Abstract
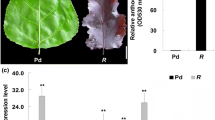
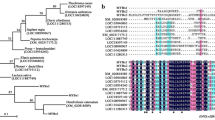
The purple kale (Brassica Oleracea var. acephala f. tricolor) is a mutation in kales, giving the mutant phenotype of brilliant purple color in the interior. Total anthocyanin analysis showed that the amount of anthocyanins in the purple kale was up to 1.73 mg g−1 while no anthocyanin was detected in the white kale. To elucidate the molecular mechanism of the anthocyanin biosynthesis in the purple kale, we analyzed the expression of structural genes and some transcription factors associated with anthocyanin biosynthesis in the purple cultivar “Red Dove” and the white cultivar “White Dove”. The result showed that nearly all the anthocyanin biosynthetic genes showed higher expression levels in the purple cultivar than in the white cultivar, especially for DFR and ANS, they were barely detected in the white cultivar. Interestingly, the fact that a R2R3 MYB transcription factor named BoPAP1 was extremely up-regulated in the purple kale and induced by low temperature attracted our attention. Further sequence analysis showed that BoPAP1 shared high similarity with AtPAP1 and BoMYB1. In addition, the anthocyanin accumulation in the purple kale is strongly induced by the low temperature stress. The total anthocyanin contents in the purple kale under low temperature were about 50-fold higher than the plants grown in the greenhouse. The expression of anthocyanin biosynthetic genes C4H, F3H, DFR, ANS and UFGT were all enhanced under the low temperature. These evidences strongly suggest that BoPAP1 may play an important role in activating the anthocyanin structural genes for the abundant anthocyanin accumulation in the purple kale.




Similar content being viewed by others
References
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48:958–970
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. The Plant Cell Online 12:2383–2394
Burr FA, Burr B, Scheffler BE, Blewitt M, Wienand U, Matz EC (1996) The maize repressor-like gene intensifier1 shares homology with the r1/b1 multigene family of transcription factors and exhibits missplicing. The Plant Cell Online 8:1249–1259
Chiu LW, Zhou X, Burke S, Wu X, Prior RL, Li L (2010) The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol 154:1470–1480
Christie PJ, Alfenito MR, Walbot V (1994) Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194:541–549
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Feng S, Wang Y, Yang S, Xu Y, Chen X (2010) Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232:245–255
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Goodrich J, Carpenter R, Coen ES (1992) A common gene regulates pigmentation pattern in diverse plant species. Cell 68:955–964
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780
Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76:543–553
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinforma 5:150–163
Mano H, Ogasawara F, Sato K, Higo H, Minobe Y (2007) Isolation of a regulatory gene of anthocyanin biosynthesis in tuberous roots of purple-fleshed sweet potato. Plant Physiol 143:1252–1268
Middleton E, Kandaswami C, Harborne J (1993) The flavonoids: advances in research since 1986, vol 21. Chapman and Hall, London, pp 619–652
Mori K, Sugaya S, Gemma H (2005) Decreased anthocyanin biosynthesis in grape berries grown under elevated night temperature condition. Sci Hortic 105:319–330
Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J 49:641–654
Piero ARL, Puglisi I, Rapisarda P, Petrone G (2005) Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J Agric Food Chem 53:9083–9088
Quattrocchio F, Wing JF, Leppen HTC, Mol JNM, Koes RE (1993) Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. The Plant Cell Online 5:1497–1512
Rapisarda P, Fanella F, Maccarone E (2000) Reliability of analytical methods for determining anthocyanins in blood orange juices. J Agric Food Chem 48:2249–2252
Schmidt S, Zietz M, Schreiner M, Rohn S, Kroh LW, Krumbein A (2010) Genotypic and climatic influences on the concentration and composition of flavonoids in kale (Brassica oleracea var. sabellica). Food Chem 119:1293–1299
Schwinn K, Venail J, Shang Y, Mackay S, Alm V, Butelli E, Oyama R, Bailey P, Davies K, Martin C (2006) A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. The Plant Cell Online 18:831–851
Spelt C, Quattrocchio F, Mol JNM, Koes R (2000) Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. The Plant Cell Online 12:1619–1632
Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216–1232
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
Ubi BE, Honda C, Bessho H, Kondo S, Wada M, Kobayashi S, Moriguchi T (2006) Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature. Plant Sci 170:571–578
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Yuan Y, Chiu LW, Li L (2009) Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta 230:1141–1153
Zhang Y, Zheng S, Liu Z, Wang L, Bi Y (2010) Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J Plant Physiol 47:934–945
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 30871709 and 31000911) and Fundamental Research Funds for the Central Universities (No. CDJXS11232244).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Quiros.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, B., Hu, Z., Zhang, Y. et al. A putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (Brassica Oleracea var. acephala f. tricolor). Plant Cell Rep 31, 281–289 (2012). https://doi.org/10.1007/s00299-011-1162-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-011-1162-3