Abstract
Background
Aglaonema commutatum ‘Red Valentine’, as a foliage ornamental plant, is widely used for interior and exterior decoration because of its easy cultivation and management. However, reduced proportion of red foliage during large-scale production of A. commutatum seedlings is a frequent occurrence, which has considerable implications on the plant’s ornamental and market value. However, the molecular mechanisms underlying this phenomenon remain unclear.
Results
To explore the molecular basis of the variation in leaf color of A. commutatum Red Valentine, we performed transcriptome sequencing with the Illumina platform using two different varieties of A. commutatum, namely Red Valentine and a green mutant, at three different stages of leaf development. We annotated 63,621 unigenes and 14,186 differentially expressed genes by pairwise comparison. Furthermore, we identified 26 anthocyanin biosynthesis structural genes. The transcript per million (TPM) values were significantly higher for Red Valentine than for the green mutant in all three developmental stages, consistent with the high anthocyanin content of Red Valentine leaves. We detected positive transcription factors that may be involved in the regulation of anthocyanin biosynthesis using BLAST and through correlation analysis. Downregulation of these transcription factors may downregulate the expression of anthocyanin genes. We obtained full-length cDNA of the anthocyanin biosynthesis and regulatory genes and constructed phylogenetic trees to ensure accuracy of the analysis.
Conclusions
Our study provides insights into the molecular mechanisms underlying leaf variation in A. commutatum Red Valentine and may be used to facilitate the breeding of ornamental cultivars with high anthocyanin levels.
Similar content being viewed by others
Background
Flower, fruit, and leaf colors of agricultural crops are important traits of commercial and ornamental value and are determined by natural plant pigments, including anthocyanins, betalains, chlorophylls, and carotenoids. Among them, anthocyanins are the major pigments that confer red, violet, or blue color [1]. In plants, anthocyanins play vital roles in seed dispersal and stress responses [2, 3]; in humans, they serve as antioxidants and protect against cardiovascular diseases, cancers, and several chronic diseases. Moreover, they are widely used in the food, medicine, and cosmetic industries [4,5,6].
Studies have shown that the anthocyanin biosynthesis pathway is a ubiquitous branch of the flavonoid biosynthesis pathway in plants. Genes regulating anthocyanin biosynthesis and transport have been identified in numerous plant species [7,8,9]. This pathway starts from the phenylpropanoid pathway, involving three major genes: PAL (phenylalanine ammonia-lyase), C4H (trans-cinnamate 4-monooxygenase), and 4CL (4-coumarate-CoA ligase 2). The flavonoid biosynthesis pathway includes two classes of genes, namely early (EBGs) and late biosynthesis genes (LBGs) [10]. Chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3-hydroxylase (F3’H), and flavonol synthase (FLS) participate in early biosynthesis. These genes regulate the biosynthesis of flavonols and other flavonoid compounds, whereas dihydroflavonol-4-reductase (DFR), anthocyanidin synthase (ANS), and UDP-glucose flavonoid 3-O-glucosyltransferase (UFGT) regulate the biosynthesis of anthocyanins. Subsequently, water-soluble anthocyanins are transported into the vacuole for stable storage [8].
Transcriptional regulation is important for regulating the expression profiles of anthocyanin biosynthesis genes in several plant species, and it is primarily regulated by three types of transcription factors (TFs): the R2R3-MYB gene family, bHLH TFs, and WD40 repeat proteins [11]. EBGs are regulated by R2R3-MYB genes, whereas LBGs are activated by a transcriptional activation complex consisting of MYB, bHLH, and WD40 proteins (MBW) [10, 12]. In Arabidopsis thaliana, PAP1 is a key R2R3-MYB TF that regulates anthocyanins, and its overexpression can significantly induce anthocyanin accumulation, resulting in dark purple leaves [7]. Other R2R3-MYB TFs (PAP2, MYB113, and MYB114), bHLH gene family members (TT8, GL3, and EGL3) and the WD40 gene (TTG1) have been identified as the key TFs involved in anthocyanin biosynthesis [13, 14]. Important TFs that regulate anthocyanin structural genes have also been found in some ornamental plant species, including in Gerbera hybrida (GMYB10 and GMYC1 [15];), Chrysanthemum morifolium (CmMYB6 and CmbHLH [16, 17];), ornamental kale (BoTTG1 and BoTT8 [14, 18];), Lilium spp. (LhMYB6, LhMYB12, and LhbHLH1 [19, 20];), and Anthurium andraeanum (AaMYB2 and AabHLH1) [21, 22].
The name Aglaonema is used for collectively referring to a group of perennial flowering plants belonging to the family Araceae. These plants are native to tropical Asian countries, including India, Thailand, Vietnam, the Philippines, Malaysia, and Indonesia. Aglaonema commutatum is a plant species widely used for interior decoration and as a courtyard ornamental plant because of its easy cultivation and management. Its leaves can have a variety of colors and beautiful stripes, making it increasingly popular in the market as an excellent foliage and fresh-cut ornamental plant [23]. At present, large-scale production of A. commutatum seedlings is mainly performed through tissue culture. However, during the tissue culture process, cases of reduced proportions of red foliage have been observed; in some cases, the foliage changed from red to green. This phenomenon has considerable implications for the ornamental and market value of this species. In recent years, studies on A. commutatum have mainly focused on tissue culture [24,25,26,27], stress resistance, breeding [27], and interspecies genetic relationships at the phenotypic and biochemical level. On the other hand, relatively fewer studies have examined the mechanisms underlying leaf color variations in A. commutatum. Moreover, little is known about the mechanisms regulating anthocyanin biosynthesis in A. commutatum.
Here, the effect of developmental stages on the leaf color of two varieties of A. commutatum (Red Valentine and a green mutant) was examined at the biochemical and genomic levels. The total anthocyanin and chlorophyll contents and anthocyanin monomer compositions of leaves of the two varieties were examined at three leaf developmental stages (S1–S3). Additionally, the TFs and structural genes regulating anthocyanin biosynthesis were identified using RNA-seq technology.
Results
Leaf color changes during different leaf development stages
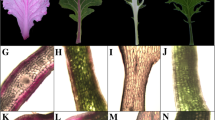
Except for the leaf color variation between the two A. commutatum varieties, no significant differences in the leaf shape and plant size were observed between the two varieties (Fig. 1A). Some changes in leaf appearance were observed during S1 to S2, namely leaf uncurling and color change from white to light pink and green. At maturity, the leaf color changed to deep red and dark green, and the leaf surface was shiny, indicating waxy coating. In the green mutant, red patches were concentrated in the middle of the leaves (Fig. 1B). To determine the distribution of pigments inside the leaf tissues, anatomical observations of leaf lamina of the two varieties at the three developmental stages were performed. Deep red pigmentation was observed only in the palisade tissues, and chlorophylls were observed in both the sponge and palisade tissues (Fig. 1C).
Morphological characteristics of Aglaonema commutatum leaves. A Pigmentation and phenotypes of A. commutatum Red Valentine and the green mutant. B Leaf phenotypes: stage 1, white curly leaves with small amount of pigment (7 d); stage 2, uncurled leaves exhibiting light pink coloration (28 d); stage 3: mature leaves exhibiting dark red coloration with oily surface (35 d). R1, R2, and R3: stages 1, 2, and 3 of A. commutatum Red Valentine, respectively; G1, G2, and G3: stages 1, 2 and 3 of the green mutant, respectively. The scale bars are 1 cm. C Anatomical observation of pigments in the lamina of A. commutatum Red Valentine and a green mutant. The scale bars are 100 μm
Leaf chlorophyll and anthocyanin contents
To check whether the color change was governed by the change in total anthocyanin and chlorophyll contents, the contents of the two pigments in the leaves at the three developmental stages were measured. A significant increase in both leaf anthocyanin and chlorophyll contents was observed in both varieties of A. commutatum from S1 to S2. However, the anthocyanin content of Red Valentine leaves was significantly higher than that of green mutant leaves in all three development stages. The leaf anthocyanin content of Red Valentine was approximately 5.58-fold higher than that of green mutant leaves at S3 (Fig. 2A). In contrast, the chlorophyll content of green mutant leaves was significantly higher than that of Red Valentine leaves in the three developmental stages. The chlorophyll content in green mutant leaves was approximately 5.57-fold higher than that in Red Valentine leaves at S3 (Fig. 2B). Furthermore, the leaf anthocyanin/chlorophyll ratio of the mutant at the three developmental stages was not significantly affected by the developmental stages, with the highest value (0.01) observed at S3. However, in Red Valentine, a significant increase in leaf anthocyanin/chlorophyll ratio was observed from S2 (0.07) to S3 (0.39), indicating that the mutants did not accumulate anthocyanins as much as the Red Valentine did (Fig. 2C).
Furthermore, HPLC was used to determine the composition of anthocyanin monomers in the two varieties at S3. Three anthocyanin monomers, including cyanidin, paeonidin, and pelargonin, were identified in the leaves. Among them, cyanidin was the most abundant, accounting for 98.1 and 95.8% pigments in the leaves of Red Valentine and the mutant, respectively, indicating that it was responsible for the red color of the leaves. Trace amounts of paeonidin and pelargonin were also detected (Fig. 2D). Our data implied that the composition and proportion of anthocyanin monomers contributed to the color changes in mature leaves, as no significant difference in the ratio of the two monomers was observed between the two varieties.
Library construction and de novo assembly
A total of 18 high quality RNA samples were extracted from the leaves of the two A. commutatum varieties at the three developmental stages for further transcriptome analysis using Illumina sequencing with the HiSeq 6000 sequencer. The raw sequencing data were checked for quality and subjected to data filtering. The number of clean reads for each library ranged from 22,791,579 to 28,919,060, with a mapped ratio between 66.71 and 76.25%. The GC content ranged from 50.54 to 54.95%, and the Q30 base percentage was ≥94.71%. A total of 63,621 unigenes with an N50 length of 1340 bp were generated (Table 1).
Functional annotation and classification
A total of 63,621 assembled unigenes were BLAST-searched against the NCBI non-redundant protein (NR), Nt, SwissProt, Kyoto Encyclopedia of Genes and Genomes (KEGG), KOG, Pfam, and Gene Ontology (GO) databases using BLASTx, and 34,554 unigenes were annotated (Fig. 3A). The unigenes were annotated and aligned in the NR database. Quercus suber had the highest matching degree (10.62%), followed by Elaeis guineensis (8.40%) and Phoenix dactylifera (7.57%). Approximately 42.29% of the unigene sequence did not match the protein sequences of the other species (Fig. 3B). Results of the GO analysis showed that the genes were mainly involved in metabolic processes, including cell part and compound binding, which were the major metabolic processes. Results of the KEGG analysis showed that the unigenes were involved in translation, carbohydrate metabolism, and folding, sorting, and degradation pathways (Supplementary Fig. S2).
Functional annotation and classification. A Gene ontology (GO) classification of unigenes. B Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway classification. C Numbers of functional annotation of all unigenes in six databases. D Species distribution of unigenes matching the main species obtained using BLASTx in the NR database
Identification and KEGG classification of differentially expressed genes (DEGs)
The transcript per million (TPM) values of the unigenes were used to identify DEGs between the leaves of Red Valentine and the green mutant at the three developmental stages using the DEGseq R package. A total of 14,186 DEGs (p-adjust ≤0.05; FC ≥ 2) were identified by pairwise comparison (G1 vs. G2, G2 vs. G3, R1 vs. R2, and R2 vs. R3). A Venn diagram of the DEGs showed that 843 DEGs were common to the four comparison groups (Fig. 4A). Among the four comparisons, the number of DEGs in S2 vs. S3 was higher than that in S1 vs. S2. The changes in gene expression are shown in Fig. 4B. Additionally, the KEGG pathway enrichment analysis showed that the DEGs were significantly enriched in glyoxylate and dicarboxylate metabolism, photosynthesis, phenylpropanoid biosynthesis, fatty acid elongation, flavonoid biosynthesis, and amino sugar and nucleotide sugar metabolism (Fig. 4C).
Expression patterns of genes involved in anthocyanin biosynthesis
The expression pattern of the unigenes was determined using the TPM values. The expression profiles of the genes encoding enzymes related to anthocyanin biosynthesis in Red Valentine leaves showed that the phenylpropanoid pathway was regulated by three groups of genes, including five PAL unigenes, two C4H, and 4CL unigenes. Two CHS, one CHI, and one F3H unigenes were expressed in the early biosynthesis stage. Accordingly, three DFR, one ANS, and five UFGTs unigenes were expressed during the later biosynthesis stage. Consistent with the high anthocyanin content of Red Valentine leaves, the TPM values of the anthocyanin structural unigenes were higher in the Red Valentine leaves than in the leaves of the green mutant at all three developmental stages. From stage R1 to stage R2, a significant increase (10-fold) in the expression profiles of most anthocyanin related genes (22 out of 26) was observed. Although the fold change was slightly lower than that observed from stage R1 to R2, the gene expression profiles also exhibited an upward trend from stage R2 to stage R3. Contrarily, a significant decrease in the gene expression profiles was observed in the leaves of the green mutant, with extremely low TPM values at all three developmental stages (Fig. 5). Full-length cDNA of the genes was obtained from RNA-seq data, and rapid amplification of cDNA ends (RACE), PCR, and phylogenetic tree construction were performed to ensure the accuracy of the analysis.
Identification of TFs related to anthocyanin biosynthesis
Anthocyanin biosynthesis genes are mainly regulated by three different families of TFs, namely MYB, bHLH, and WDR. Thus, the TFs regulating anthocyanin biosynthesis in the leaves of A. commutatum were examined. A total of 1068 TFs belonging to 32 TF families were identified by aligning to the PlantTFDB database using BLASTX (Fig. 6). The top 10 annotated TFs included the MYB_superfamily, AP2/ERF, C2C2, bZIP, bHLH, C3H, B3_superfamily, NAC, WRKY, and LBD (AS2/LOB), each of which comprised over 48 unigenes in both varieties of A. commutatum. To identify MYB and WDR TFs related to anthocyanin biosynthesis, a functional search in the RNA-seq annotation file was performed, and the following unigene information was obtained. TRINITY_DN91028_c1_g1 (Unigene ID) was annotated to be a putative flavonoid or anthocyanin regulator (A. andraeanum). The gene AaMYB1 in combination with bHLH genes has been reported to be associated with anthocyanin biosynthesis through biolistic transformation in a Cymbidium orchid [28]. Furthermore, TRINITY_DN62889_c2_g2 was annotated to have the highest homology with AaMYB2 (A. andraeanum), which was proven to primarily contribute to the regulation of anthocyanin biosynthesis in the spathes and leaves of A. andraeanum [22]. The results of qRT-PCR confirmed the profile of these two unigenes in relation to anthocyanin biosynthesis in A. commutatum. Moreover, TRINITY_DN84708_c1_g1 had a high similarity with TTG1, which is involved in anthocyanin biosynthesis regulation in many plant species [29, 30].
Regarding the bHLH TFs, we were unable to identify the bHLH unigenes involved in anthocyanin biosynthesis in the two varieties of A. commutatum based on the RNA-seq annotation data. Therefore, based on the expression profiles of the anthocyanin biosynthesis genes, we searched based on the expression profiles of anthocyanin biosynthesis genes. Red Valentine 26 putative anthocyanin biosynthesis genes (Date A) (Supplementary Table S1) and all bHLH TFs (Date B) were first normalized to reduce the error caused by exact TPM means. A correlation analysis was performed between the mean values of Date A (which could represent the anthocyanin biosynthesis gene expression level as a whole) and Date B to determine the relationship between each bHLH TF and the expression profiles of the anthocyanin biosynthesis genes, which were ranked in descending order (Table 2). The top 10 bHLH unigenes were aligned against the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using BLASTX to obtain detailed information of the unigenes. We found that TRINITY_DN107490_c2_g1, TRINITY_DN8 7484_c1_g1, and TRINITY_DN97087_c0_g10 had the highest correlation coefficients with the basic helix-loop-helix TF 1 (A. andraeanum) annotation. AabHLH1 was reported to participate in the regulation of the flavonoid metabolic pathway [21], suggesting that it may also be involved in the regulation of anthocyanin biosynthesis in Red Valentine. This gene was cloned using RACE PCR, and we found that the three unigenes belong to one gene named AcbHLH1. qRT-PCR revealed the gene expression patterns of AcbHLH1, which were consistent with the anthocyanin structural genes in the leaves of the two A. commutatum varieties. The CDS sequence of these TFs was cloned by sequence information and RACE PCR (Supplementary Table S2) for the phylogenetic tree analysis (Supplementary Fig. S3) and further studies, and they were named AcMYB1, AcMYB2, AcTTG1 and AcbHLH1.
qRT-PCR analysis of anthocyanin-related DEGs
To validate the RNA-seq data, qRT-PCR assays were performed using gene-specific primers for the DEGs and TFs involved in anthocyanin biosynthesis. In total, nine unigenes of anthocyanin biosynthesis and identified TFs in the two varieties of A. commutatum were selected for qRT-PCR. The results of the qRT-PCR analysis generally showed expression patterns similar as the TPM values, indicating that the expression data obtained by RNA-seq were reliable (Fig. 7; Supplementary Table S3). Melting curves of the selected gene are shown in Supplementary Fig. S4.
Comparative expression of candidate genes in the Red Valentine variety. Real-time qPCR of PAL1 (TRINITY_DN98590_c0_g3); PAL3 (TRINITY_DN89524_c3_g6); C4H (TRINITY_DN95047_c0_g1); 4CL (TRINITY_DN95116_c0_g1); CHS1 (TRINITY_DN78614_c3_g1); UFGT2 (TRINITY_DN90166_c0_g1); F3’H1 (TRINIY_DN103984_c0_g1); DFR1 (TRINITY_DN75107_c2_g1); DFR3 (TRINITY_DN99635_c0_g4); AcbHLH; AcMYB1; AcMYB2 in A. commutatum Red Valentine leaves. The columns represent means ± standard deviations from three independent biological replicates
Discussion
Transcriptome analysis of A. commutatum red valentine at the different leaf developmental stages
Even though several studies have been conducted on Red Valentine, they were mostly focused on phenotypic and biochemical characteristics [23,24,25,26,27], whereas none have investigated the molecular mechanisms behind the red coloration of Red Valentine foliage. The red colored leaves of Red Valentine are of considerable ornamental value and this trait is of interest to plant breeders. In the present study, we performed RNA-seq analysis of the leaves of A. commutatum Red Valentine and a green mutant to reveal their gene expression changes at three developmental stages and to identify the key genes involved in their leaf color variations. A total of 196.15 Gb clean data were obtained from 18 RNA-seq libraries. A total of 63,621 unigenes with an average length of 934 bp and an N50 length of 1340 bp were assembled. This number was similar to that in other Araceae species, such as Amorphophallus konjac [31], Pinellia ternata [32], and Caladium hortulanum [33]. The percentage of filtered Q30 and the GC content were higher than 94.71 and 50.54%, respectively. Xiong et al. [34] performed a transcriptome analysis of variegated citrus plants and reported a Q30 score higher than 94.19%. In another study on the transcriptome of calla lily spathes, the GC content was between 50.5 and 51.05% [35]. Thus, the quality of the RNA-seq data in the present study was good for further analyses.
Variation in leaf anthocyanin and chlorophyll contents
Leaf color changes in ornamental plants are associated with the biosynthesis of pigments. Anthocyanin is a major pigment in plants and confers red, blue, and purple colors to the leaves, with chlorophyll being responsible for green leaf coloration [1, 36, 37]. In the Red Valentine variety of A. commutatum, we found that anthocyanins accumulated mainly in the palisade tissues, whereas chlorophylls were present in both the sponge and palisade tissues. At S3, the anthocyanin content of Red Valentine leaves was approximately 5.58-fold higher than that of the green mutant leaves. Contrarily, the chlorophyll content of the green mutant leaves was approximately 5.57-fold higher than that of Red Valentine leaves, which was in accordance with the leaf color phenotype of the two varieties. A previous HPLC analysis showed that cyanidin was the major anthocyanin component in the ever-red leaves of Malus cv. ‘Royalty’ [38]. Through metabolome analyses, cyanidin was proved to be the main contributor of the purple leaf phenotype in Brassica napus [39]. In the present study, cyanidin was found to be the major anthocyanin in the leaves of A. commutatum; however, the composition and proportion of anthocyanin monomers did not significantly affect the leaf color of A. commutatum. The results of our examination of the phenotypic and biochemical basis of changes in leaf color through the three developmental stages are vital for further molecular studies.
Genes involved in anthocyanin and chlorophyll biosynthesis and degradation
RNA-seq technology is a powerful tool for revealing the relationship between different biological processes and gene expression, as well as for identifying key genes under various conditions [40]. Transcriptome analysis has been used to explore the mechanism underlying leaf color variation in many plant species, such as ornamental kale [41], Paeonia qiui [42], sweet basil [43], and Acer truncatum [44]. In the present study, we identified 283,130 unigenes in the leaves of A. commutatum. After filtering out low-expression unigenes, we identified 26 differentially expressed putative anthocyanin structural unigenes. Full-length cDNA of the genes was obtained, and phylogenetic trees were constructed to get more functional information for future studies. In accordance with the phenotype and anthocyanin content, the TPM values of most anthocyanin genes were significantly higher in the leaves of Red Valentine than in the leaves of the green mutant at the three developmental stages. Particularly, the TPM values of CHS (TRINITY_DN78614_c3_g2), CHI (TRINITY_ DN98854_c4_g1), DFR (TRINITY_DN75107_c2_g1, TRINITY_DN99635_c0_g4), and UFGT (TRINITY_ DN91923_c1_g1, TRINITY_DN90166_c0_g1) in Red Valentine leaves were more than 5-fold higher than those in the leaves of the green mutant at developmental stage S3.
Furthermore, we identified 31 unigenes involved in chlorophyll biosynthesis and 15 unigenes involved in chlorophyll degradation in the leaves of A. commutatum. The TPM values of these genes (Supplementary Table S4) were used to elucidate the gene expression pattern during the three developmental stages. Among the unigenes, the expression of 10 chlorophyll biosynthesis unigenes in the leaves of Red Valentine was 2-fold higher than that in the leaves of the mutant at the same developmental stage, indicating that these genes play vital roles in chlorophyll biosynthesis and accumulation. However, no significant differences in the expression profiles of the other chlorophyll biosynthesis and degradation unigenes were observed between the two varieties. Further research should be conducted to elucidate how chlorophyll biosynthesis and degradation genes influence these leaf color changes.
TFs related to anthocyanin biosynthesis
Previous studies have revealed that anthocyanin biosynthesis is transcriptionally modulated by TFs, including R2R3-MYB, bHLH, and WD40. Among them, MYB TFs alone or in combination with MBW complexes (MYB, bHLH, and WD40) can control anthocyanin synthesis and accumulation [10, 45]. In the present study, two MYB TFs, namely AcMYB1 and AcMYB2, were identified as candidate anthocyanin biosynthesis regulators. These TFs are homologs of AaMYB1 and AaMYB2 in A. andraeanum, which were reported to be involved in anthocyanin regulation. Our phylogenetic analysis showed that the two TFs belonged to the same anthocyanin subgroup. Overexpression of AcMYB2 resulted in the red leaf phenotype of transgenic plants, indicating its anthocyanin regulatory activity in A. commutatum Red Valentine (Supplementary Fig. S5). Transposon insertion and methylation of the promoter on R2R3-MYB TFs regulating anthocyanin biosynthesis have been reported in Phalaenopsis orchids [46], apple [47, 48], and pear [49]; they could directly control MYB TF expression, consequently affecting anthocyanin accumulation. The candidate unigenes exhibited significantly higher expression profiles in the Red Valentine variety than in the green mutant. Therefore, we speculated that transposon insertion or methylation of the AcMYB2 promoter may occur during leaf development, consequently decreasing or stopping the expression of AcMYB2, resulting in green mutants. Further studies should investigate the role of these key TFs in the regulation of leaf color.
Additionally, several other TFs have been reported to participate in anthocyanin biosynthesis. In Arabidopsis and B. napus, BnWRKY41–1 was found to have a similar role as that of AtWRKY41 in anthocyanin biosynthesis regulation [50]. In pear, ERFs can interact with PpMYB114 to regulate the light-dependent anthocyanin pathway in fruits [51]. NAC TFs were found to be potential regulators of anthocyanin biosynthesis in apple [52] and litchi [53] during fruit ripening. In the present study, a total of 1068 TFs belonging to 32 TF families were identified by aligning them to the PlantTFDB database using BLASTX. Among them, TRINITY_DN60809_c4_g1 (WRKYs), TRINITY_DN106460_c0_g1 (WRKYs), TRINITY_DN62456_c3_g4 (ERFs), and TRINITY_DN101050_c1_g1 (NACs) were screened as the candidate TFs regulating the anthocyanin pathway because their expression pattern was similar to that of anthocyanin-related genes.
Effects of plant hormones on leaf color variation
Plant hormones play critical roles in stimulating anthocyanin accumulation. For example, abscisic acid (ABA) and jasmonates (JAs) can induce anthocyanin accumulation by regulating anthocyanin-related genes. In Litchi chinensis, LcABF1/2/3 were reported to be important ABA-dependent regulators in both chlorophyll degradation and anthocyanin biosynthesis [54]. After silencing a key gene in the ABA biosynthesis pathway of sweet cherry, the fruits showed a colorless phenotype [55]. Besides, the molecular mechanism underlying JA-regulated anthocyanin accumulation was first reported in Arabidopsis [56]. Jasmonate-ZIM-domain proteins disrupted the formation of WD-repeat/bHLH/MYB complexes to repress JA-regulated anthocyanin accumulation [57]. In the present study, a total of 354 unigenes were annotated to plant hormone signal transduction in the KEGG databases. Among them, more than 43 unigenes were identified as DEGs by pairwise comparison. The abundant gene expression and sequence data provided in the present study lays the foundation for exploring the molecular processes underlying the role of plant hormones in the leaf variation of A. commutatum.
Conclusions
The present study demonstrated that high chlorophyll and low anthocyanin contents were responsible for the green leaf coloration of the A. commutatum green mutant. Furthermore, 26 anthocyanin biosynthesis structural genes and four key regulatory TFs, namely AcMYB1, AcMYB2, AsTTG1, and AcbHLH1, were identified. Downregulation of these TFs may downregulate the expression of anthocyanin biosynthesis genes. Further molecular studies are needed to elucidate the role of each TF and structural gene in anthocyanin biosynthesis in Red Valentine. The findings of the present study provide insights into anthocyanin candidate genes, which can facilitate the breeding and development of ornamental cultivars and contribute to the understanding of leaf color variation in A. commutatum.
Methods
Plant materials
In the present study, 1-year-old tissue culture seedlings of A. commutatum ‘Red Valentine’ and a green mutant were selected as experimental materials (Fig. 1). The plants were purchased from the Guangzhou Flowers Research Centre (Guangzhou, China; NCBI:txid1210880) and grown in a greenhouse covered with a 60% shade cloth at the South China Botanical Garden, Chinese Academy of Sciences (Guangzhou, China). The use of this plant material was licensed and approved by the Guangzhou Flowers Research Centre. The temperature and relative humidity of the greenhouse ranged from 15 to 34 °C and 75 to 99%, respectively. Different leaf developmental stages were defined as follows: stage 1 (S1), white curly leaves with small amount of pigment (7 d); stage 2 (S2), uncurled leaves exhibiting light pink coloration (28 d); stage 3 (S3): mature leaves with characteristic dark coloration and oily surface (35 d). All samples were stored at − 80 °C until further analysis in May 2019.
Anthocyanin and chlorophyll extraction and observation
Total anthocyanin content of A. commutatum was determined using the pH difference method [58] with modifications. Briefly, methanolic extracts of fresh leaves were prepared. To improve the extraction efficiency, fresh leaves (1 g) were subjected to ultrasonic agitation for 0.5 h. Subsequently, the sample was incubated with 15 mL of methanol (containing 0.05% HCl) in darkness for 12 h at 0 °C. Absorbance at 510 nm and 700 nm was measured in triplicate using a microplate reader (Tecan Infinity, Switzerland). For quantifying anthocyanin monomers, total anthocyanins extracted from leaf samples were hydrolyzed using 3 mol/L muriatic acid for 1 h at 97 °C in darkness, after which the solution was dried using a vacuum thickener (Concentrator plus; Eppendorf) to obtain anthocyanin monomer powder. The powder was re-dissolved in acidic methanol (0.05% HCl) for HPLC analysis, with cyanidin chloride, peonidin chloride, delphinidin chloride, pelargonidin chloride, petunidin chloride, and malvidin chloride (Phyproof, Germany) as internal standards. The anthocyanin content was determined by linear regression and expressed in mg/g of fresh leaf weight. Total leaf chlorophyll content was determined following the procedures described in Lichtenthaler [59]. Briefly, approximately 1 g of leaf sample was extracted with 1 ml of methanol, and the absorbance was measured at 652 nm, 665 nm, and 470 nm using a microplate reader (Tecan Infinity).
Freehand sectioning of the leaves was performed for anatomical observation of the pigments and their localization in the leaf tissues of the two varieties at the three developmental stages using a digital microscope (DVM6; Leica, USA).
RNA extraction, cDNA library construction, and sequencing
Total RNA was isolated from the leaves using an RNA kit (Polysaccharides & Polyphenolics-rich) (Hua Yueyang, Beijing, China) with RNase-free DNase I (TaKara) to eliminate genomic DNA contamination. Total RNA integrity and purity were determined using the 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) and quantified using the ND-2000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Wilmington, DE, USA). Only high-quality RNA samples were used for constructing the sequencing library. RNA purification, reverse transcription, library construction, and sequencing were performed by Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) (Supplementary Fig. S1). Independent triplicate whole-leaf materials from Red Valentine (R) and a green mutant (G) at developmental stages S1, S2, and S3 were used to construct 18 cDNA libraries, which were named as R1_1, R1_2, R1_3, R2_1, R2_2, R2_3, R3_1, R3_2, R3_3, G1_1, G1_2, G1_3, G2_1, G2_2, G2_3, G3_1, G3_2, and G3_3. Libraries were prepared using an Illumina TruSeqTM RNA sample preparation kit (Illumina, San Diego, CA, USA) and sequenced on a flow cell using the Illumina HiSeq™ 6000 sequencing platform (Illumina).
de novo assembly and annotation
High quality reads from the 18 libraries were assembled de novo using Trinity (http://trinityrnaseq.sourceforge.net/). To annotate the unigenes, the transcripts alignment was subjected against public databases, including the NCBI protein NR (http://www.ncbi.nlm.nih.gov), Clusters of Orthologous Groups (http://www.ncbi.nlm.nih.gov/COG), and KEGG databases (http://www.genome.jp/kegg), using BLASTx. To determine the processes the unigenes are involved in, GO function annotation was performed using the BLAST2GO software (http://www.blast2go.com/b2ghome). Metabolic pathway analysis was performed using the KEGG database [60].
Identification of differentially expressed genes (DEGs) and functional enrichment analysis
To identify the DEGs between the two varieties, the expression level of each transcript was calculated according to the TPM method. RSEM (http://deweylab.biostat.wisc.edu/rsem/) [61] was used to quantify the gene abundance. Differential expression analysis was performed using DESeq2 [62], with a Q value ≤0.05. Unigenes with fold change > 2 or < − 2 and a Q value ≤0.05 were considered significantly differentially expressed. The DEGs were used for GO function enrichment analysis and KEGG pathway analysis using Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) [63].
Quantitative RT-PCR analysis
Total RNA (the same sample used for RNA-seq) was reverse-transcribed to first-strand cDNA for qRT-PCR using a TransScript® One-Step gDNA Removal cDNA Synthesis SuperMix (Transgen, Beijing, China). Diluted cDNA was amplified with specific primers (listed in Additional file) and reacted with PerfectStart Green qPCR SuperMix (Transgen) on a LightCycler 480 II (Roche) under the following conditions: 45 cycles at 94 °C for 5 s, 57 °C for 15 s, and 72 °C for 10 s. The gene expression levels were normalized using A. commutatum Red Valentine actin gene as the internal control and calculated using the 2-ΔΔCT method. Three biological replicates were used for the analysis.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and the supplementary information files. The sequence data was deposited in the NCBI database under the BioProject PRJNA793608.
Abbreviations
- CDS:
-
Coding sequence
- FC:
-
Fold change
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- RACE:
-
Rapid-amplification of cDNA ends
- TPM:
-
Transcript per million reads
References
Tanaka Y, Ohmiya A. Seeing is believing: engineering anthocyanin and carotenoid biosynthetic pathways. Curr Opin Biotech. 2008;19(2):190–7.
Huang XW, Zou XB, Zhao JW, Shi JY, Zhang XL, Holmes M. Measurement of total anthocyanins content in flowering tea using near infrared spectroscopy combined with ant colony optimization models. Food Chem. 2014;164:536–43.
Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54(4):733–49.
Ha US, Bae WJ, Kim SJ, Yoon BI, Hong SH, Lee JY, et al. Anthocyanin induces apoptosis of DU-145 cells in vitro and inhibits xenograft growth of prostate Cancer. Yonsei Med J. 2015;56(1):16–23.
Hou DX. Potential mechanisms of cancer chemoprevention by anthocyanins. Curr Mol Med. 2003;3(2):149–59.
Martin C, Butelli E, Petroni K, Tonelli C. How can research on plants contribute to promoting human health? Plant Cell. 2011;23(5):1685–99.
Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12(12):2383–93.
Zhao J, Dixon RA. The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 2010;15(2):72–80.
Zhao L, Gao LP, Wang HX, Chen XT, Wang YS, Yang H, et al. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct Integr Genomic. 2013;13(1):75–98.
Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181(3):219–29.
Ramsay NA, Glover BJ. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10(2):63–70.
Xu WJ, Grain D, Le Gourrierec J, Harscoet E, Berger A, Jauvion V, et al. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 2013;198(1):59–70.
Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156(3):1349–62.
Zhang B, Hu ZL, Zhang YJ, Li YL, Zhou S. Chen GP: a putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (brassica Oleracea var. acephala f. tricolor). Plant Cell Rep. 2012;31(2):281–9.
Elomaa P, Uimari A, Mehto M, Albert VA, Laitinen RAE, Teeri TH. Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol. 2003;133(4):1831–42.
Liu XF, Xiang LL, Yin XR, Grierson D, Li F, Chen KS. The identification of a MYB transcription factor controlling anthocyanin biosynthesis regulation in Chrysanthemum flowers. Sci Hortic-Amsterdam. 2015;194:278–85.
Xiang LL, Liu XF, Li X, Yin XR, Grierson D, Li F, et al. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS One. 2015;10(11).
Guo N, Han S, Zong M, Wang GX, Zheng SN, Liu F. Identification and differential expression analysis of anthocyanin biosynthetic genes in leaf color variants of ornamental kale. BMC Genomics. 2019;20(1):564.
Nakatsuka A, Yamagishi M, Nakano M, Tasaki K, Kobayashi N. Light-induced expression of basic helix-loop-helix genes involved in anthocyanin biosynthesis in flowers and leaves of Asiatic hybrid lily. Sci Hortic-Amsterdam. 2009;121(1):84–91.
Yamagishi M, Shimoyamada Y, Nakatsuka T, Masuda K. Two R2R3-MYB genes, homologs of Petunia AN2, regulate anthocyanin biosyntheses in flower Tepals, Tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol. 2010;51(3):463–74.
Li CH, Qiu J, Huang SR, Yin JM, Yang GS. AaMYB3 interacts with AabHLH1 to regulate proanthocyanidin accumulation in Anthurium andraeanum (Hort.)-another strategy to modulate pigmentation. Hortic Res-Engl. 2019;6:14.
Li CH, Qiu J, Yang GS, Huang SR, Yin JM. Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum (Hort.). Plant Cell Rep. 2016;35(10):2151–65.
Bao-Gui DU, Liang CH, Chen YX, Lan TWJ. Study on the factors influencing the bud induction of Aglaonemum cultured in vitro; 2013.
Chen J, Henley RW, Henny RJ, Caldwell RD, Robinson CA. A Simple Leaf-assay Method for Evaluating Aglaonema Sensitivity to Chilling Temperatures; 1998.
Zhang JX, Wu KL, Li Q, Zeng SJ, Duan JJPPC. In vitro Rapid Propagation of Aglaonema costatum N. E. Br; 2009.
Zhou Z, Zhang G, Liu C, Li Z, University NJG, Biology A: In vitro culture of Aglaonema commutatum Schott ‘Red Valentine’. 2018.
Budiarto KJ. Conventional propagation of several aglaonema accessions using split single-bud stem cutting; 2011.
Albert NW, Arathoon S, Collette VE, Schwinn KE, Jameson PE, Lewis DH, et al. Activation of anthocyanin synthesis in Cymbidium orchids: variability between known regulators. Plant Cell Tiss Org. 2010;100(3):355–60.
deVetten N, Quattrocchio F, Mol J, Koes R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997;11(11):1422–34.
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11(7):1337–49.
Gao H, Zhao Y, Huang L, Huang Y, Chen J, Zhou H, et al. Comparative analysis of buds transcriptome and identification of two florigen gene AkFTs in Amorphophallus konjac. Sci Rep. 2022;12(1):6782.
Ma G, Zhang M, Xu J, Zhou W, Cao L. Transcriptomic analysis of short-term heat stress response in Pinellia ternata provided novel insights into the improved thermotolerance by spermidine and melatonin. Ecotoxicol Environ Saf. 2020;202:110877.
Cao Z, Deng Z. De Novo Assembly, Annotation, and characterization of root transcriptomes of three Caladium cultivars with a focus on necrotrophic pathogen resistance/defense-related genes. Int J Mol Sci. 2017;18(4).
Xiong B, Qiu X, Huang S, Wang X, Zhang X, Dong T, et al. Physiological and transcriptome analyses of photosynthesis and chlorophyll metabolism in variegated Citrus (Shiranuhi and Huangguogan) seedlings. Sci Rep. 2019;9(1):15670.
Fang Y, Lei T, Wu YM, Jin XH. Mechanism underlying color variation in Calla lily spathes based on transcriptomic analysis. J Am Soc Hortic Sci. 2021;146(6):387-+.
Iwashina T. Contribution to flower colors of flavonoids including anthocyanins: a review. Nat Prod Commun. 2015;10(3):529–44.
Lightbourn GJ, Griesbach RJ, Novotny JA, Clevidence BA, Rao DD, Stommel JR. Effects of anthocyanin and carotenoid combinations on foliage and immature fruit color of Capsicum annuum L. J Hered. 2008;99(2):105–11.
Tian J, Peng Z, Zhang J, Song T, Wan H, Zhang M, et al. McMYB10 regulates coloration via activating McF3’H and later structural genes in ever-red leaf crabapple. Plant Biotechnol J. 2015;13(7):948–61.
Li H, Du Y, Zhang J, Feng H, Liu J, Yang G, et al. Unraveling the mechanism of purple leaf formation in Brassica napus by integrated metabolome and transcriptome analyses. Front Plant Sci. 2022;13:945553.
Vilperte V, Lucaciu CR, Halbwirth H, Boehm R, Rattei T, Debener T. Hybrid de novo transcriptome assembly of poinsettia (Euphorbia pulcherrima Willd. Ex Klotsch) bracts. BMC Genomics. 2019;20(1).
Ren J, Liu Z, Chen W, Xu H, Feng H. Anthocyanin degrading and chlorophyll accumulation Lead to the formation of bicolor leaf in ornamental kale. Int J Mol Sci. 2019;20(3).
Luo J, Duan J, Huo D, Shi Q, Niu L, Zhang Y. Transcriptomic analysis reveals transcription factors related to leaf anthocyanin biosynthesis in Paeonia qiui. Molecules. 2017;22(12).
Torre S, Tattini M, Brunetti C, Guidi L, Gori A, Marzano C, et al. De novo assembly and comparative transcriptome analyses of red and green morphs of sweet basil grown in full sunlight. PLoS One. 2016;11(8):e0160370.
Si F, Wang X, Du X, Wang J, Tao J, Qiao Q, et al. Transcriptome sequencing and screening of anthocyanin-related genes in the leaves of Acer truncatum Bunge. Biochem Genet. 2022;60(6):1845–64.
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53(5):814–27.
Hsu CC, Su CJ, Jeng MF, Chen WH, Chen HH. A HORT1 retrotransposon insertion in the PeMYB11 promoter causes harlequin/black flowers in Phalaenopsis orchids. Plant Physiol. 2019;180(3):1535–48.
Jiang SH, Wang N, Chen M, Zhang R, Sun QG, Xu HF, et al. Methylation of MdMYB1 locus mediated by RdDM pathway regulates anthocyanin biosynthesis in apple. Plant Biotechnol J. 2020;18(8):1736–48.
Ma CQ, Jing CJ, Chang B, Ya JY, Liang BW, Liu L, et al. The effect of promoter methylation on MdMYBl expression determines the level of anthocyanin accumulation in skins of two non-red apple cultivars. BMC Plant Biol. 2018;18(1):108.
Wang ZG, Meng D, Wang AD, Li TL, Jiang SL, Cong PH, et al. The methylation of the PcMYB10 promoter is associated with green-skinned sport in max red Bartlett pear. Plant Physiol. 2013;162(2):885–96.
Duan S, Wang J, Gao C, Jin C, Li D, Peng D, et al. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018;268:47–53.
Ni J, Bai S, Zhao Y, Qian M, Tao R, Yin L, et al. Ethylene response factors Pp4ERF24 and Pp12ERF96 regulate blue light-induced anthocyanin biosynthesis in ‘Red Zaosu’ pear fruits by interacting with MYB114. Plant Mol Biol. 2019;99(1–2):67–78.
Zhang S, Chen Y, Zhao L, Li C, Yu J, Li T, et al. A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol. 2020;40(3):413–23.
Jiang G, Li Z, Song Y, Zhu H, Lin S, Huang R, et al. LcNAC13 physically interacts with LcR1MYB1 to Coregulate anthocyanin biosynthesis-related genes during Litchi fruit ripening. Biomolecules. 2019;9(4).
Hu B, Lai B, Wang D, Li J, Chen L, Qin Y, et al. Three LcABFs are involved in the regulation of chlorophyll degradation and anthocyanin biosynthesis during fruit ripening in Litchi chinensis. Plant Cell Physiol. 2019;60(2):448–61.
Shen X, Zhao K, Liu L, Zhang K, Yuan H, Liao X, et al. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 2014;55(5):862–80.
Shan X, Zhang Y, Peng W, Wang Z, Xie D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot. 2009;60(13):3849–60.
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, et al. The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23(5):1795–814.
Wrolstad RE, Culbertson JD, Cornwell CJ, Mattick LR. Detection of adulteration in blackberry juice concentrates and wines. J Assoc Off Ana Chem. 1982;65(6):1417–23.
HKJME L. Chlorophylls and carotenoids. Pigments Photosynthetic Membranes. 1987;148.
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. Bmc Bioinformatics. 2011;12:323.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12).
Xie C, Mao XZ, Huang JJ, Ding Y, Wu JM, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–22.
Acknowledgements
We would like to express our gratitude to all the colleagues in our laboratory for their insightful conversations and technical assistance.
Funding
This work was funded by the Guangdong Key Technology Research and Development Program (2018B020202003) and the Guangdong Modern Agricultural Industry Technology System Program (2018LM2176, 2021KJ121).
Author information
Authors and Affiliations
Contributions
LJ, LF, and SZ conceived the research project and designed the study. LJ and LF performed the experiments. LJ, LF, LL, and GM analyzed the data. LJ, LF, and SZ wrote the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The plants (NCBI:txid1210880) were purchased from the Guangzhou Flowers Research Centre (Guangzhou, China), and the use of this plant material was licensed and approved by the Guangzhou Flowers Research Centre. This study did not involve animal or human testing.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Fig. S1.
Functional categories of all unigenes in A. commutatum “Red Valentine” RNA-seq database. A. Gene Ontology (GO) classification of unigenes. All annotated unigenes were divided into three functional GO categories: biological process (BP), cellular component (CC) and molecular function (MF); B. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway classification. Supplementary Fig. S2. Melting curves of the reference genes actin (ACT) and 12 qRT-PCR genes involved in anthocyanin biosynthesis and regulation. PAL: phenylalanine ammonia-lyase; C4H: trans-cinnamate 4-monooxygenase; 4CL: 4-coumarate--CoA ligase 2; CHS: chalcone synthase; UFGT: UDP-flavonoid glucosyl transferase; F3’H: flavanone 3-hydroxylase; DFR: dihydroflavonol 4-reductase.Supplementary Fig. S3. Phylogenetic trees of transcription factors and structural gene related to anthocyanin biosynthesis in A. commutatum “Red Valentine”. The phylogenetic trees were constructed using Neighbor-jonining of MEGA software (Molecular Evolutionary Genetics Analysis version 7.0, Kumar et al., 2016) with 1000 bootstrap replicates. CHI: chalcone isomerase; F3H: flavanone 3-hydroxylase; ANS: anthocyanidin synthase.Supplementary Fig . S4. The phenotype of AsMYB2 transgenic tobacco with 35s promotor. A. Red leaf phenotype of transgenic plants overexpressing AsMYB2 against control plants. B. T-DNA region of the vector used in transgenic plants. LB: LB T-DNA repeat; NOST: NOS terminator; BlpR: phosphinothricin acetyltransferase, NOSP:NOS promoter, 35S: CaMV 35S promoter; mGFP: green fluorescent protein; EcoR I, Xba I: restriction site; RB: RB T-DNA repeat. Supplementary Table S1. Primers used in reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis. Supplementary Table S2. Data used in transcription factor identification. 26 putative anthocyanin biosynthesis unigenes were identified in RNA-seq database, and then normalized the exact PKM value in two varieties at there stages to reduce the error caused by the exact means using the free online platform of Majorbio Cloud Platform (www.majorbio.com), the normalized data were list in the table. Supplementary Table S3. The PKM value of unigenes connected with chlorophyll biosynthesis and degradation. Supplementary Table S4. Protein sequence of transcription factors and structural gene related to anthocyanin biosynthesis.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, J., Wu, K., Li, L. et al. Transcriptomic analysis reveals biosynthesis genes and transcription factors related to leaf anthocyanin biosynthesis in Aglaonema commutatum. BMC Genomics 24, 28 (2023). https://doi.org/10.1186/s12864-022-09107-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-09107-1