification of candidate genes related to root traits in maize. Euphytica. 2018;214(12):1–15.
Wang Y, Wang Y, Wang X, Deng D. Integrated meta-QTL and genome-wide association study analyses reveal candidate genes for maize yield. J Plant Growth Regul. 2020;39(1):229–38.
Zhao X, Peng Y, Zhang J, Fang P, Wu B. Mapping QTLs and meta-QTLs for two inflorescence architecture traits in multiple maize populations under different watering environments. Mol Breed. 2017;37(7):1–18.
Khahani B, Tavakol E, Shariati JV. Genome-wide meta-analysis on yield and yield-related QTLs in barley (Hordeum vulgare L.). Mol Breed. 2019;39(4):1–16.
Said JI, Lin Z, Zhang X, Song M, Zhang J. A comprehensive meta QTL analysis for fiber quality, yield, yield related and morphological traits, drought tolerance, and disease resistance in tetraploid cotton. BMC Genomics. 2013;14(1):776.
International Wheat Genome Sequencing Consortium [IWGSC], Appels R, Eversole K, Stein N, Feuillet C, Keller B, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361(6403):eaar7191.
Quraishi UM, Pont C, Ain QU, Flores R, Burlot L, Alaux M, et al. Combined genomic and genetic data integration of major agronomical traits in bread wheat (Triticum aestivum L.). front. Plant Sci. 2017;1843:8.
Adams TM, Olsson TSG, Ramírez-González RH, Bryant R, Bryson R, Campos PE, et al. Rust expression browser: An open source database for simultaneous analysis of host and pathogen gene expression profiles with expVIP. BMC Genomics. 2021;22(1):166.
Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, et al. The transcriptional landscape of polyploid wheat. Science. 2018;361(6403):eaar6089.
Saini DK, Srivastava P, Pal N, Gupta PK. Meta-QTLs, ortho-meta-QTLs and candidate genes for grain yield and associated traits in wheat (Triticum aestivum L.). Theor Appl Genet. 2022;135(3):1049–81.
Zhang K. Construction of wheat (Triticum aestivum L.) molecular genetic map and QTL analysis. PhD Thesis. 2008. https://doi.org/10.7666/d.y1374914.
Mason RE, Mondal S, Beecher FW, Pacheco A, Jampala B, Ibrahim AMH, et al. QTL associated with heat susceptibility index in wheat (Triticum aestivum L.) under short-term reproductive stage heat stress. Euphytica. 2010;174(3):423–36.
Mason RE, Mondal S, Beecher FW, Hays DB. Genetic loci linking improved heat tolerance in wheat (Triticum aestivum L.) to lower leaf and spike temperatures under controlled conditions. Euphytica. 2011;180(2):181–94.
An M, Yang D, Li M, Zhang G, Li W. Construction of genetic linkage map and marker polymorphism analysis in wheat RIL population. J Gansu Agric Univ. 2012;47(6):74–80.
Wang L. Construction of wheat molecular genetic map and QTL analysis for agronomic and quality traits. PhD Thesis. 2012. https://doi.org/10.7666/d.y2117062.
Zarei L, Farshadfar E, Cheghamirza K, Desiderio F, Cattivelli L. QTL mapping of morphological traits associated with drought adaptation in a Iranian mapping population of durum wheat. In: Porceddu E, Damania AB, Qualset CO, editors. Proceedings of the International Symposium on Genetics and breeding of durum wheat. Bari: CIHEAM; 2014. p. 355-361.
Chang X, Li F, Zhang Z, Zhang X, Liu L, Yang X, et al. Mapping QTLs for flag leaf length, width and area in wheat. Acta Bot Bor-Occid Sin. 2014;34(5):896–901.
Fan X, Cui F, Zhao C, Zhang W, Yang L, Zhao X, et al. QTLs for flag leaf size and their influence on yield-related traits in wheat (Triticum aestivum L.). Mol Breed. 2015;35(1):24.
Yan X, Shi Y, Liang Z, Yang B, Li X, Wang S, et al. QTL mapping for morphological traits of flag leaf in wheat. Acta Agric Nucl Sin. 2015;29(7):1253–9.
Zhao P, Xu F, Jiang W, Qi P, Li C, Bai H, et al. Quantitative trait loci analysis of flag leaf length, width and chlorophyll content of spring wheat. J Triticeae Crops. 2015;35(5):603–8.
Wu Q, Chen Y, Fu L, Zhou S, Chen J, Zhao X, et al. QTL mapping of flag leaf traits in common wheat using an integrated high-density ssr and SNP genetic linkage map. Euphytica. 2016;208(2):337–51.
Xie Q, Mayes S, Sparkes DL. Preanthesis biomass accumulation of plant and plant organs defines yield components in wheat. Eur J Agron. 2016;81:15–26.
Lian J, Zhang D, Wu B, Song X, Ma W, Zhou L, et al. QTL mapping of flag leaf traits using an integrated high-density 90k genotyping chip. J Triticeae Crops. 2016;36(6):689–98.
Lu X, Bai H, Dong J, Hui J, Sun Y, Cai Z, et al. QTL mapping for size traits of flag leaf in spring wheat. J Triticeae Crops. 2016;36(12):1587–93.
Fu J, Dang Z, Li B, Zhao W, Zhao W, Yang W, et al. QTL mapping for flag leaf size and spike related traits in wheat (Triticum aestivum L.). J Triticeae Crops. 2017;37(6):713–20.
El-Feki WM, Byrne PF, Reid SD, Haley SD. Mapping quantitative trait loci for agronomic traits in winter wheat under different soil moisture levels. Agronomy. 2018;8(8):133.
Zhao C, Bao Y, Wang X, Yu H, Ding A, Guan C, et al. QTL for flag leaf size and their influence on yield-related traits in wheat. Euphytica. 2018;214(11):209.
Liu M, Zhang M, Zhang Q, Liu X, Guo Y, Sun J, et al. Genetic analysis of a major stable QTL QFLW-5B for wheat flag leaf width. J Triticeae Crops. 2019;39(12):1399–405.
Ma J, Tu Y, Zhu J, Luo W, Liu H, Li C, et al. Flag leaf size and posture of bread wheat: genetic dissection, QTL validation and their relationships with yield-related traits. Theor Appl Genet. 2020;133(1):297–315.
Khanna-Chopra R, Singh K, Shukla S, Kadam S, Singh NK. QTLs for cell membrane stability and flag leaf area under drought stress in a wheat RIL population. J Plant Biochem Biotechnol. 2020;29(2):276–86.
Hu J, Wang X, Zhang G, Jiang P, Chen W, Hao Y, et al. QTL mapping for yield-related traits in wheat based on four RIL populations. Theor Appl Genet. 2020;133(3):917–33.
Wang Y, Zhao L, Dong Z, Ren Y, Zhang N, Chen F. QTL mapping for plant height and flag leaf traits in common wheat. J Triticeae Crops. 2019;39(7):761–7.
Tu Y, Liu H, Liu J, Tang H, Mu Y, Deng M, et al. QTL mapping and validation of bread wheat flag leaf morphology across multiple environments in different genetic backgrounds. Theor Appl Genet. 2021;134(1):261–78.
Yan X, Wang S, Yang B, Zhang W, Cao Y, Shi Y, et al. QTL mapping for flag leaf-related traits and genetic effect of QFLW-6A on flag leaf width using two related introgression line populations in wheat. PLoS One. 2020;15(3):e0229912.
Yao J, Zhang C, Song X, Xu X, Xing Y, Lu D, et al. QTL analysis of wheat spike length and flag leaf length based on 90k assay. J Triticeae Crops. 2020;40(11):1283–9.
Li X, Wang S, Ni S. QTL mapping for traits of flag leaf and seedling in wheat. J Triticeae Crops. 2021;41(5):532–7.
Yang Y, Amo A, Wei D, Chai Y, Zheng J, Qiao P, et al. Large-scale integration of meta-QTL and genome-wide association study discovers the genomic regions and candidate genes for yield and yield-related traits in bread wheat. Theor Appl Genet. 2021;134(9):3083–109.
Mirzaghaderi G, Mason AS. Broadening the bread wheat D genome. Theor Appl Genet. 2019;132(5):1295–307.
Kumar A, Simons K, Iqbal MJ, de Jiménez MM, Bassi FM, Ghavami F, et al. Physical mapping resources for large plant genomes: radiation hybrids for wheat D-genome progenitor aegilops tauschii. BMC Genomics. 2012;13(1):1–15.
Wang J, Liu H, Zhao C, Tang H, Mu Y, Xu Q, et al. Mapping and validation of major and stable QTL for flag leaf size from tetraploid wheat. Plant Genome. 2022;15(4):e20252.
Wang Y, Qiao L, Yang C, Li X, Zhao J, Wu B, et al. Identification of genetic loci for flag-leaf-related traits in wheat (Triticum aestivum L.) and their effects on grain yield. Front. Plant Sci. 2022;13:990287.
Tsukaya H. Mechanism of leaf-shape determination. Annu Rev Plant Biol. 2006;57:477–96.
Tsukaya H. Leaf shape: genetic controls and environmental factors. Int J Dev Biol. 2005;49(5-6):547–55.
Siddiqui MN, Teferi TJ, Ambaw AM, Gabi MT, Koua P, Léon J, et al. New drought-adaptive loci underlying candidate genes on wheat chromosome 4B with improved photosynthesis and yield responses. Physiol Plant. 2021;173(4):2166–80.
Liu Y, Tao Y, Wang Z, Guo Q, Wu F, Yang X, et al. Identification of QTL for flag leaf length in common wheat and their pleiotropic effects. Mol Breed. 2018;38(1):1–11.
Wang C, Zhang L, Li Y, Ali Buttar Z, Wang N, Xie Y, et al. Single nucleotide mutagenesis of the TaCHLI gene suppressed chlorophyll and fatty acid biosynthesis in common wheat seedlings. Front Plant Sci. 2020;11:97.
Baute J, Polyn S, De Block J, Blomme J, Van Lijsebettens M, Inzé D. F-box protein fbx92 affects leaf size in arabidopsis thaliana. Plant Cell Physiol. 2017;58(5):962–75.
Saini K, Markakis MN, Zdanio M, Balcerowicz DM, Beeckman T, De Veylder L, et al. Alteration in auxin homeostasis and signaling by overexpression of pinoid kinase causes leaf growth defects in arabidopsis thaliana. Front Plant Sci. 2017;8:1009.
Gaut BS. Evolutionary dynamics of grass genomes. New Phytol. 2010;154(1):15–28.
Kao Y-T, Gonzalez KL, Bartel B. Peroxisome function, biogenesis, and dynamics in plants. Plant Physiol. 2018;176(1):162–77.
Su T, Li W, Wang P, Ma C. Dynamics of peroxisome homeostasis and its role in stress response and signaling in plants. Front Plant Sci. 2019;10:705.
Zhang X, Wang Y, Zhu X, Wang X, Zhu Z, Li Y, et al. Curled flag leaf 2, encoding a cytochrome p450 protein, regulated by the transcription factor roc5, influences flag leaf development in rice. Front Plant Sci. 2020;11:616977.
Schenck CA, Maeda HA. Tyrosine biosynthesis, metabolism, and catabolism in plants. Phytochemistry. 2018;149:82–102.
Amesz J. The function of plastoquinone in photosynthetic electron transport. BBA-Bioenergetics. 1973;301(1):35–51.
Norris SR, Barrette TR, DellaPenna D. Genetic dissection of carotenoid synthesis in arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell. 1995;7(12):2139–49.
Li W, Wu C, Hu G, Xing L, Qian W, Si H, et al. Characterization and fine mapping of a novel rice narrow leaf mutant nal9. J Integr Plant Biol. 2013;55(11):1016–25.
Zhang J, Liu X, Li S, Cheng Z, Li C. The rice semi-dwarf mutant sd37, caused by a mutation in CYP96B4, plays an important role in the fine-tuning of plant growth. PLoS One. 2014;9(2):e88068.
Yu D, Ranathunge K, Huang H, Pei Z, Franke R, Schreiber L, et al. Wax crystal-sparse leaf1 encodes a β–ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta. 2008;228(4):675–85.
Darvasi A, Soller M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav Genet. 1997;27(2):125–32.
Guo B, Sleper DA, Lu P, Shannon JG, Nguyen HT, Arelli PR. QTLs associated with resistance to soybean cyst nematode in soybean: Meta-analysis of QTL locations. Crop Sci. 2006;46(2):595–602.
Maccaferri M, Zhang J, Bulli P, Abate Z, Chao S, Cantu D, et al. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3-Genes Genom Genet. 2015;5(3):449–65.
Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, et al. Biomercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics. 2004;20(14):2324–6.
Sosnowski O, Charcosset A, Joets J. Biomercator v3: An upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics. 2012;28(15):2082–3.
Goffinet B, Gerber S. Quantitative trait loci: A meta-analysis. Genetics. 2000;155(1):463–73.
Veyrieras JB, Goffinet B, Charcosset A. MetaQTL: A package of new computational methods for the meta-analysis of QTL mapping experiments. BMC Bioinformatics. 2007;8(1):1–16.
Pal N, Saini DK, Kumar S. Meta-QTLs, Ortho-MQTLs and candidate genes for the traits contributing to salinity stress tolerance in common wheat (Triticum aestivum L.). Physiol Mol Biol Plants. 2021;27(12):2767–86.
Zhang L, Dong C, Chen Z, Gui L, Chen C, Li D, et al. Wheatgmap: A comprehensive platform for wheat gene mapping and genomic studies. Mol Plant. 2021;14(2):187–90.
Borrill P, Ramirez-Gonzalez R, Uauy C. expVIP: A customizable RNA-seq data analysis and visualization platform. Plant Physiol. 2016;170(4):2172–86.
Wagner GP, Kin K, Lynch VJ. A model based criterion for gene expression calls using RNA-seq data. Theory Biosci. 2013;132(3):159–64.
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202.
Acknowledgements
Not applicable.
Funding
This work was financially supported by the Key Sci & Tech Special Project of Gansu Province (22ZD6NA010), the Key Research and Development Program of Gansu Province, China (21YF5NA089), the Industrial Support Plan of Colleges and Universities in Gansu Province (2022CYZC-44), and the Research Program Sponsored by State Key Laboratory of Aridland Crop Science, China (GHSJ 2020-Z4).
Author information
Authors and Affiliations
Contributions
BK conducted analysis and wrote the draft of manuscript. JM, PZ, TC, YL and ZC collected and analyzed the original QTL data. FS provided scientific feedback and edited the draft. DY conceived and designed the experiments, reviewed and edited the writing of the manuscript. All authors revised and approved the final manuscript.
Corresponding author
Ethics declarations
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Description of 64 MQTLs identified for flag leaf size. Table S2. The GWAS analysis on flag leaf related traits used in this study. Table S3. The details of 97 rice known genes. Table S4. The 41 wheat homolog genes of 35 rice known genes located in MQTL. Table S5. The information of 2221 genes identified in 64 MQTL regions. Table S6. Summary of 134 putative candidate genes exhibiting significant expression (TPM > 2) within MQTLs.
Additional file 2: Figure S1.
Frequency of candidate genes in each of 16 different proteins associated with flag leaf traits.
Additional file 3.
The information of consensus map.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kong, B., Ma, J., Zhang, P. et al. Deciphering key genomic regions controlling flag leaf size in wheat via integration of meta-QTL and in silico transcriptome assessment. BMC Genomics 24, 33 (2023). https://doi.org/10.1186/s12864-023-09119-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09119-5