Abstract
Background
African swine fever (ASF) is a lethal hemorrhagic disease affecting domestic pigs resulting in up to 100% mortality rates caused by the ASF virus (ASFV). The locally-adapted pigs in South-western Kenya have been reported to be resilient to disease and harsh climatic conditions and tolerate ASF; however, the mechanisms by which this tolerance is sustained remain largely unknown. We evaluated the gene expression patterns in spleen tissues of these locally-adapted pigs in response to varying infective doses of ASFV to elucidate the virus-host interaction dynamics.
Methods
Locally adapted pigs (n = 14) were experimentally infected with a high dose (1x106HAD50), medium dose (1x104HAD50), and low dose (1x102HAD50) of the highly virulent genotype IX ASFV Ken12/busia.1 (Ken-1033) isolate diluted in PBS and followed through the course of infection for 29 days. The in vivo pig host and ASFV pathogen gene expression in spleen tissues from 10 pigs (including three from each infective group and one uninfected control) were analyzed in a dual-RNASeq fashion. We compared gene expression between three varying doses in the host and pathogen by contrasting experiment groups against the naïve control.
Results
A total of 4954 differentially expressed genes (DEGs) were detected after ASFV Ken12/1 infection, including 3055, 1771, and 128 DEGs in the high, medium, and low doses, respectively. Gene ontology and KEGG pathway analysis showed that the DEGs were enriched for genes involved in the innate immune response, inflammatory response, autophagy, and apoptosis in lethal dose groups. The surviving low dose group suppressed genes in pathways of physiopathological importance. We found a strong association between severe ASF pathogenesis in the high and medium dose groups with upregulation of proinflammatory cytokines and immunomodulation of cytokine expression possibly induced by overproduction of prostaglandin E synthase (4-fold; p < 0.05) or through downregulation of expression of M1-activating receptors, signal transductors, and transcription factors. The host-pathogen interaction resulted in induction of expression of immune-suppressive cytokines (IL-27), inactivation of autophagy and apoptosis through up-regulation of NUPR1 [5.7-fold (high dose) and 5.1-fold (medium dose) [p < 0.05] and IL7R expression. We detected repression of genes involved in MHC class II antigen processing and presentation, such as cathepsins, SLA-DQB1, SLA-DOB, SLA-DMB, SLA-DRA, and SLA-DQA in the medium and high dose groups. Additionally, the host-pathogen interaction activated the CD8+ cytotoxicity and neutrophil machinery by increasing the expression of neutrophils/CD8+ T effector cell-recruiting chemokines (CCL2, CXCL2, CXCL10, CCL23, CCL4, CXCL8, and CXCL13) in the lethal high and medium dose groups. The recovered pigs infected with ASFV at a low dose significantly repressed the expression of CXCL10, averting induction of T lymphocyte apoptosis and FUNDC1 that suppressed neutrophilia.
Conclusions
We provide the first in vivo gene expression profile data from locally-adapted pigs from south-western Kenya following experimental infection with a highly virulent ASFV genotype IX isolate at varying doses that mimic acute and mild disease. Our study showed that the locally-adapted pigs induced the expression of genes associated with tolerance to infection and repression of genes involved in inflammation at varying levels depending upon the ASFV dose administered.
Similar content being viewed by others
Introduction
African swine fever (ASF) is an important notifiable transboundary disease that impedes the pig value chain and threatens global pig productivity and food security [1]. The aetiologic agent is the African swine fever virus (ASFV), a large double-stranded DNA virus. ASFV was first reported in Kenya in 1921 [2] and has continued to spread to Europe and Asia, causing global food security concerns. ASF is a contagious disease transmitted by direct contact between infected pigs and susceptible ones, through contact with infectious secretions and excretions, fomites [3], or tick vectors Ornithodoros spp. [4,5,6]. ASFV persists in tissues [7] and the environment, transmitting over long distances through swill containing pork and pork products and fomites such as contaminated material, vehicles, or visitors to pig premises [3]. ASF causes persistent outbreaks in endemic and non-endemic regions [8]. Currently, no effective vaccine or treatment is available and the control measures entail strict biosecurity measures like slaughtering infected and exposed animals (stamping out), movement and trade restrictions [9,10,11,12,13]. Ongoing efforts on ASF vaccine design focus on targeted gene deletion to attenuate virulent viruses or develop subunit vaccines targeting known protective antigens. However, these face a bottleneck in optimal delivery systems, limited knowledge of protective antigens [14], and a high risk of converting the attenuated virus to a virulent strain [11].
Presently, ASFV is the only member of the family Asfarviridae, genus Asfivirus [15], whose genome is a double-stranded DNA of about 170–195 kb that encodes 150–195 genes that code for viral proteins responsible for normal cellular metabolic activity, DNA replication, repair, and modulation of host immunity and multigene families [16]. Replication of ASFV occurs in (i) the swine macrophages and monocytes, where it causes an overproduction of cytokines leading to induction of apoptosis in pig cells [9] and (ii) soft ticks (Ornithodoros spp.) in which the virus replicates and is transmitted to other susceptible swine hosts following tick bites [17]. In domestic pigs, clinical manifestations vary depending on the virus strain. Virulent ASF virus strains result in 100% mortality. The acute form of ASF develops over seven days, compared with 10–20 days for the sub-acute form of the disease in which the virus is shed up to 70 days from the oropharynx [18]. The varying clinical forms pose a challenge in diagnosis and persist in surviving pigs, which may serve as reservoirs for future outbreaks [19]. The moderate-to-low virulence ASF virus isolates trigger persistent viral infections, resulting in ASF persistence in endemic areas [1, 20]. Since ASFV primarily affects cells of the mononuclear phagocytic system [21], the interaction between ASFV and the host macrophages thus influences the pathognomonic outcome [22]. Animals with acute ASF display fever and a tendency to crowd, loss of appetite, inactivity, apathy and early leukopenia induced by lymphopenia and changes in monocyte numbers [23]. Affected pigs show erythema mainly affecting the skin of the ears, tail, distal extremities, chest, abdomen and perianal area, and vascular lesions and cyanosis of the skin tend to be more apparent in exotic pig breeds. Other symptoms include vomiting, abdominal pain, constipation, and diarrhoea that was initially mucoid but may later become bloody [24].
The fact that ASFV can persist in its natural hosts comprising warthogs, bush pigs, soft ticks and recovered domestic pigs, confirmed that the virus has effective mechanisms to evade detection by the host defense system [6, 25]. The key pathway modulated by the virus to evade host immunity is the macrophage signaling pathway resulting in altered expression of the immunomodulatory genes and the calcineurin-dependent pathways [6, 25, 26]. Studies have shown that some pigs are very susceptible to ASF, while others are asymptomatic carriers of the virus, despite infection with similar virus genotypes [7, 25, 27, 28]. The locally-adapted pigs are favorites among smallholder farmers in many parts of the world [29, 30], including in South-western Kenya, where ASFV persistence in tissues of apparently healthy pigs has been reported [7]. The level of protection has been attributed to antibody-dependent cytotoxicity that offers limited protection only against homologous ASFV challenge, delaying the onset of clinical signs and viremia [31,32,33]. A recent study by Franzoni and colleagues showed that virulent ASFV isolates had evolved mechanisms to upset activated macrophage response, promoting viral survival and dissemination in the host and pathogenesis [34]. To date, numerous viral proteins have been identified as immunogenic [35], but the mechanisms by which they elicit an effective immune response in surviving animals remain poorly described. Understanding how ASFV persists in hosts is needed to design better therapeutic strategies [6, 36]. But gaps still exist in the characterization of the host-pathogen interactions, which may yield new insights into how to induce a protective immune response.
RNA-Seq is now extensively applied in differential expression studies yielding vast amounts of data matched to a reference genome [37,38,39]. In pigs, RNA-Seq studies have compared breeds for development and meat quality using brain, liver, and tonsil tissues [40,41,42,43] and immunological determination in tonsils, blood and lymph nodes [40, 44, 45]. There are minimal transcriptomic studies that assess other pig organs, such as the spleen [46], with none hitherto assessing pig spleen gene expression following ASFV infection. The spleen tissue is a secondary lymphoid organ with various immunologic functions alongside hematopoiesis and clearance of red blood cells [47, 48]. Its structure allows it to filter blood containing pathogens and abnormal cells with a high prospect of interacting with antigen-presenting cells (APCs) and related lymphocytes [47, 48].
The current study aimed to determine the in vivo host and pathogen gene expression profile in spleen tissues of locally-adapted Kenyan pigs, following experimental challenge with varying (low, medium, and high) infective doses of a highly virulent ASFV isolate, the Ken12/busia.1 responsible for ASF outbreaks in East Africa. The ASFV Ken12/busia.1 isolate was originally isolated from the spleen of a pig sacrificed in the former Busia district (now Busia County) in Western Kenya during a longitudinal survey of ASF and has been confirmed to belong to Genotype IX [49]. The gene expression profiles between the infective dose groups show common and unique patterns between the medium and high dose groups than with the low dose groups with upregulation of pig host genes associated with macrophages, NK cells, and viral genes associated with modification of host immunity.
Results
Clinical outcomes and pathology
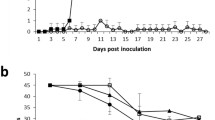
The clinical signs recorded were intermittent fever, high body temperature (> 40 °C), depression, and anorexia. The pigs in the low dose group did not show any ASF clinical signs or gross pathology throughout the experiment and were euthanized at the termination of the experiment (29 dpi: Fig. 1). From 7 dpi, all locally-adapted pigs in the medium and high dose groups showed acute ASF clinical signs, including depression, anorexia, recumbence, accelerated and labored breathing, diarrhea, and slight ataxia, and were euthanized humanely from 7 dpi (n = 1 in high dose), 9 dpi (n = 2; 1 each from high and medium dose), 10 dpi (n = 3; 2 from high dose and 1 from medium dose), 11 dpi (n = 1 from medium dose) and 17 dpi (n = 3 from medium dose). The locally-adapted pigs have black pigment, and as a result, skin lesions could not be scored. We also observed recumbence, reduced feed intake, weight loss from 4 dpi, and foul-smelling watery diarrhea. The highest temperature (39.6 °C) was recorded at 7 dpi from the first pig that succumbed at 7 dpi from the high dose group (Supplementary Table 1 and Fig. 1- survival analysis). From 9 dpi, all the remaining locally-adapted pigs showed febrile temperature reactions (40.5 °C to 41.4 °C). Survival rates between high and medium doses were not significantly different (p = 0.075) from that between the high and low (p = 0.013) and medium compared to that of low (p = 0.017), which were significant. All the spleen samples in the infection groups collected post-euthanasia tested positive by ASFV qPCR, while in the high- and medium dose groups, ASFV positivity by PCR was detected much earlier in blood collected from 4 dpi. Postmortem examination of the organs showed hemorrhagic lymph nodes and fluid accumulation in the abdominal, thoracic, and pericardial cavities.
RNA-Seq data quality check, mapping to pig host and ASFV pathogen reference genomes
Total RNA from 10 porcine splenic tissues was used for RNA-seq (3 each from medium, high, and low dose groups, and one animal from the control group). The total number of reads attained from the 10 locally-adapted pigs is summarized in Table 1. After QC, deduplication, and trimming off of multi-mapped reads, we retained an average of 45.3 M reads from the low dose pig samples, 12.89 M reads from the medium dose pig samples, and 9.66 M reads from the high dose pig samples (Table 1). There were no significant variations in the number of reads between the replicates.
Following the trimming of adaptors, deduplication, and removal of poor-quality reads, on average, 72.58% of the reads mapped to the pig genome (Fig. 2A). Pig HB_1066 from the medium dose group had very low mapping rates to the pig reference genome (5.75%) and was removed from the analysis. On average, of the trimmed data, 0.15% (min 0.01% and max 0.5%) of the reads mapped to the ASFV genome (Fig. 2B). The pigs challenged with the high- and medium doses had higher ASFV mapping rates, with pig HB_1069 from the medium dose group having the highest ASFV mapping rate of 0.5% (Fig. 2B).
Mapping rates (A) to the pig and (B) to the ASF virus genomes. Created with Datawrapper on https://www.datawrapper.de/; accessed on 2nd March 2022
Differential gene expression in locally-adapted pig spleen tissues
Compared to the control, we detected the expression of 15,543, 13,755 and 14,544 known pig genes in the low, medium, and high dose groups, respectively (Table 1 and Fig. 3). The complete list of pig genes detected by RNA-Seq is in Supplementary Table 2. We observed a wide variation in the gene count in transcripts per Million (TPM) of the expressed genes in the three infection groups, possibly due to differences in sample collection time points since these pigs reached humane end-point at different times (see survival curve, Fig. 1). We then selected genes with at least a 2-fold increase in gene expression relative to the control genes in at least two pigs as the top expressed pig genes. A summary of select differentially expressed genes (DEGs) is shown in Table 2. Of the 3055 DEGs detected in the high dose group, 1711 were upregulated while 1344 were downregulated (Fig. 3A), while the medium dose group had 896 DEGs upregulated and 875 downregulated. In the low dose group, 105 genes were upregulated while 23 were downregulated (Fig. 3A). In the high and medium dose groups, the top-upregulated pig host genes were associated with response to infection due to a highly pathogenic ASFV isolate. These DEGs could be divided into five groups, namely a) genes found on the macrophages, b) genes associated with natural killer cells, c) genes involved in ASFV infection, d) genes linked with the lymphocytes, e) other genes not linked to virus infection or immunity such as DRAM2, and SOGA1, are reported to be associated with autophagy. Other genes such as TIMP1, LTF, CHP2, FOSL1, and FOXF1 play critical roles in viral pathogenesis [Table 2] [50, 51].
DEGs between three dose groups (A) in the pig host and (B) in the ASFV genes, and (C) Volcano plots for DEGs between high dose and control groups of pigs and (D) Volcano plots for DEGs between medium dose and control groups of pigs. (E) Volcano plots for DEGs between low dose and control groups of pigs. The dotted line across the volcano plots shows the cutoff for DEGs > 95% confidence (p < 0.05). (F) A heatmap of the DEGs
Additionally, the Forkhead box (FOX) family of transcription factors (FOXF1, FOXS1, FOXM1, FOXK1, FOXP4, and FOXC1) play a role in cell differentiation and proliferation and are implicated in cancer and drug resistance [77]. These FOX transcription factors were all upregulated in the medium and high dose groups but not detected in the low dose group. The CSF3R is a type 1 cytokine receptor that binds the granulocyte colony-stimulating factor (G-CSF). G-CSF is a cytokine that is required for granulocyte proliferation and differentiation. IFITM3 was significantly upregulated (over 3-fold), and it plays a critical role in immunity during viral infection in that it directly engages and shuttles incoming virus particles to lysosomes. We also detected the expression of ATF4, a transcription factor that upregulates genes involved in amino acid import, antioxidative stress response, and regulation of autophagy [78, 79].
Regarding genes associated with resistance/tolerance to ASF [25], the RELA proto-oncogene, NF-kB subunit, or transcription factor p65 (RELA) was detected at higher counts (2846 TPM) in the low dose group than the high and medium doses at 257 and 269 TPM, respectively (Supplementary Table 2). RELA was downregulated at − 1.94-fold (padj = 1.98E-04) and − 2.34-fold (padj = 4.71E-05) in the high- and medium dose groups but was not differentially expressed in the low dose group.
In terms of the genes associated with macrophage regulation [80], there was downregulation of the macrophage surface marker gene, CD164 (decreased by 1.15-fold) in pigs that survived a low dose ASFV challenge. HMOX1 codes for heme oxygenase-1, and its expression decreased by 1.89-fold and 1.46-fold in the high- and medium dose, respectively. The macrophage-associated genes were also upregulated in the medium and high dose groups, including S100A4/A6/A8/A9/A13/A16 that play a role in Calcium-binding, innate immune response, and leukocyte migration associated with inflammatory response [81,82,83]. The angiopoietin-like protein (ANGPTL1) genes were highly differentially expressed at 1.61-fold and 5.02-fold in high- and medium dose groups. CD244 is involved in the activation of NK cells leading to cell-mediated cytotoxicity [65] was downregulated in the high and medium dose groups by over 1.65 and 4.7-fold, respectively. Another differentially downregulated gene in medium and high dose groups was CD36 at 3.89 and 4.91-fold change, respectively. CD36 is a scavenger receptor expressed in multiple cell types, mediates lipid uptake, immunological recognition, inflammation, molecular adhesion, and apoptosis, and binds thrombospondin-1 (TSP-1), resulting in attenuation of angiogenesis and induction of apoptosis/blocking the vascular endothelial growth factor receptor 2 (EGFR2) pathway in tumor microvascular endothelial cells [84]. We detected an upregulation of TSP-1 in the high and medium dose groups by 1.92- and 3.25-fold change, respectively.
Antimicrobial peptides (AMPs) are molecules that possess a broad-spectrum activity against bacteria, fungi, protozoa, and viruses found in insects, amphibians, and mammals with antimicrobial, immunomodulatory and regulatory activity gut microbiota [85]. We detected AMPs of pharmaceutical value, such as protegrin-4 (PG4), a peptide isolated from porcine leucocytes [75]. NPG4 was significantly upregulated in the medium dose (7.1-fold) and high dose (3.44-fold). The other upregulated genes were those involved in the host response to virus infection, such as PPP1R15A (protein phosphatase 1 regulatory subunit 15A), which codes for GADD34 complex, a host protein involved in the dephosphorylation of P-eIF2α (Eukaryotic Initiation Factor 2) by an interferon-induced double-stranded RNA-activated protein kinase (PKR) in a prominent cellular antiviral pathway [62, 86]. Another essential gene expressed in macrophages is the SpiC, which we found to be the most downregulated gene in the high- and medium dose groups at 6.15-fold and 7.19-fold, respectively. SpiC plays a role in the downregulation of pro-inflammatory cytokines while promoting iron efflux by regulating ferroportin expression in activated macrophages [63].
CD163, a hemoglobin-specific receptor found on the cell surface of macrophages, is implicated in iron recycling and inflammatory response [52, 53], was downregulated. In contrast, RSG16, which restricts the pro-inflammatory response of monocytes [54], was significantly upregulated by over 4-fold in the medium and high dose groups. Expression of Prostaglandin E synthase (PGES) was upregulated in the medium and high dose groups. PGES plays a crucial role in inflammation by converting prostaglandin (PG) H2 to PGE2 [87]. The matrix metalloproteinase (MMP) family catalyze proteolytic activities that result in the breakdown of the extracellular matrix [88]. MMPs thus play a key role in tumor invasion, neoangiogenesis, and metastasis [88]. The matrix metallopeptidase 17 (MMP17; or MT4-MMP) was highly upregulated (4.1-fold) in the low dose group compared to the high dose (1.84-fold). MMP8 was upregulated in the medium and high dose groups (5.96- and 3.87-fold, respectively).
Nine cytokines showed significant differential expression from the high and medium dose groups (Table 3); eight of these were upregulated, and one was down-regulated. Interleukin-6 (IL-6), VEGFA and IL27 were among the most upregulated genes by 5.67-, 3.81- and 2.2-fold, respectively. Seventeen cytokine receptors were differentially expressed, with IL1RL1 being the most highly differentially expressed (5-fold), followed by TNFRSF11A (2.67-fold). TNFRSF9 (TNF receptor superfamily member 9), also known as 4-1BBL or CD137, was most down-regulated by up to 3.11-fold in the medium dose. The interleukin cytokines IL6, IL27, and IL17B, were upregulated in the medium and high dose groups resulting in hemorrhagic fever by cytokine storm. There was an up-regulation of the proinflammatory cytokines involved in the apoptotic processes such as TNF (1.5-fold change), members of the TWEAK family such as TNFSF12 (1.53-fold change), and TNFSF13 (1.63-fold change). The IFN-ω has cross-species antiviral activities and was significantly downregulated in the high dose group (1.62-fold change). The immune-suppressive cytokine, IL27, was significantly upregulated in the high dose (2.2-fold) and medium dose groups (2.61-fold). We detected upregulation of the Interleukin-1 receptor (IL-1R) in the medium and high dose groups by over 5-fold in the high dose group and 4.01-fold in the medium dose group. We detected expression of interleukin-1 receptor-associated kinases (IRAKs) in the high dose group only where IRAK1 and IRAK3 were downregulated while expression of IRAK4 was upregulated (Table 3).
In the spleen, chemokines are essential in modulating adaptive immune response by promoting the initial priming of lymphocytes and guiding their differentiation and phenotype. There were nine differentially expressed chemokines (Table 4). The C-C ligand 2 (CCL2) and C-X-C motif chemokine ligand 2 (CXCL2), CXCL10, CCL23, CCL4, and CXCL8 were significantly upregulated in the medium dose group, while in the high dose group, CCL2, CCL4, CXCL2 and CXCL10 were significantly upregulated. CCL21, CCL26 and CXCL13 were significantly downregulated in both the high and medium dose groups (Table 4). CCL4 and CXCL10, the chemo-attractants for immune response, were upregulated in the medium and high dose groups, while CXCL10 was downregulated in the low dose group. CCL2 was the most upregulated chemokine (4.03- and 4.11-fold), while CCL26 and CXCL13 were the most downregulated in the high (5.28- and 2.5-fold) and CCL21 and CCL26 in the medium dose groups (4.30- and 4.17-fold). CXCL2 and CXCL8 are involved in recruiting neutrophils. The ELR+ (glutamic acid – leucine – arginine) CXC chemokines CXCL2 were significantly upregulated in the medium (2.68-fold) and high (3.44-fold) dose groups. CXCR2 signaling is essential for neutrophil release from the bone marrow into the blood.
Antigen processing and presenting cells were downregulated in the medium and high dose groups. The differentially expressed genes involved in MHC antigen processing and presentation are listed in Table 5. The expression of SLA-DMB, SLA-DQA, SLA-DRA, SLA-DRB, and SLA-DOB were down-regulated in the medium and high dose groups. The cathepsin S was downregulated among the medium and high dose groups, compromising the antigen presentation by MHC class II molecules. Cathepsin S (CTSS) gene is involved in processing antigens before loading to MHC class II and was significantly downregulated (2.10-fold change). The proteasome activators (PSMC, PSMD, PSME, PSMF) were upregulated in the medium and low dose groups.
In total, we detected 10 autophagy-related genes in the medium and high dose study groups and only one was detected in the low dose group. Seven of these were downregulated, namely ATG4C, DRAM2, DCT, EPG5, APAF1, NBR1 and FUNDC1 (Table 6). Autophagy-associated cell death is inhibited by the nuclear protein 1 (NUPR1), a transcriptional regulator gene that was significantly upregulated in the high and medium dose groups by over 5-fold (Tables 2 and 6). In the high and medium dose groups, we also detected the upregulation of pro-apoptosis and an autophagy inducer gene, BCL2 interacting protein 3 (BNIP3). Twelve [12] other autophagy and cell death regulating genes were detected, of which 10 were upregulated, including FAIM2, GAS2L1, GAS2L2, GAS8, MAD1L1, MAD1L2, GADD45G, GADD45B, GAS7, and NUPR1. Five of these were significantly upregulated (FAIM2, GAS2L2, GAS8, MAD1L1, and GADD45G). GAS2L3 (Growth arrest-specific 2 like 3) and APAF1 were downregulated, with GAS2L3 being significantly downregulated (Table 6).
In total, we detected 57 signal transduction and transcription genes required for macrophage activation in the medium and high dose groups (Supplementary Table 3). Twenty-seven of them were upregulated, with 9 being significantly upregulated (> 2-fold change). Of the 30 downregulated signal transduction and transcription genes, nine (USP34, USP44, USP45, USP37, MAP2K6, MAP3K2, IRF4, MEF2C, and MEF2B) were significantly downregulated (> 2-fold change). Two CCAAT enhancer-binding proteins (or CEBPs) were upregulated in the medium and high dose groups, namely CEBPD and CEBPB. TAB3 expression was detected, and five other key immune transcription factors (FOSB, FOSL1, FOSL2, IRF7, JUNB, IRF4, and IRF8) were all upregulated except for IRF4 and IRF8, which were downregulated in the high and medium dose groups (Supplementary Table 3). Also detected was the expression of ten ubiquitin-specific peptidases (USP) and three suppressors of cytokine signaling (SOCS), namely USP1, USP14, USP20, USP24, USP25, USP33, USP34, USP35, USP37, USP4, USP44, USP45, USP47, USP48, USP7, USP9X, SOCS3, SOCS4 and SOCS7. The expression of SOCS3, SOCS7, USP14, USP20, and USP35 was upregulated, while the rest were downregulated in the high dose group.
In total, we detected the expression of 20 mitogen-activated protein kinase (MAPK), namely MAP2K3, MAP2K4, MAP2K6, MAP3K1, MAP3K10, MAP3K14, MAP3K2, MAP3K5, MAP3K6, MAP3K9, MAP4K2, MAP4K4, MAPK1, MAPK6, MAPK7, MAPK8, MAPK8IP3, MAPK9, MAPKAP1, and MAPKAPK2. Of these, MAPK7, MAP3K6, MAP3K10, MAP4K4, MAP2K3, MAP3K14, MAP3K9, MAPK8IP3, MAP3K5 and MAPKAPK2 were upregulated, while MAP3K1, MAP2K4, MAPK1, MAPK6, MAPK8, MAPK9, MAP3K2, MAP2K6, MAP4K2, and MAPKAP1 were downregulated. MAPK cascades are signaling pathways that regulate cellular processes, such as proliferation, differentiation, apoptosis and stress responses crucial for cancer development and progression [89].
Small nucleolar RNAs (SnoRNAs) exhibit oncogenic and tumor-suppressive actions vital in lung cancer tumorigenesis and progression by participating in the invasion of growth suppressors and cell death, activation of invasion and metastasis angiogenesis, and continued proliferative signaling [90]. In this study, we detected differential expression of several small SnoRNAs in all three study groups at 10.32- and − 2.54-fold across the groups. 42, 33, and 30 SnoRNAs were differentially expressed in the high, medium and low dose groups, respectively. The downregulated SnoRNAs were 2 in the high dose group and 3 in the low dose group. The small nucleolar RNA, C/D box 45A, was downregulated in both the medium and high dose groups.
ASFV gene expression in infected spleen tissues
In total, we detected the expression of 172 (high dose), 174 (in medium dose), and 167 (low dose) known ASFV genes and multigene families (MGFs) in the spleen samples analyzed from the locally-adapted pigs (Table 1). The complete list of ASFV genes detected is found in Supplementary Table 4. A total of 44 known MGFs were detected in reference to the Ken06.Bus ASFV genome Supplementary Table 5. We observed a wide variation in the gene count in Transcripts per Million (TPM) of the expressed genes in the different pigs studied, probably due to variations in the time the pigs were euthanized and variations in the number of infected macrophages at the time of euthanasia. In ASFV, higher gene counts of MGFs were detected in pigs from the high- and medium dose groups (Supplementary Table 5). The uncharacterized protein (C84L), viral DNA polymerase (G1211R), polyprotein pp220 (CP2475L), and a hypothetical protein (ASFV_G_ACD_00190) were the top four genes in the low dose group with 204, 123, 109, and 101 TPM being detected. In the medium dose group, MGF_100-1 L, Uncharacterized protein (E184L), and the structural protein p72 (B646L) were expressed in 2097, 1533, and 1271 TPM, respectively. The top 10 genes with the highest counts in TPM were MGF 100-1 L, E184L, B646L, B385R, I196L, MGF 360-4 L, NP1450L, MGF 360-1La, I215L, and K145R (Table 7). The gene L83L interacts with the host IL-1R and was detected in high dose (13 TPM), medium dose (45 TPM) and low dose (6 TPM) [Supplementary Table 4]. E184L, B646L, and MGF_100-1 L were expressed at 1004, 1001, and 862 TPM in the high dose group. The top three genes in the medium and high dose were the same, with differences in the number of TPM.
MGFs demonstrate divergence in sequence, indicating they have evolved over long periods and thus offer a selective advantage to the virus [91]. This study detected several MGFs and the top 10 MGFs detected were MGF_100-1 L, MGF_360-4 L, MGF_360-1La, MGF_360-6 L, MGF_360-21R, MGF_360-15R, MGF_100-3 L, MGF_505-4R, MGF_505-1R and MGF_110-7 L (Supplementary Table 5). These MGFs were detected at very low gene counts (in TPM) in the low dose group (Supplementary Table 5 and Fig. 4) compared to the medium and high dose groups (Fig. 4A). The highest gene counts in the low dose group were for MGF 505-4R (62 TPM), MGF 360-6 L (40 TPM), and MGF 360-15R (34 TPM). The highest gene counts in the high and medium dose groups were for MGF 100-1 L (862, 2097 TPM), MGF 360-4 L (376, 2097 TPM), and MGF_360-1La (297, 864 TPM). The MGFs (MGF 360-10 L, 11 L, 12 L, 13 L, 14 L, and MGF505-1R, 2R, 3R) have been associated with virulent ASFV isolates [91] and were detected in all the three study groups affirming the virulence of the ken12/busia.1 ASFV isolate (Supplementary Table 5).
A Gene counts (in TPM) of ASFV Multigene families (MGFs), in high, medium and low dose groups, created with Datawrapper on https://www.datawrapper.de/; accessed on 2nd March 2022, (B) Venn diagram of shared DEGs in the pig host and (C) Venn diagram of shared DEGs in the ASFV pathogen
We then selected genes with a 2-fold increase in gene expression relative to the control genes in the control animal (Table 8). The highest mean expression levels were detected in E184L, MGF 100-1 L and NP1450L, all in the high and medium dose groups. MGF 100-1 L was recently shown to be highly expressed in ASF surviving pigs [92]. We detected ASFV structural, non-structural, and host regulatory genes and genes of unknown function (Supplementary Table 3). We detected MGF 360–15R (A276R) expression that is vital in blocking early innate immune responses by inhibiting the induction of IFN-β [93]. Also detected were ASFV structural genes such as CP204L, which encodes p30, an immunodominant phosphoprotein of the virion and a preferred target for ASFV serological detection of infection. Another structural protein also detected is P72 (B646L), a major capsid protein involved in virus entry and a major molecular marker for distinguishing and genotyping ASFV isolates. Additionally, the structural protein P54 (E183L) was also differentially expressed. P54 binds the LC8 chain of dynein, and is involved in virus entry, and is also required to recruit envelope precursors. We detected the other structural genes, such as KP177R (p22), A04R (histone-like), A151R (pA151R), and EP402R, which are similar to the pig host CD2 protein that is required to bind the host red blood cells to infected cells and extracellular particles resulting in haemadsorption to infected cells [16]. EP402R had fewer gene counts in the low dose group (36 TPM) compared to the medium (234 TPM) and high dose [94] groups. The expression of ASFV A238L was repressed in the high and medium dose groups compared to the low dose groups with gene counts of 18, 2, and 0 TPM, respectively. Another ASFV gene critical in host-pathogen interaction is A224L, an apoptosis inhibitor, which was detected at high amounts in the high and medium dose groups (172 and 303 TPM, respectively) compared to the low dose groups (13 TPM); however, it was not differentially expressed (Supplementary Table 4).
Additional genes differentially expressed were those that code for immunodominant ASFV proteins, namely E184L, CP312R, K205R, and K145R [45]. Other genes detected in higher counts in the medium and high dose groups are associated with late ASFV Infection, such as NP1450L and EP1242L [101]. Another late ASFV infection gene detected was S273R, which codes for SUMO-1-specific proteases, that cleave the viral polyproteins pp62 and pp220 [102]. S273R was detected at very low gene counts (< 20 TPM) in all the study pigs (Supplementary Table 4).
Overlaps in differential gene expression in the spleen
Of the 4954 DEGs, overlaps in differential gene expression between the low-, medium-, and high dose groups were detected. In the pig host, 992 (20%) genes were shared between medium and high dose groups, and 22 (< 1%) genes were shared between high and low dose groups (Fig. 4B and Supplementary Table 7). A total of 63 (1%) DEGs were shared between the three study groups. Some of the uniquely expressed genes in the pig host in the high dose group were CCL4, Calnexin, GAS2L2, IFN-ω, IRAK1, IRAK3, IRAK4, MAD1L1, MAD1L2, NBR1, SLA-DRA, ATG4C, ATGD and SOGA1 (Fig. 4A). In the medium dose group, CCL21, CCL26, CXCL8, FAIM2, GAS2L3, SLA-DRB1, SLA-DOB, and TNFSF13 were among the uniquely expressed genes. While in the low dose group, the expression of CD164, FUNDC1, HIG1A, SNORA12, and SNORD49A were unique to this study group. In the ASFV pathogen, 15 DEGs were shared between the medium and high dose groups (Fig. 4C). Two genes were unique to the high dose [CP204L (p32%2C p30) and K78R)], while the medium dose group had one unique ASFV DEG [I215L (k13L)].
Mapping pig genes to KEGG pathways
The pig and human gene atlases were used to link host gene expression with cells and tissues. The pathways containing the most significant number of genes represented were functionally characterized using the DAVID gene enrichment tool to report KEGG pathways. The medium dose group had the highest number of significantly enriched pathways (n = 11; Table 9). The pathways with the highest gene counts were linked to immune response functions primarily associated with the host immune response to viral infection. In the medium dose group, pathway hsa04060, cytokine-cytokine receptor interaction, had the highest number of highly upregulated genes (n = 24), and the upregulated genes were ssc04657 (IL-17 signaling pathway), ssc04061, (viral protein interaction with cytokine and cytokine receptor) and ssc04657 (IL-17 signaling pathway). The above three pathways contained highly upregulated genes that represented the class 1 helical cytokines (IL6, IL27, CSF3 and IL15RA), IL17-like cytokines (IL17B and ILA7RA), the CC- and CXC-subfamily of chemokines (CCL2, CXCL2, CXCL10, CCL23, CCL4, CXCL8, and CXCL13) and TNF family (TWEAK and TNFR1).
In the surviving low dose group, four pathways were significantly enriched in which all represented genes were significantly downregulated: ssc04610 (complement and coagulation cascades), ssc05171 (Coronavirus disease, COVID-19), ssc04611 (platelet activation), ssc04613 (neutrophil extracellular trap formation). The following genes were significantly downregulated in the complement and coagulation cascade, platelet activation and neutrophil extracellular trap formation pathway: FGG (fibrinogen gamma chain) plays a crucial role in pathophysiologic processes, such as inflammation and thrombosis [102] was highly downregulated by 5.47-fold. Other genes in the complement and coagulation cascades were FGA (fibrinogen alpha chain) downregulated at 4.96-fold and fibrinogen beta chain suppressed by over 3.7-fold. In the COVID-19 pathway, the CXCL10 chemokines were highly downregulated in the low dose group by over 2.56-fold.
The Hippo signaling pathway that controls animal organ size through cell proliferation and apoptosis regulation, including cell proliferation, apoptosis, and various stress responses [103], was significantly enriched in the medium and high dose groups (Supplementary Figs. 1 and 2). The tumor promotor Hippocalcin 1 (HPCAL1) was differentially expressed by 1.54 and 1.93-fold change in the medium and high dose groups, respectively, resulting in the observed organomegaly [103] among pigs in the high and medium dose groups following a postmortem. The hypoxia-inducible pathways were also significantly enriched, such as the HIF-1 signaling pathway that mediates adaptive responses to oxygen deprivation [104], typical in ASF infection.
In the low dose group, the enriched pathways were in response to viral carcinogenesis, thermogenesis, antigen processing and presentation, protein synthesis and metabolic activities (Supplementary Figs. 3 A, B and C). All the genes in the KEGG enriched terms in the low dose group were downregulated. HIG1 hypoxia inducible domain family member 1A (HIGD1A) gene was downregulated in the surviving low dose group only by 1.02-fold.
Functional annotation
When comparing the enriched GO terms, we detected 139 terms, of which 115 represented the biological processes (CC), 3 were cellular components (CC), and 21 molecular functions (MF) [Supplementary Table 6]. The GO terms: response to external stimulus (GO:0009605), extracellular region (GO:0005576), and cellular response to an organic substance (GO:0071310) had the highest gene counts in the three infective groups studied. The GO terms response to external stimulus (GO:0009605), extracellular region (GO:0005576), cellular response to an organic substance (GO:0071310), defense response (GO:0006952), the biological process involved in interspecies interaction between organisms (GO:0044419), response to external biotic stimulus (GO:0043207), response to other organisms (GO:0051707), and inflammatory response (GO:0006954) were detected in the high and medium dose groups only. In the low dose, there were less than 5 gene counts represented in these GO terms: extracellular region (GO:0005576), signaling receptor binding (GO:0005102), extracellular space (GO:0005615), cytokine receptor binding (GO:0005126), heme-binding (GO:0020037), hydrogen peroxide metabolic process (GO:0042743), tetrapyrrole binding (GO:0046906). The last three terms were detected only in the low dose group.
The host gene expression within cells and tissues was linked using the pig and human gene atlases. The transcriptome profiles were primarily associated with immunity, consistent with the upregulation of genes in monocytes, macrophages and lymphocytes. In the high dose group, only three molecular mechanisms were significantly enriched for transmembrane signaling receptor activity and molecular transducer activity (Fig. 5A and B). Interestingly, the genes represented here were downregulated (Fig. 5C), including signal transduction and transcription regulatory molecules shown in Supplementary Table 3. The cytokine regulators and signal transduction molecules are critical in stimulating apoptosis and inflammation [26, 37].
Top enriched KEGG pathways and GO annotations of the DEGs identified from the high dose group. A. Overrepresented gene ratios of significantly expressed genes activated or suppressed in the high dose group. B. Enrichment distribution of the overrepresented terms in the high dose group. C. Network of interacting genes in the top three enriched GO terms
The enriched terms were immune responses to infection and biotic stimulus in the medium dose group (Fig. 6). The genes represented by these terms were predominantly upregulated, with a few being downregulated (Fig. 6C). The upregulated genes for the immune response include LTF, CCL3L, TNFS13, IL6, and TGFB3, all over 3-fold, while CD244 was downregulated by 4.7-fold (Fig. 6C).
Top enriched KEGG pathways and GO annotations of the common DEGs identified from the medium dose group. A. Overrepresented gene ratios of significantly expressed genes activated or suppressed in the medium dose group. B. Enrichment distribution of the overrepresented terms in the medium dose group. C. Network of interacting genes in the top three enriched GO terms color-coded by their fold change value
Another significantly activated gene in the low dose group was CD164 (endolyn), which encodes a transmembrane sialomucin. This cell adhesion molecule regulates the proliferation, adhesion, and migration of hematopoietic progenitor cells and is also a significant contributor to tumorigenesis in normal human cells [80]. The encoded protein by CD164 also interacts with the C-X-C chemokine receptor type 4 and may regulate muscle development [105]. CD164 is enhanced by the FOXK2 gene (also known as interleukin enhancer-binding Factor 1) transcriptional regulator involved in glucose metabolism, aerobic glycolysis, and autophagy. When all GO terms were considered, we detected suppression of a set of genes involved in protein translation, phosphatase activity, and replication peptide metabolic process (Fig. 7).
Top enriched KEGG pathways and GO annotations of the common DEGs identified from the low dose group. A. Overrepresented gene ratios of significantly expressed genes activated or suppressed in the low dose group. B. Enrichment distribution of the overrepresented terms in the low dose group. C. Network of interacting genes in the top three enriched GO terms color-coded by their fold change value
Discussion
ASFV infections result in varying clinical outcomes depending on the virulence of the isolate but also the amount of infective dose used [45, 106]. As in other viral infections, the host responds by triggering biological processes, generally through gene expression, to counteract the effect of invading pathogens, including apoptosis, autophagy, and stress-induced unfolded protein response (UPR), inhibiting ASFV virus replication. Several theories exist to elucidate the molecular mechanisms of the differences in ASFV pathogenesis, such as hemorrhagic fever, top among which is the cytokine storm triggered by overexpression of proinflammatory cytokines by monocytes and macrophages, typical of ASFV infections [23, 26]. Our study used a virulent ASFV isolate from Kenya to infect locally-adapted pigs (anecdotally reported to tolerate ASF) at varying doses (high, medium, and low) to produce an effective dose that mimics severe and mild disease and determine the variations in expression levels of the genes in these locally-adapted pigs and the potential molecular mechanisms of severe and mild ASF disease.
Our results confirm that the amount of infective material determines the clinical outcomes ranging from mild clinical signs (in the low dose group of pigs) to acute hemorrhagic fever and death (as in the medium and high dose groups). The pigs in the high and medium dose groups showed severe symptoms of ASF from 6 dpi and were humanely euthanized from 7 to 17 dpi (Fig. 1). Conversely, the pigs in the low dose group did not develop severe disease and survived to the end of the experiment (29 dpi). We determined the ASFV dose to induce lethal ASF disease as 104 HAD50/ml, while a low dose of 102 HAD50/ml mimicked exposure to ASF and the presence of the virus in tissues as reported in field situations [7, 8, 28]. The limitation of this study is in the wide variations in the number of infected cells arising from differences in the time of euthanasia and thus differences in sample collection time points for the three different study groups ranging from 7 to 29 dpi.
The study reports the transcriptomic profile of spleen tissues following an experimental challenge with three different doses of ASFV. The spleen filters blood containing abnormal cells and pathogens and facilitates potential interactions between antigen-presenting cells (APCs) and cognate lymphocytes [48, 107]. The spleen allows the analysis of changes in gene expression in a mixed population of lymphatic system cells yielding a snapshot of the interactions between the host cells and pathogens. We identified several biological functions and pathways associated with response to viral infection, immune response, protein, and carbohydrate metabolism at varying levels between the low, medium, and high dose groups. Notably, in the surviving low dose group, the significant KEGG pathways consisted of highly downregulated genes (FGA, FGB and FGG) which are implicated in inflammation and thrombosis [108], indicating that survival entailed significant suppression of these pathophysiological processes.
Interferons (IFNs) form part of the first line of defense against viral infections and play a substantial role in the early immune response cells [109]. IFNs provide host defense against viral infections by inducing the expression of numerous IFN-stimulated genes (ISGs), activating host antiviral immunity [26, 110]. We detected significant upregulation of TNF inflammatory cytokines (Table 3). More TNF transcripts were detected in the medium and high dose groups, indicating that TNF plays a role in disease severity and that ASFV regulates the immune response by manipulating cytokine expression in host cells [109]. Prostaglandin E (PGE2), which modulates the immune system by regulating the expression of cytokines [87], was upregulated by over 4-fold in the medium and high dose groups.
There was significant upregulation of interferon-inducible transmembrane (IFITM) proteins. IFITMs are ISG products that confer cellular resistance to several viruses [111,112,113,114]. IFITMs are thought to function by either (i) blocking viral entry and release from cells by altering membrane rigidity and curvature, thus suppressing viral membrane fusion [111, 112, 115, 116] or (ii) by inhibiting viral replication mechanisms [111]. IFITM1–3 are expressed by T cells and regulate CD4+ T helper cell differentiation in a T-cell-intrinsic manner affirming their role in adaptive immunity. For example, IFITM3 was shown to play a critical role in the early phase of viral entry during influenza A infection by clustering on virus-containing endosomes and lysosomes a couple of hours after infection [117, 118]. Given that ASFV enters into the host macrophages by dynamin-dependent and clathrin-mediated endocytosis and macropinocytosis [119], the upregulation of IFITM2 and 3 alters the distribution of early and late lysosomes and endosomes in the absence of IFITM1 during ASFV infection [120]. IFITM1 and IFITM3 genes were significantly upregulated in the medium dose group, indicating the alteration of the endosomal compartment in an attempt to inhibit virus entry.
ASFV infection causes neutrophilia in pigs: a higher neutrophil count in the blood than the normal reference range typical during infections and inflammation [121]. We detected upregulated expression of colony-stimulating factor 3 receptor (CSF3), also called the receptor for granulocyte colony-stimulating factor (G-CSF), that contributes to the proliferation and granulocyte differentiation of myeloid progenitor cells [122]. CSF3 or G-CSF is a neutrophil growth-promoting cytokine that was significantly upregulated in the medium and high dose groups. The neutrophil recruiting chemokine, CXCL2 [123], was significantly upregulated in the medium and high dose groups. The other chemokines detected in this study (Table 4) involved in monocyte, T and NK cell recruitment were CCL2, CCL4 and CCL23, which were upregulated in the medium and high dose groups, resulting in an increased number of cells susceptible to infection [124]. The expression of CCL21 and CCL26, which have a role in the recruitment of CD8+ effector T cells and Th2 T lymphocytes, was suppressed, while the expression of chemokines involved in neutrophil trafficking was induced (Table 4). Induction of CXCL10 may result in lymphocyte priming toward the Th1 phenotype or induction of T lymphocyte apoptosis. Interestingly, the CXCL10 was significantly downregulated in the surviving low dose group indicating suppression of neutrophilia. These results corroborate the reports that ASFV-infected pigs develop neutrophilia, a critical antiviral response [26, 45].
IL-17B is expressed in neutrophils, germinal center B cells, neurons, stromal cells, and gut epithelium [125]. The upregulation of cytokines of the IL-17 family mediates signaling through the ACT1-dependent pathway, causing activation of pro-inflammatory factors, for example, nuclear factor (NF)-κB, associated with innate immune signaling [125]. Reports show that IL-17B promoted the TNF-α-induced production of G-CSF and IL-6 in the fibroblasts and induced the expression of inflammatory cytokines of IL-1α, IL-6, and IL-23. Sun et al. showed that intraperitoneal injection of rhIL-17B caused indirect recruitment of neutrophils to the peritoneal cavity and induced the migration of CXCR4+ or CXCR5+ GC B cells due to CXCL12 and CXCL13 [126]. We detected upregulation of expression of IL-17B in the medium and high dose groups, while CXCL12 was downregulated, contributing to our understanding of the ASFV pathogenesis.
ASFV infection has been shown to upregulate the expression of immunosuppressive cytokines to put a strain on the host immune response. Previous studies have shown that IL27, a suppressor of the immune response of Th1 and Th17 cells, is downregulated during ASFV infection [127]. However, IL27 was significantly upregulated in our study by over 2-fold in the medium and high dose groups. Interleukin 1 receptor antagonist (IL1RN), an endogenous pyrogen, causes an inflammatory response and promotes B cell proliferation and differentiation when combined with IL-1R [128]. IL-1R was upregulated in the medium and high dose groups, which may result in the exacerbated inflammatory response in these two groups. We detected the high expression of the ASFV L83L gene in the high and medium dose groups but only lowly (in TPM) in the low dose group. The ASFV L83L gene encodes a protein that interacts with the host IL-1β and its deletion did not affect IL-1 production, indicating that the host could be using an alternative mechanism for IL-1 production [129]. The interleukin-1 receptor-associated kinases (IRAKs) are crucial mediators of toll-like receptor and interleukin-1 receptor signaling processes [94]. The expression of IRAKs was detected only in the high dose group, where IRAK4 and IRAK3 were suppressed while the expression of IRAK1 was upregulated. Recent studies have shown that the actions of IRAK-1 participate in the stimulation of NF-kappaB-dependent transcriptional events [130].
It has been shown previously that the variation between the ASFV genomes of different isolates is primarily a result of the increase (gain) or decrease (loss) of members of the multigene families, namely MGF-100, MGF-110, MGF-300, MGF-360 and MGF-505/530 families [131, 132]. Our study detected, on average, 172 ASFV genes and multigene families (Supplementary Table 4 and 5) with higher gene count in TPM detected in the MGF 100-1 L, E184L, and B646L genes. The MGF 360 has been shown to have a high gene copy number and is highly variable, containing 22 paralogous genes in total [16]. We did not detect a critical gene, DP71L, which codes for a protein that functions as a cofactor for the pig host protein phosphatase 1 (PP1), a phosphatase that dephosphorylates P-eIF2α [62, 86]. However, upregulation of the host PPP1R15A explains why the loss of DP71L does not affect host cell translation during ASFV infection. PPP1R15A codes for the GADD34 complex, a host protein that dephosphorylates P-eIF2α by an interferon-induced double-stranded RNA-activated protein kinase (PKR) in a prominent cellular antiviral pathway [62, 86]. Since PPP1R15A was significantly upregulated in high and medium dose groups, it likely provided a mechanism to ensure P-eIF2α is maintained in a dephosphorylated state during ASFV infection [45]. Following infection, P-eIF2α initiates the transcription of ATF4, which then upregulates the expression of UPR-associated genes, including PPP1R15A and an inducer of apoptosis called CHOP. Our study detected upregulation of ATF4 in the medium and high dose groups, which further affirms the apoptosis in ASFV pathogenesis.
ASFV, like other viruses, can manipulate immune responses to promote infection, such as the delayed onset of apoptosis and autophagy, by expressing several genes that evade the host response [133, 134]. The highly conserved and immunodominant ASFV proteins, namely E184L, CP312R, K205R, and K145R [45], were significantly upregulated in the high and medium dose group. Autophagy and apoptosis have a critical role in the innate and adaptive immune response against viral infection [135] and in delivering antigens for MHC Class II epitope processing [136]. Our data indicate that host gene expression following ASFV infection could suppress apoptosis, particularly autophagy-associated apoptosis. Up-regulated genes overrepresented the apoptosis processes, and there was a significant overrepresentation of down-regulated genes involved in autophagy. NUPR1 suppresses metabolic stress-induced autophagy-associated programmed cell death by apoptosis and programmed necrosis [64] and was significantly up-regulated in the high dose group 5-fold. We also detected the upregulation of a pro-apoptosis and an autophagy inducer gene called BCL2 interacting protein 3 (BNIP3), further confirming that apoptosis and autophagy were enhanced in the high and medium dose groups. Further, GADD45B is known to suppress apoptosis and autophagy and it was significantly upregulated in the high and medium dose groups. Another crucial regulatory gene in the UPR pathway, PPP1R15A, was significantly upregulated after ASFV infection. Other signaling transducers were downregulated in the high and medium dose groups, resulting in delayed apoptosis induction by high TNF expression.
Autophagy-related genes such as GAS2 and GAS2L3 were downregulated, contrasting observations in ex vivo macrophages [26]. We also found the suppression of expression of one autophagy-related gene called FUNDC1 in the low dose group. FUNDC1 is a crucial mitochondrial outer-membrane protein that is a receptor for hypoxia-induced mitophagy [137]. Hypoxia-induced mitophagy is a specialized process that occurs in hypoxic cells whereby the dysfunctional mitochondria are selectively removed [138]. Thus, suppression of FUNDC1 results in a defect in hypoxia-induced mitophagy. FUNDC1 has been shown to work with DRP1 and calnexin [CALN] (suppressed at 1.36-fold in the high dose group) during hypoxic conditions in mammalian cells [138]. Thus, detection of suppression of FUNDC1 in low dose groups confirms that the pigs in this group likely survived the ASFV infection by suppressing hypoxia-induced mitophagy.
Signal transducers and transcription factor genes are essential in M1 macrophage polarization. Expression of TAB3 and the other five critical immune transcription factors, FOSB, FOS, IRF7, JUNB, IRF4, and IRF8, were all upregulated apart from IRF4 and IRF8, which were downregulated in the high and medium dose groups (Supplementary Table 3). FOS and JUN are early response genes whose expression is induced by cell-extrinsic and cell-intrinsic signals like during viral infection [139]. Contrary to previous reports [26], our study showed that JUN and FOS were upregulated in the medium and high dose groups.
Cytokines, growth factors, and hormones utilize the Janus kinase–signal transducer and activator of transcription (JAK-STAT) pathway to transmit their information into the cell nucleus. At the same time, the cytokine signaling inhibitors such as ubiquitin-specific peptidases (USP) and Suppressors of Cytokine Signaling (SOCS) downregulate interferon responses. SOCS protein family inhibits STAT activation by many JAK-STAT activating receptors [140]. Our results corroborate further that ASFV immune evasion could be via interference with the NF-κB, JAK, and IRF signaling pathways.
ASFV encodes genes that allow it to evade the host defense systems and modulate host cell function. A224L is an ASFV gene that inhibits apoptosis, which is vital in host-pathogen interaction [21]. Though not differentially expressed, we detected more A224L transcripts in the high and medium dose groups (172 and 303 TPM, respectively) compared to the low dose groups (13 TPM), indicating that the virus was attempting to evade the apoptosis process, especially at a higher viral dose. The expression of ASFV A238L gene was suppressed in the high and medium dose groups compared to the surviving low dose group. The ASFV gene, A238L, is a known inhibitor of host gene transcription through inhibition of NF-kB and calcineurin phosphatase [141, 142] that has been frequently detected in surviving pigs and modulates the host immune responses [92, 143, 144], as was the case in this study. In the pig host, the RELA proto-oncogene [NF-kB subunit, or p65 (RELA)] was detected at higher counts (2846 TPM) in the low dose group than the high and medium doses at 257 and 269 TPM, respectively (Supplementary Table 2). RELA has been associated with resistance/tolerance to ASF [25], and the fact that it was downregulated in the high dose (1.94-fold) and medium dose (2.34-fold) confirms the lethality of this dose in these reportedly resilient pigs.
The ASFV CD2v (EP153R) activates NF-κB, subsequently inducing IFN signaling and apoptosis in swine macrophages [110]. The EP153R gene is vital in the haemadsorption of red blood cells and suppresses the expression of MHC class I molecules by impairing the exocytosis process without affecting the synthesis of MHC antigens [145]. Additionally, proteases found on macrophages called Cathepsins process antigens before loading on MHC class II molecules [146]. We determined that antigen processing and presenting cells were downregulated in the medium and high dose groups. Cathepsin S (CTSS) was downregulated in both the medium and high dose groups (Table 5). The swine SLA-DMA and SLA-DMB are required in epitope loading onto MHC Class II molecules by removing the invariant chain in the groove of the MHC molecules, while SLA-DOA and SLA-DOB inhibit this process [147]. The SLA-DMB, SLA-DQA, SLA-DRA, SLA-DRB, and SLA-DOB were down-regulated in the medium and high dose groups in contrast to a previous study where SLA-DOA and SLA-DOB were up-regulated following ASFV infection [26]. The MHC class II genes were downregulated in the medium and high dose group of pigs, indicating that the MHC class II activity is compromised during ASFV infection. The TNF-like weak inducer of apoptosis (TWEAK) is a member of the TNFSF ligands [148]. TNFSF9 (4-1BBL/CD137) and TNFRSF11A were downregulated in the high and medium dose groups. TNFSF9 contributes to the clonal expansion, survival, and development of T cells induces proliferation in peripheral monocytes and enhances T cell apoptosis induced by TCR/CD3 triggered activation [149]. TNFRSF11A also regulates CD28 co-stimulation to promote Th1 cell responses. Additionally, TRAF adaptor proteins have been shown to bind to this receptor and transduce the signals leading to activation of NF-kB. On the other hand, TNFSF11 are important regulators of interactions between T cells and dendritic cells [150]. In this regard, ASFV infection inhibits MHC class II antigen presentation and retards T cells activation by antigen-presenting cells such as macrophages [26].
Overall, our study provides the first gene expression data from locally-adapted Kenyan pigs following experimental infection with a highly virulent ASFV genotype IX isolate, Ken/busia.1 (Ken-1033), at varying doses to mimic acute and mild disease. Our results indicate a strong correlation between severe ASF pathogenesis in the high and medium dose groups with upregulation of proinflammatory TNF cytokines, IL17B and IL6, while expression of IFN-ω, IL16, IL10RA, IL21R, TNFRSF9, TNFRSF11A, and TNFRSF21 was suppressed. We showed that the ASFV pathogen increases its replication and transcription machinery to evade the host immune response, while the pig host induces immunological and stress response pathways to counter the viral attack. Additionally, ASFV infection suppressed genes involved in MHC antigen processing and presentation by down-regulation of Cathepsins S (CTSS), SLA-DQB1, SLA-DOB, SLA-DMB, SLA-DRA, and SLA-DRA SLA-DQA, TNFSF11 and TNFSF13, and up-regulation of SLA-DOA and SLA-DOB. Conversely, we detected suppression of the critical genes involved in inflammation and neutrophilia in the surviving low dose groups. However, the spleen samples sequenced in this study comprised a mixed population of infected and uninfected bystander cells and thus, the changes in host gene expression are also likely attributed to a bystander effect rather than a direct effect on the infected cells alone [151].
Conclusions
Our study corroborates most previously available data on host responses and adds to the growing body of knowledge about the cytokines, chemokines, and regulatory factors involved in local and systemic ASFV pathogenesis and immune evasion. Our study shows that the locally-adapted pigs induced expression of protective genes associated with tolerance to infection and repression of genes involved in inflammation at varying levels. Further, the array of differentially expressed genes detected augments our knowledge of ASF pathogenesis critical in understanding the intricate host-pathogen interaction, especially during exposure to varying ASFV doses as in field situations. The survival of the low dose group strongly indicates the high likelihood that the local pig breeds are tolerant to ASFV at a 102HAD50 low dose. However, these pigs may serve as carriers and sources of acute new infections, further contributing to the persistence of ASFV in swine populations [152]. We recommend further studies that compare ASF pathogenesis in response to low doses of ASFV between locally-adapted pigs and exotic pig breeds to underpin genetic variations, if any.
Materials and method
Ethics statement
All animal experiments reported in this study were approved by Institutional Animal Care and Use Committee (IACUC) and Institutional Biosafety Committee (IBC) of the International Livestock Research Institute (ILRI), Kenya (Reference: VN_IACUC-2011-04). All methods were carried out in accordance with the approved protocol and relevant regulations. All the pigs were maintained, sampled, and euthanized humanely by the institutional veterinarian. All methods are reported in accordance with ARRIVE guidelines for the reporting of animal experiments.
Experimental design, study animals and viral isolate
A total of 14 locally-adapted pigs were used in this study in a biosecurity level II animal facility at ILRI, Nairobi. The pigs were 6-months old African breeds (also termed herein as locally-adapted) from Homa Bay County in South-western Kenya of both genders (male and female); purchased from local farmers who reared them in a free-range management system. The pigs were transported to the ILRI Nairobi farm facility, quarantined and acclimatized for 21 days.
The ASFV virus was isolated from a spleen tissue detected in Sagalame, Busia County, Kenya, and has been extensively characterized and determined to be genotype IX [GenBank ID: KM000146.1] [49]. The spleen tissue was homogenized and passaged in peripheral blood monocytic cells (PBMCs) to obtain a pure isolate of the Ken12/Busia.1 ASFV. The culture supernatant was freeze-thawed severally to lyse the cells and expose the virus. The virus was diluted ten-fold to the required doses used in the experiment. The dilutions were based on an end-point virus titration of the original material on macrophages derived from PBMCs. Before the ASFV challenge, blood samples were collected from all the studied pigs and ASFV diagnostics targeting the conserved VP72 capsid protein-coding region of the ASFV genome were conducted by qPCR [153]. A negative qPCR result qualified the pigs for inclusion in the experiment.
After a 21-day acclimatization period, the pigs were moved into a BSL2 facility at ILRI Nairobi and randomly assigned in the respective groups then inoculated intramuscularly with 1 ml of the ASFV Ken12/busia.1 isolate as follows: uninfected control pig (sterile PBS; n = 1; pig #1); an experimental low dose (102HAD50/ml) ASFV infection group (n = 3; pigs #2, #3 and #4); an experimental medium dose (104HAD50/ml) ASFV infection group (n = 5; pigs #5, #6, #7, #8 and #9); and experimental high dose (106HAD50/ml) ASFV infection group (n = 5; pigs #10, #11, #12, #13 and #14). While the intramuscular (IM) route does not represent the natural infection, it is reported to be the most reliable route of challenge, allowing high infection incidence, allowing more control of infective dose and timing of challenge [21, 154]. Back titration was carried out to confirm the administered dose. All the pigs were housed in a separate room based on infective dosage and gender, isolated from the uninfected control pig to avoid contact and, consequently, ASFV transmission.
The pigs were monitored daily for symptoms of the disease, and their body temperatures were recorded. Pig blood was collected every other day from the jugular vein using BD Vacutainer® needles gauge 20 (Becton, Dickinson and Company, New Jersey, USA) into 10 ml BD Vacutainer® glass serum tubes and 10 ml 15% EDTA tubes. Whole blood in heparin was harvested per pig for haemadsorption assays, whole blood in EDTA from each pig for use in virus detection and infection monitoring by qPCR, and 5 mL of whole blood to harvest serum. All the samples were well labeled and transported in a cool box to the laboratory for storage at − 80 °C and further processing. Back titration of the viral innocula showed that the pigs received the required dose per group. The pigs that reached a humane end-point were anesthetized by intramuscular injection with Xylazine + Ketamine (Merck KGaA, Darmstadt, Germany) at a dose of 2 mg/kg + 25 mg/kg. Sedated animals were humanely euthanased using Sodium pentobarbital 390 mg + Sodium phenytoin 50 mg/ml (Euthasol®, Le Vet. Pharma, The Netherlands) injected intravenous into the jugular vein at a dose of 0.22 ml/kg. A post-mortem was carried out, and all organs and tissue samples were collected and stored in 50 ml Falcon® tubes at − 80 °C until further analyses.
Total RNA isolation, library construction, and sequencing
Total RNA was isolated from 10 porcine spleen samples (3 each for the high, medium and low dose groups, and one uninfected control). Total RNA was extracted from 10 mg of spleen tissue using a modified protocol described previously [155] that combines tissue lysis with TRIzol™ (Thermo Fisher Scientific, USA) and silica-column purification using the RNeasy® Mini Kit (Qiagen, Germany) with on-column DNase I treatment step (Thermo Fisher Scientific, USA). The pure RNA was eluted from the columns using 40 μl of nuclease-free water. The RNA quality was assessed with the 2100 Bioanalyzer and Eukaryote Total RNA Nano Kit (Agilent Technologies, Inc., CA, USA). The total RNA quantity was determined using an ssRNA assay on the Qubit® 2.0 fluorometer (Thermo Fisher Scientific, USA). The total RNA purity was determined on a NanoDrop™ spectrophotometer (Thermo Fisher Scientific, USA), considering that the absorbance ratio at 260 nm and 280 nm close to 2.0 indicates highly pure RNA. The stranded RNA-Seq libraries were prepared with the KAPA RNA HyperPrep Kit with RiboErase (Roche, USA) to deplete the rRNA following the manufacturer’s instructions. Each stranded library was paired-end sequenced (2 × 150 bp) in an Illumina NovaSeq 6000 S4 platform at Quick Biology Inc. (Pasadena, CA, USA).
Statistical and bioinformatic analyses
The quality of the paired-end reads generated from the Illumina NovaSeq 6000 System was checked using FastQC v 0.11.7 [156]. The sequencing adapters and low-quality reads were removed using Trimmomatic v 0.38 [157] using a sliding window approach and trimming the reads once the average quality within the window fell below the given threshold. We removed duplicates in BBMap v38.67 (Bushnell, 2014). The preprocessed reads were simultaneously mapped to the pig host (Sus scrofa build 11.1) and ASFV pathogen (Ken06.Bus; Genbank ID KM111295.1) reference genomes [158] using STAR v 2.5.3a [159]. The resulting alignments were used to generate gene counts using featureCounts, part of the Subread v1.6.2 suite [160]. Differential gene expression analysis was performed using DESeq2 [161] in R v.3.6 [162]. To obtain the gene counts in transcripts per million, we transformed the raw counts by normalizing them based on gene lengths and sequencing depth.
Data exploration using principal component analysis allowed us to detect and remove outlier samples from downstream analysis. Lowly expressed genes, those with a low number of gene counts per sample, were also removed at a threshold of 100 gene counts. The count data were transformed by the varianceStabilizingTransformation function in DESeq2 [161], which normalizes the raw counts using size factors. A list of significant and differentially expressed genes was detected for each set using the Benjamini-Hochberg (B.H.) multiple testing procedure on the p-values to obtain adjusted p-values (threshold 0.05). The log2 fold change was used to distinguish between downregulated and upregulated genes and the resulting genes were visualized using the Bioconductor package EnhancedVolcano [163]. To identify and visualize differentially expressed genes with overlaps in the different conditions, we used VennDiagram [164]. Genes showing differential expression were identified using ‘Severity’ (low, medium, and high) as factors using the following contrasts: High dose vs. Control, Medium dose vs. Control, and Low dose vs. Control.
Gene ontology (GO) enrichment and mapping to the KEGG pathways
The Database of Annotation Visualization and Integrated Discovery (DAVID) v 6.750 [165] was used to annotate the differentially expressed transcripts. The significant DEGs were mapped to the KEGG pathways, and the reported gene names were listed. The DAVID gene enrichment tool was used to detect the KEGG pathways and GO terms that were overrepresented within the top DEGs. P-values were calculated using the Fisher exact test based on the fraction of genes that mapped to a specific pathway compared to the number of background genes associated with the pathway.
Availability of data and materials
The RNA-seq data generated in this study are accessible on the National Center for Biotechnology Information (NCBI) database under the BioProject PRJNA823664 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA823664).
References
Penrith ML. African swine fever. Onderstepoort J Vet Res. 2009;76(1):91–5.
Eustace Montgomery R. On A Form of Swine Fever Occurring in British East Africa (Kenya Colony). J Comp Pathol Ther. 1921;34:159–91. https://doi.org/10.1016/S0368-1742(21)80031-4.
Mur L, Martinez-Lopez B, Costard S, de la Torre A, Jones BA, Martinez M, et al. Modular framework to assess the risk of African swine fever virus entry into the European Union. BMC Vet Res. 2014;10:145.
Ravaomanana J, Jori F, Vial L, Pérez-Sánchez R, Blanco E, Michaud V, et al. Assessment of interactions between African swine fever virus, bushpigs (Potamochoerus larvatus), Ornithodoros ticks and domestic pigs in north-western Madagascar. Transbound Emerg Dis. 2011;58(3):247–54.
Penrith ML, Vosloo W, Jori F, Bastos ADS. African swine fever virus eradication in Africa. Virus Res. 2013;173(1):228–46.
Dixon LK, Abrams CC, Bowick G, Goatley LC, Kay-Jackson PC, Chapman D, et al. African swine fever virus proteins involved in evading host defence systems. Vet Immunol Immunopathol. 2004;100:117–34.
Abworo EO, Onzere C, Amimo JO, Riitho V, Mwangi W, Davies J, et al. Detection of African swine fever virus in the tissues of asymptomatic pigs in smallholder farming systems along the Kenya–Uganda border: Implications for transmission in endemic areas and ASF surveillance in East Africa. J Gen Virol. 2017;98(7):1806–14.
Chambaro HM, Sasaki M, Sinkala Y, Gonzalez G, Squarre D, Fandamu P, et al. Evidence for exposure of asymptomatic domestic pigs to African swine fever virus during an inter-epidemic period in Zambia. Transbound Emerg Dis. 2020;67(6):2741–52.
Franzoni G, Graham SP, Giudici SD, Bonelli P, Pilo G, Anfossi AG, et al. Characterization of the interaction of African swine fever virus with monocytes and derived macrophage subsets. Vet Microbiol. 2017;198:88–98.
Hübner A, Petersen B, Keil GM, Niemann H, Mettenleiter TC, Fuchs W. Efficient inhibition of African swine fever virus replication by CRISPR/Cas9 targeting of the viral p30 gene (CP204L). Sci Rep. 2018;8(1):1449.
Chen W, Zhao D, He X, Liu R, Wang Z, Zhang X, et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci China Life Sci. 2020;63(5):623–34.
Costard S, Wieland B, De Glanville W, Jori F, Rowlands R, Vosloo W, et al. African swine fever: how can global spread be prevented? Philos Trans R Soc B Biol Sci. 2009;364(1530):2683–96.
Busch F, Haumont C, Penrith ML, Laddomada A, Dietze K, Globig A, et al. Evidence-based African swine fever policies: do we address virus and host adequately? Front Vet Sci. 2021;8:637487.
Goatley LC, Reis AL, Portugal R, Goldswain H, Shimmon GL, Hargreaves Z, et al. A pool of eight virally vectored African swine fever antigens protect pigs against fatal disease. Vaccines. 2020;8(2):234.
Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. 2018;46(D1):D708–17.
Dixon LK, DDGAG C, Netherton CL, Upton C. African swine fever virus replication and genomics. Virus Res. 2012;173(1):1–12 Available from: http://www.ncbi.nlm.nih.gov/pubmed/23142553.
Penrith ML, Bastos AD, Etter EMC, Beltrán-Alcrudo D. Epidemiology of African swine fever in Africa today: sylvatic cycle versus socio-economic imperatives. Transbound Emerg Dis. 2019;66:672–86.
Guinat C, Reis AL, Netherton CL, Goatley L, Pfeiffer DU, Dixon L. Dynamics of African swine fever virus shedding and excretion in domestic pigs infected by intramuscular inoculation and contact transmission. Vet Res. 2014;45(1):93.
Arias M, de la Torre A, Dixon L, Gallardo C, Jori F, Laddomada A, et al. Approaches and perspectives for development of African swine fever virus vaccines. Vaccines. 2017;5(4):35.
Costard S, Mur L, Lubroth J, Sanchez-Vizcaino JM, Pfeiffer DU. Epidemiology of African swine fever virus. Virus Res. 2013;173:191–7.
Dixon LK, Sánchez-Cordón PJ, Galindo I, Alonso C. Investigations of pro- and anti-apoptotic factors affecting African swine fever virus replication and pathogenesis. Viruses. 2017;9:241.
Sánchez-Cordón PJ, Montoya M, Reis AL, Dixon LK. African swine fever: a re-emerging viral disease threatening the global pig industry. Vet J Bailliere Tindall Ltd. 2018;233:41–8.
Gómez-Villamandos JC, Bautista MJ, Sánchez-Cordón PJ, Carrasco L. Pathology of African swine fever: the role of monocyte-macrophage. Virus Res. 2013;173:140–9.
Penrith M-L, Nyakahuma D, Nations F; AO of the U. Recognizing African Swine Fever: A Field Manual. FAO Animal Health Manual. 2000. https://www.fao.org/3/x8060e/X8060E.HTM. Accessed 22 Mar 2022.
Palgrave CJ, Gilmour L, Lowden CS, Lillico SG, Mellencamp MA, Whitelaw CBA. Species-specific variation in RELA underlies differences in NF- B activity: a potential role in African swine fever pathogenesis. J Virol. 2011;85(12):6008–14.
Zhu JJ, Ramanathan P, Bishop EA, O’Donnell V, Gladue DP, Borca MV. Mechanisms of African swine fever virus pathogenesis and immune evasion inferred from gene expression changes in infected swine macrophages. PLoS One. 2019;14(11):e0223955.
Uttenthal A, Braae UC, Ngowi HA, Rasmussen TB, Nielsen J, Johansen MV. ASFV in Tanzania: asymptomatic pigs harbor virus of molecular similarity to Georgia 2007. Vet Microbiol. 2013;165(1–2):173–6.
Patrick BN, Machuka EM, Githae D, Banswe G, Amimo JO, Ongus JR, et al. Evidence for the presence of African swine fever virus in apparently healthy pigs in south-Kivu Province of the Democratic Republic of Congo. Vet Microbiol. 2020;240:108521.
Mujibi FD, Okoth E, Cheruiyot EK, Onzere C, Bishop RP, Fèvre EM, et al. Genetic diversity, breed composition and admixture of Kenyan domestic pigs. PLoS One. 2018;13(1):e0190080.
Ieda N, Van Bui Q, Nguyen NTD, Lapar L, Marshall K. Characterization of smallholder pig breeding practices within a rural commune of north Central Vietnam. Trop Anim Health Prod. 2015;47:1005–16.
Gómez Román VR, Murray JC, Weiner LM. Antibody-Dependent Cellular Cytotoxicity (ADCC). In Antibody Fc. Elsevier; 2014. p. 1–27. https://doi.org/10.1016/B978-0-12-394802-1.00001-7.
Sereda AD, Imatdinov AR, Dubrovskaya OA, Kolbasov DV. Mechanisms of immune response and prospects for DNA vaccines against African swine fever. Sel’skokhozyaistvennaya Biol. 2017;52(6):1069–82.
Sereda AD, Imatdinov AR, Dubrovskaya OA, Kolbasov DV. Vaccines against african swine fever (review). Sel’skokhozyaistvennaya Biologiya. 2017. https://doi.org/10.15389/agrobiology.2017.6.1069rus.
Franzoni G, Graham SP, Sanna G, Angioi P, Fiori MS, Anfossi A, et al. Interaction of porcine monocyte-derived dendritic cells with African swine fever viruses of diverse virulence. Vet Microbiol. 2018;216:190–7.
Gallardo C, Blanco E, Rodríguez JM, Carrascosa AL, Sanchez-Vizcaino JM. Antigenic properties and diagnostic potential of African swine fever virus protein pp62 expressed in insect cells. J Clin Microbiol. 2006;44(3):950–6.
Arias M, Jurado C, Gallardo C, Fernández-Pinero J, Sánchez-Vizcaíno JM. Gaps in African swine fever: analysis and priorities. Transbound Emerg Dis. 2018;65:235–47.
Salguero FJ, Gil S, Revilla Y, Gallardo C, Arias M, Martins C. Cytokine mRNA expression and pathological findings in pigs inoculated with African swine fever virus (E-70) deleted on A238L. Vet Immunol Immunopathol. 2008;124(1–2):107–19.
Che T, Li D, Jin L, Fu Y, Liu Y, Liu P, et al. Long non-coding RNAs and mRNAs profiling during spleen development in pig. PLoS One. 2018;13(3):e0193552.
McGettigan PA. Transcriptomics in the RNA-seq era. Curr Opin Chem Biol. 2013;17:4–11.
Fernández de Marco M, Salguero FJ, Bautista MJ, Núñez A, Sánchez-Cordón PJ, Gómez-Villamandos JC. An immunohistochemical study of the tonsils in pigs with acute African swine fever virus infection. Res Vet Sci. 2007;83:198–203.
Gan L, Xie L, Zuo F, Xiang Z, He N. Transcriptomic analysis of Rongchang pig brains and livers. Gene. 2015;560:96–106.
Sodhi SS, Song KD, Ghosh M, Sharma N, Lee SJ, Kim JH, et al. Comparative transcriptomic analysis by RNA-seq to discern differential expression of genes in liver and muscle tissues of adult Berkshire and Jeju native pig. Gene. 2014;546(2):233–42.
Xu Y, Qian H, Feng X, Xiong Y, Lei M, Ren Z, et al. Differential proteome and transcriptome analysis of porcine skeletal muscle during development. J Proteome. 2012;75(7):2093–108.
Fernandes LT, Tomás A, Bensaid A, Sibila M, Sánchez A, Segalés J. Microarray analysis of mediastinal lymph node of pigs naturally affected by postweaning multisystemic wasting syndrome. Virus Res. 2012;165(2):134–42.
Jaing C, Rowland RRRRRRR, Allen JEJE, Certoma A, Thissen JBJB, Bingham J, et al. Gene expression analysis of whole blood RNA from pigs infected with low and high pathogenic African swine fever viruses. Sci Rep. 2017;7:10115.
Zhang Y, Xue L, Xu H, Liang W, Wu Q, Zhang Q, et al. Global analysis of alternative splicing difference in peripheral immune organs between Tongcheng pigs and large white pigs artificially infected with PRRSV in vivo. Biomed Res Int. 2020;2020:4045204.
Lewis SM, Williams A, Eisenbarth SC. Structure and function of the immune system in the spleen. Sci Immunol. 2019;4:eaau6085.
Mebius RE, Kraal G. Structure and function of the spleen. Nat Rev Immunol. 2005;5:606–16.
Onzere CK, Bastos AD, Okoth EA, Lichoti JK, Bochere EN, Owido MG, et al. Multi-locus sequence typing of African swine fever viruses from endemic regions of Kenya and eastern Uganda (2011–2013) reveals rapid B602L central variable region evolution. Virus Genes. 2018;54(1):111–23.
Wudu M, Ren H, Hui L, Jiang J, Zhang S, Xu Y, et al. DRAM2 acts as an oncogene in non-small cell lung cancer and suppresses the expression of p53. J Exp Clin Cancer Res. 2019;38(1):72.
Mo JJ, Zhang W, Wen QW, Wang TH, Qin W, Zhang Z, et al. Genetic association analysis of ATG16L1 rs2241880, rs6758317 and ATG16L2 rs11235604 polymorphisms with rheumatoid arthritis in a Chinese population. Int Immunopharmacol. 2021;93:107378.
Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman H-J, Law SKA, et al. Identification of the haemoglobin scavenger receptor. Nature. 2001;409:198–201.
Yang H, Zhang J, Zhang X, Shi J, Pan Y, Zhou R, et al. CD163 knockout pigs are fully resistant to highly pathogenic porcine reproductive and respiratory syndrome virus. Antivir Res. 2018;151:63–70.
Suurväli J, Pahtma M, Saar R, Paalme V, Nutt A, Tiivel T, et al. RGS16 restricts the pro-inflammatory response of monocytes. Scand J Immunol. 2015;81(1):23–30.
Vijayan V, Wagener FADTG, Immenschuh S. The macrophage heme-heme oxygenase-1 system and its role in inflammation. Biochem Pharmacol. 2018;153:159–67.
Chiang SK, Chen SE, Chang LC. A dual role of heme oxygenase-1 in cancer cells. Int J Mol Sci. 2019;20:39.
Okada K, Arai S, Itoh H, Adachi S, Hayashida M, Nakase H, et al. CD68 on rat macrophages binds tightly to S100A8 and S100A9 and helps to regulate the cells’ immune functions. J Leukoc Biol. 2016;100(5):1093–104.
Touw IP, Van De Geijn GJM. Granulocyte colony-stimulating factor and its receptor in normal myeloid cell development, leukemia and related blood cell disorders. Front Biosci. 2007;12:800–15.
Isenberg JS, Roberts DD. THBS1 (thrombospondin-1). Atlas Genet Cytogenet Oncol Haematol. 2020;24(8):291–9.
Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem. 2011;286(9):7257–66.
Spence JS, He R, Hoffmann HH, Das T, Thinon E, Rice CM, et al. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat Chem Biol. 2019;15(3):259–68.
Crespillo-Casado A, Chambers JE, Fischer PM, Marciniak SJ, Ron D. PPP1R15A-mediated dephosphorylation of eIF2a is unaffected by sephin1 or guanabenz. Elife. 2017;6:e26109.
Alam Z, Devalaraja S, Li M, To TKJ, Folkert IW, Mitchell-Velasquez E, et al. Counter regulation of Spic by NF-κB and STAT signaling controls inflammation and Iron metabolism in macrophages. Cell Rep. 2020;31(13):107825.
Santofimia-Castaño P, Lan W, Bintz J, Gayet O, Carrier A, Lomberk G, et al. Inactivation of NUPR1 promotes cell death by coupling ER-stress responses with necrosis. Sci Rep. 2018;8(1):16999.
Agresta L, Hoebe KHN, Janssen EM. The emerging role of CD244 signaling in immune cells of the tumor microenvironment. Front Immunol. 2018;9:2809.
Hurtado C, Granja AG, Bustos MJ, Nogal ML, González De Buitrago G, De Yébenes VG, et al. The C-type lectin homologue gene (EP153R) of African swine fever virus inhibits apoptosis both in virus infection and in heterologous expression. Virology. 2004;326(1):160–70.
Velazquez-Salinas L, Verdugo-Rodriguez A, Rodriguez LL, Borca MV. The role of interleukin 6 during viral infections. Front Microbiol. 2019;10:1057.
Li Y, Zhou D, Ren Y, Zhang Z, Guo X, Ma MK, et al. Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy. 2019;15(3):478–92.
Song G, Xu S, Zhang H, Wang Y, Xiao C, Jiang T, et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J Exp Clin Cancer Res. 2016;35(1):148.
Kell DB, Heyden EL, Pretorius E. The biology of Lactoferrin, an Iron-binding protein that can help defend against viruses and Bacteria. Front Immunol. 2020;11:1221.
Jin Q, Kong B, Yang X, Cui B, Wei Y, Yang Q. Overexpression of CHP2 enhances tumor cell growth, invasion and metastasis in ovarian cancer. In Vivo (Brooklyn). 2007;21(4):593–8.
Yáñez M, Gil-Longo J, Campos-Toimil M. Calcium binding proteins. Adv Exp Med Biol. 2012;740:461–82.
Vallejo A, Perurena N, Guruceaga E, Mazur PK, Martinez-Canarias S, Zandueta C, et al. An integrative approach unveils FOSL1 as an oncogene vulnerability in KRAS-driven lung and pancreatic cancer. Nat Commun. 2017;8:14294.
Bolte C, Flood HM, Ren X, Jagannathan S, Barski A, Kalin TV, et al. FOXF1 transcription factor promotes lung regeneration after partial pneumonectomy. Sci Rep. 2017;7(1):10690.
Ranade SS, Ramalingam R. A review on bioactive porcine peptide, Protegrin-1. Int J Pept Res Ther. 2020;26:1493–501.
Schuligoi R, Grill M, Heinemann A, Peskar BA, Amann R. Sequential induction of prostaglandin E and D synthases in inflammation. Biochem Biophys Res Commun. 2005;335(3):684–9.
Lam EWF, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13:482–95.
Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, et al. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010;29(31):4424–35.
Vanhoutte D, Schips TG, Vo A, Grimes KM, Baldwin TA, Brody MJ, et al. Thbs1 induces lethal cardiac atrophy through PERK-ATF4 regulated autophagy. Nat Commun. 2021;12(1):3928.
Wang CC, Hueng DY, Huang AF, Chen WL, Huang SM, Chan JYH. CD164 regulates proliferation, progression, and invasion of human glioblastoma cells. Oncotarget. 2019;10(21):2041–54.
Donato R, Sorci G, Giambanco I. S100A6 protein: functional roles. Cell Mol Life Sci. 2017;74:2749–60.
Wang Y, Liu C, Fang Y, Liu X, Li W, Liu S, et al. Transcription analysis on response of porcine alveolar macrophages to Haemophilus parasuis. BMC Genomics. 2012;13(1):68.
Pan S, Hu Y, Hu M, Xu Y, Chen M, Du C, et al. S100A8 facilitates cholangiocarcinoma metastasis via upregulation of VEGF through TLR4/NF-κB pathway activation. Int J Oncol. 2020;56(1):101–12.
Wang J, Li Y. CD36 tango in cancer: Signaling pathways and functions. Theranostics. 2019;9:4893–908.
Valdez-Miramontes CE, De Haro-Acosta J, Aréchiga-Flores CF, Verdiguel-Fernández L, Rivas-Santiago B. Antimicrobial peptides in domestic animals and their applications in veterinary medicine. Peptides. 2021;142:170576.
Fusade-Boyer M, Dupré G, Bessière P, Khiar S, Quentin-Froignant C, Beck C, et al. Evaluation of the antiviral activity of Sephin1 treatment and its consequences on eIF2α phosphorylation in response to viral infections. Front Immunol. 2019;10:134.
Sander WJ, O’Neill HG, Pohl CH. Prostaglandin E2 as a modulator of viral infections. Front Physiol. 2017;8:89.
Yu J, He Z, He X, Luo Z, Lian L, Wu B, et al. Comprehensive analysis of the expression and prognosis for MMPs in human colorectal Cancer. Front Oncol. 2021;11:771099.
Guo Y, Pan W, Liu S, Shen Z, Xu Y, Hu L. ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med. 2020;19:1997–2007.
Mourksi NEH, Morin C, Fenouil T, Diaz JJ, Marcel V. snoRNAs Offer Novel Insight and Promising Perspectives for Lung Cancer Understanding and Management. Cells. 2020;9:541.
Netherton CL, Connell S, Benfield CTO, Dixon LK. The genetics of life and death: virus-host interactions underpinning resistance to African swine fever, a viral hemorrhagic disease. Front Genet. 2019;10:402.
Bosch-Camós L, López E, Navas MJ, Pina-Pedrero S, Accensi F, Correa-Fiz F, et al. Identification of promiscuous African swine fever virus t-cell determinants using a multiple technical approach. Vaccines. 2021;9(1):29.
Correia S, Ventura S, Goodbourn S, Parkhouse RME. 55: African swine fever virus include multiple mechanisms for the manipulation of interferon responses. Cytokine. 2013;63(3):256.
Rhyasen GW, Starczynowski DT. IRAK signalling in cancer. Br J Cancer. 2015;112:232–7.
Song J, Li K, Li T, Zhao G, Zhou S, Li H, et al. Screening of PRRSV- and ASFV-encoded proteins involved in the inflammatory response using a porcine iGLuc reporter. J Virol Methods. 2020;285:113958.
O’Donnell V, Holinka LG, Krug PW, Gladue DP, Carlson J, Sanford B, et al. African swine fever virus Georgia 2007 with a deletion of virulence-associated gene 9GL (B119L), when administered at low doses, leads to virus attenuation in swine and induces an effective protection against homologous challenge. J Virol. 2015;89(16):8556–66.
Jia N, Ou Y, Pejsak Z, Zhang Y, Zhang J. Roles of African swine fever virus structural proteins in viral infection. J Vet Res. 2017;61(2):135–43.
Cackett G, Portugal R, Matelska D, Dixon L, Werner F. African swine fever virus and host response: transcriptome profiling of the Georgia 2007/1 strain and porcine macrophages. J Virol. 2022;96(5):e0193921.
Bao J, Wang Q, Lin P, Liu C, Li L, Wu X, Chi T, Xu T, Ge S, Liu Y, Li J, Wang S, Qu H, Jin T, Wang Z. Genome comparison of African swine fever virus China/2018/Anhui XCGQ strain and related European p72 Genotype II strains. Transbound Emerg Dis. 2019;66(3):1167–76. https://doi.org/10.1111/tbed.13124.
Alejo A, Matamoros T, Guerra M, Andrés G. A proteomic atlas of the African swine fever virus particle. J Virol. 2018;92(23):1167–76.
Yáñez RJ, Boursnell M, Nogal ML, Yuste L, Viñuela E. African swine fever virus encodes two genes which share significant homology with the two largest subunits of DNA-dependent RNA polymerases. Nucleic Acids Res. 1993;21(10):2423–7.
Andrés G, Alejo A, Simón-Mateo C, Salas ML. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J Biol Chem. 2001;276(1):780–7.
Zhang D, Liu X, Xu X, Xu J, Yi Z, Shan B, et al. HPCAL1 promotes glioblastoma proliferation via activation of Wnt/β-catenin signalling pathway. J Cell Mol Med. 2019;23(5):3108–17.
Shan L, Zhou X, Liu X, Wang Y, Su D, Hou Y, et al. FOXK2 elicits massive transcription repression and suppresses the hypoxic response and breast Cancer carcinogenesis. Cancer Cell. 2016;30(5):708–22.
Huang AF, Chen MW, Huang SM, Kao CL, Lai HC, Chan JYH. CD164 regulates the tumorigenesis of ovarian surface epithelial cells through the SDF-1α/CXCR4 axis. Mol Cancer. 2013;12(1):115.
Sánchez-Cordón PJ, Chapman D, Jabbar T, Reis AL, Goatley L, Netherton CL, et al. Different routes and doses influence protection in pigs immunised with the naturally attenuated African swine fever virus isolate OURT88/3. Antivir Res. 2017;138:1–8.
Lewis SM, Williams A, Eisenbarth SC. C E L L U L a R I M M U N O L O G Y structure and function of the immune system in the spleen. Sci Immunol. 2019;4:eaau6085.
Farrell DH. Pathophysiologic roles of the fibrinogen gamma chain. Curr Opin Hematol. 2004;11:151–5.
Wang S, Zhang J, Zhang Y, Yang J, Wang L, Qi Y, et al. Cytokine storm in domestic pigs induced by infection of virulent African swine fever virus. Front Vet Sci. 2020;7:601641 Available from: https://www.frontiersin.org/articles/10.3389/fvets.2020.601641/full.
Chaulagain S, Delhon GA, Khatiwada S, Rock DL. African swine fever virus CD2v protein induces β-interferon expression and apoptosis in swine peripheral blood mononuclear cells. Viruses. 2021;13(8):1480.
Liao Y, Goraya MU, Yuan X, Zhang B, Chiu SH, Chen JL. Functional involvement of interferon-inducible transmembrane proteins in antiviral immunity. Front Microbiol. 2019;10:1097.
Hachim MY, Al Heialy S, Hachim IY, Halwani R, Senok AC, Maghazachi AA, et al. Interferon-induced transmembrane protein (IFITM3) is upregulated explicitly in SARS-CoV-2 infected lung epithelial cells. Front Immunol. 2020;11:1372.
Yánez DC, Ross S, Crompton T. The IFITM protein family in adaptive immunity. Immunology. 2020;159:365–72.
Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza a virus. PLoS Pathog. 2011;7(1):e1001258.
Compton AA, Bruel T, Porrot F, Mallet A, Sachse M, Euvrard M, et al. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe. 2014;16(6):736–47.
Perreira JM, Chin CR, Feeley EM, Brass AL. IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol. 2013;425:4937–55.
Wrensch F, Karsten CB, Gnirß K, Hoffmann M, Lu K, Takada A, et al. Interferon-induced transmembrane protein-mediated inhibition of host cell entry of ebolaviruses. J Infect Dis. 2015;212:S210–8.
Winkler M, Wrensch F, Bosch P, Knoth M, Schindler M, Gärtner S, et al. Analysis of IFITM-IFITM interactions by a flow cytometry-based FRET assay. Int J Mol Sci. 2019;20(16):3859.
Hernaez B, Alonso C. Dynamin- and Clathrin-dependent endocytosis in African swine fever virus entry. J Virol. 2010;84(4):2100–9.
Muñoz-Moreno R, Cuesta-Geijo MÁ, Martínez-Romero C, Barrado-Gil L, Galindo I, García-Sastre A, et al. Antiviral role of IFITM proteins in African swine fever virus infection. PLoS One. 2016;11(4):e0154366.
He C, Zhang Y, Luo H, Luo B, He Y, Jiang N, et al. Identification of the key differentially expressed genes and pathways involved in neutrophilia. Innate Immun. 2020;26(4):270–84.
Bilbao-Sieyro C, Santana G, Torres-Miñana L, Rodriguez-Medina C, Saez MN, Perera M, et al. Colony-stimulating factor-3 receptor, watch out for polymorphisms. Leukemia. 2015;29:1445–6.
Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702.
Fishbourne E, Hutet E, Abrams C, Cariolet R, Le Potier MF, Takamatsu HH, et al. Increase in chemokines CXCL10 and CCL2 in blood from pigs infected with high compared to low virulence African swine fever virus isolates. Vet Res. 2013;44(1):87.
Bie Q, Jin C, Zhang B, Dong H. IL-17B: a new area of study in the IL-17 family. Mol Immunol. 2017;90:50–6.
Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). Nat Immunol. 2011;12(9):853–60.
Wang Y, Kang W, Yang W, Zhang J, Li D, Zheng H. Structure of African swine fever virus and associated molecular mechanisms underlying infection and immunosuppression: a review. Front Immunol. 2021;12:715582 Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2021.715582/full.
Rodriguez TV, Moalli F, Polentarutti N, Paroni M, Bonavita E, Anselmo A, et al. Role of toll interleukin-1 receptor (IL-1R) 8, a negative regulator of il-1r/toll-like receptor signaling, in resistance to acute Pseudomonas aeruginosa lung infection. Infect Immun. 2012;80(1):100–9.
Borca MV, O’Donnell V, Holinka LG, Ramírez-Medina E, Clark BA, Vuono EA, et al. The L83L ORF of African swine fever virus strain Georgia encodes for a non-essential gene that interacts with the host protein IL-1β. Virus Res. 2018;249:116–23 Available from: https://linkinghub.elsevier.com/retrieve/pii/S0168170218301692.
Liu G, Park Y-J, Abraham E. Interleukin-1 receptor-associated kinase (IRAK) -1- mediated NF-κ activation requires cytosolic and nuclear activity. FASEB J. 2008;22(7):2285–96.
Zhu Z, Chen H, Liu L, Cao Y, Jiang T, Zou Y, et al. Classification and characterization of multigene family proteins of African swine fever viruses. Brief Bioinform. 2021;22(4):bbaa380.
Dixon LK, Chapman DAG, Netherton CL, Upton C. African swine fever virus replication and genomics. Virus Res. 2013;173:3–14.
Dixon LK, Chapman D. African Swine Fever Virus. In Encyclopedia of Virology. Elsevier; 2008. pp. 43–51. https://doi.org/10.1016/B978-012374410-4.00358-7.
Sánchez EG, Quintas A, Nogal M, Castelló A, Revilla Y. African swine fever virus controls the host transcription and cellular machinery of protein synthesis. Virus Res. 2013;173(1):58–75.
Münz C. Autophagy proteins in viral exocytosis and anti-viral immune responses. Viruses. 2017;9:288.
Münz C. Autophagy proteins in antigen processing for presentation on MHC molecules. Immunol Rev. 2016;272:17–27.
Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14(2):177–85.
Wu W, Lin C, Wu K, Jiang L, Wang X, Li W, et al. FUNDC 1 regulates mitochondrial dynamics at the ER –mitochondrial contact site under hypoxic conditions. EMBO J. 2016;35(13):1368–84.
Bahrami S, Drabløs F. Gene regulation in the immediate-early response process. Adv Biol Regul. 2016;62:37–49.
Carow B, Rottenberg ME. SOCS3, a Major Regulator of Infection and Inflammation. Front Immunol. 2014. https://doi.org/10.3389/fimmu.2014.00058.
Silk RN, Bowick GC, Abrams CC, Dixon LK. African swine fever virus A238L inhibitor of NF-κB and of calcineurin phosphatase is imported actively into the nucleus and exported by a CRM1-mediated pathway. J Gen Virol. 2007;88(2):411–9.
Abrams CC, Chapman DAG, Silk R, Liverani E, Dixon LK. Domains involved in calcineurin phosphatase inhibition and nuclear localisation in the African swine fever virus A238L protein. Virology. 2008;374(2):477–86.
Powell PP, Dixon LK, Parkhouse RM. An IkappaB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J Virol. 1996;70(12):8527–33.
Nefedeva MV, Titov IA, Mima KA, Malogolovkin AS. Analysis of the African swine fever virus immunomodulatory proteins. Mol Genet Microbiol Virol. 2019;34(1):42–9.
Hurtado C, Bustos MJ, Granja AG, de León P, Sabina P, López-Viñas E, et al. The African swine fever virus lectin EP153R modulates the surface membrane expression of MHC class I antigens. Arch Virol. 2011;156(2):219–34.
Beers C, Burich A, Kleijmeer MJ, Griffith JM, Wong P, Rudensky AY. Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J Immunol. 2005;174(3):1205–12.
Kaufman J. Antigen processing and presentation: evolution from a bird’s eye view. Mol Immunol. 2013;55(2):159–61 Available from: https://linkinghub.elsevier.com/retrieve/pii/S0161589012004440.
Kumar M, Makonchuk DY, Li H, Mittal A, Kumar A. TNF-like weak inducer of apoptosis (TWEAK) activates Proinflammatory signaling pathways and gene expression through the activation of TGF-β-activated kinase 1. J Immunol. 2009;182(4):2439–48.
Wen T, Bukczynski J, Watts TH. 4-1BB ligand-mediated Costimulation of human T cells induces CD4 and CD8 T cell expansion, cytokine production, and the development of Cytolytic effector function. J Immunol. 2002;168(10):4897–906.
Bishop KA, Wang X, Coy HM, Meyer MB, Gumperz JE, Pike JW. Transcriptional regulation of the human TNFSF11 gene in T cells via a cell type-selective set of distal enhancers. J Cell Biochem. 2015;116(2):320–30.
Westermann AJ, Barquist L, Vogel J. Resolving host–pathogen interactions by dual RNA-seq. PLoS Pathog. 2017;13:e1006033.
Eblé PL, Hagenaars TJ, Weesendorp E, Quak S, Moonen-Leusen HW, Loeffen WLA. Transmission of African swine fever virus via carrier (survivor) pigs does occur. Vet Microbiol. 2019;237:108345.
King DP, Reid SM, Hutchings GH, Grierson SS, Wilkinson PJ, Dixon LK, et al. Development of a TaqMan® PCR assay with internal amplification control for the detection of African swine fever virus. J Virol Methods. 2003;107(1):53–61.
Blome S, Gabriel C, Schmeiser S, Meyer D, Meindl-Böhmer A, Koenen F, et al. Efficacy of marker vaccine candidate CP7-E2alf against challenge with classical swine fever virus isolates of different genotypes. Vet Microbiol. 2014;169:8–17.
Machuka EM, Muigai AWT, Amimo JO, Entfellner J-BD, Lekolool I, Abworo EO, et al. Comparative Analysis of SLA-1, SLA-2, and DQB1 Genetic Diversity in Locally-Adapted Kenyan Pigs and Their Wild Relatives, Warthogs. Vet Sci. 2021;8:180 Available from: https://www.mdpi.com/1256148.
Andrews S. FastQC. Babraham Bioinformatics. 2010. http://www.bioinformatics.babraham.ac.uk/projects/. Accessed 20 Jan 2021.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20.
Bishop RP, Fleischauer C, de Villiers EP, Okoth EA, Arias M, Gallardo C, et al. Comparative analysis of the complete genome sequences of Kenyan African swine fever virus isolates within p72 genotypes IX and X. Virus Genes. 2015;50(2):303–9.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550.
Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. R Foundation for Statistical Computing. In R Foundation for Statistical Computing, Vienna, Austria. 2015;11. https://doi.org/10.7910/DVN/UZLXSZ. Accessed 30 Sept 2021.
Blighe K, Rana S, Lewis M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. In R-Package. 2019. pp. 1–8. https://github.com/kevinblighe/EnhancedVolcano.
Chen H, Boutros PC. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35.
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3.
Acknowledgments
The authors express gratitude to the ILRI-ASF research team for their support in implementing the project.
Funding
Funds for this study were provided by the Defense Threat Reduction Agency (DTRA) grant number HDTRA1–16-1-0050. The funder had no role in the design, analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
EMM, EAO, and R.P. conceived the experiment, contributed to the experimental design and edited the manuscript. EMM conducted the experiments, carried out data analysis, and wrote the manuscript, while J.J. performed the bioinformatics analysis and edited the manuscript. JAO and AWM contributed to the experimental design and edited the manuscript. All authors reviewed and approved the manuscript for publication.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All animal experiments reported in this study were approved by Institutional Animal Care and Use Committee (IACUC) and Institutional Biosafety Committee (IBC) of the International Livestock Research Institute (ILRI), Kenya (Reference: VN_IACUC-2011-04). All methods were carried out in accordance with the approved protocol and relevant regulations. All pigs were maintained, sampled, and euthanized humanely by the institutional veterinarian. All methods are reported in accordance with ARRIVE guidelines (https://arriveguidelines.org) for the reporting of animal experiments.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Machuka, E.M., Juma, J., Muigai, A.W.T. et al. Transcriptome profile of spleen tissues from locally-adapted Kenyan pigs (Sus scrofa) experimentally infected with three varying doses of a highly virulent African swine fever virus genotype IX isolate: Ken12/busia.1 (ken-1033). BMC Genomics 23, 522 (2022). https://doi.org/10.1186/s12864-022-08754-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-08754-8