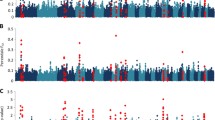
Abstract
Background
Gastrointestinal nematode infection (GNI) is the most important disease affecting the small ruminant industry in U.S. The environmental conditions in the southern United States are ideal for the survival of the most pathogenic gastrointestinal nematode, Haemonchus contortus. Host genetic variation for resistance to H. contortus allows selective breeding for increased resistance of animals. This selection process increases the prevalence of particular alleles in sheep and goats and creates unique genetic patterns in the genome of these species. The aim of this study was to identify loci with divergent allelic frequencies in a candidate gene panel of 100 genes using two different approaches (frequentist and Bayesian) to estimate Fst outliers in three different breeds of sheep and goats exposed to H. contortus.
Results
Our results for sheep populations showed SNPs under selection in C3AR1, CSF3, SOCS2, NOS2, STAT5B, TGFB2 and IL2RA genes using frequentist and Bayesian approaches. For goats, SNPs in CD1D, ITGA9, IL12A, IL13RA1, CD86 and TGFB2 genes were under selection. Common signatures of selection in both species were observed in NOS2, TGFB2 and TLR4 genes. Directional selection was present in all SNPs evaluated in the present study.
Conclusions
A total of 13 SNPs within 7 genes of our candidate gene panel related to H. contortus exposure were identified under selection in sheep populations. For goats, 11 SNPs within 7 genes were identified under selection. Results from this study support the hypothesis that resistance to H. contortus is likely to be controlled by many loci. Shared signatures of selection related to mechanisms of immune protection against H. contortus infection in sheep and goats could be useful targets in breeding programs aimed to produce resistant animals with low FEC.
Similar content being viewed by others
Background
Small ruminant industry in the US is a growing industry due to ethnic markets and increasing demand for organically produced livestock. Gastrointestinal nematode infection (GNI) is one of the most prevalent health problems in sheep and goats and represents a major productivity threat for small ruminants [1]. High disease incidence has been observed in the Southeast US regions [2] and infection with Haemonchus contortus is common throughout the year [3, 4]. This blood sucking parasite inhabits the abomasum of the host and it is responsible for weight loss, anemia and reduced performance [2].
Recent advances in genomic research have provided tools to unravel the genetics underlying phenotypic variation in complex traits [5], including resistance to GNI. Host genetic variation for GNI promises great opportunities for selective breeding of sheep and goats with increased resistance to H. contortus. Fecal egg count (FEC) is currently the method of choice to identify resistance to GNI and is the standard phenotypic measure to achieve rapid genetic progress [6]. Host resistance based on FEC is a heritable trait in both sheep and goats, with heritability estimates ranging between 0.01 to 0.65, and 0.1 to 0.33, respectively [7,8,9,10,11,12,13,14,15,16]. In accordance, breeding studies of small ruminants have revealed a FEC reduction after concurrent selective breeding of naturally resistant sheep to GIN infection [17,18,19].
Sheep and goats were the first livestock species to be domesticated by humans and were initially used mainly for meat, rather than wool or milk [20]. Natural selection and artificial selective breeding are the main driving forces shaping genetic variation across the sheep and goat genomes, and have gradually changed the phenotypes of these species. Within breeding strategies, selection increases the frequency of particular alleles at different loci in the population and creates unique genetic patterns in the DNA sequence that can be traced back and investigated for further analyses [21].
Two of the most used statistical methods for the analysis of signatures of selection are the detection of long haplotypes and the identification of differences in allele frequencies. The long haplotype detection method requires accurate allele assignment to one of the parental chromosomes (chromosome phasing) and ancestral allele identification, which sometimes can be a limitation when information about ancestors and pedigree is not available [22]. On the other hand, genetic differentiation among groups can be computed using the Fst method. This approach allows for identification of loci showing differences in allelic frequencies between two or more divergent populations, and therefore believed to be under selection. Highly genetic divergent loci between populations have more extreme Fst values (greater than 0.25) than low genetic divergent loci [23], and extreme Fst values are associated with either natural or artificial selection.
Using this approach for sheep, few loci have been identified as regions under selection for resistance or susceptibility to GNI [24], and in goats, information is even more scarce [25]. Some candidate markers within Ovar-DRA and Ovar-DRB genes have been identified as possible genetic markers associated with low H. contortus FEC in sheep and goat populations [26]. However, more knowledge is required to understand the genetic architecture underlying resistance against GNI in these species. Thus, the aim of this study was to identify immune loci (among a candidate gene panel of 100 genes) with divergent allelic frequencies in three different lines of sheep and goats, respectively, using the Fst statistic.
Results
Genotyping, quality control and population structure in sheep and goats
The sheep and goat DNA samples were sequenced with a median depth of 24x across 5000 probes (average of 50 probes/gene) The initial SNP data set consisted of 5346 SNPs for both sheep and goats. Only biallelic SNPs were identified in our populations. After quality control, the final SNP data set contained 1339 SNPs for sheep and 1020 SNPs for goats.
The plot from principal component analysis (PCA) for sheep (Fig. 1a), presented one specific cluster per breed. The first two principal components explained 25.6 and 18.4% of the total variance observed in sheep, respectively. For goats, the PCA plot (Fig. 1b) clustered the animals within breed and one specific cluster was observed per breed. PC1 and PC2 explained 21.7 and 17.2% of the total variance observed in goats, respectively.
Sheep and goat PCA plots. The PCA plots show the population structure of the breeds under the study based on the first two principal components (PC). For sheep (a), the PC1 and PC2 explain 25.6 and 18.4% of the total variation, respectively. For goats (b), the PC1 and PC2 explain 21.7 and 17.2% of the total variation observed in the populations, respectively
FEC descriptive statistics in sheep and goats
Dorper sheep had the highest FEC (1475 ± 207.4 eggs per gram of feces) across breeds with Katahdin (1087 ± 191.2) and St. Croix (969 ± 180.6) sheep, which were considered resistant in this study, presenting lower FEC. Thus, Katahdin and St. Croix breeds had 26.3 and 34.3% less eggs per gram of feces than Dorper sheep, respectively.
For goats, Boer goats had 1548 ± 173.1 eggs per gram of feces. Kiko (936 ± 159.1) and Spanish (911 ± 150.9), categorized as resistant breeds, had 39.5 and 41.1% less eggs per gram of feces than Boer goats, respectively.
Signatures of selection using Fst in sheep
A total of 18 different SNPs in CCR3, CD86, EPS15, TLR4, IL2RB, STAT2, C3AR1, SOCS2, TLR10, NOS2, CSF3, STAT5B, TGFB2, LAMC1, IL2RA and IL12RA1 genes were identified under selection using the frequentist Fst (Additional file 1: Figure S1, Additional file 2: Table S1). For Bayesian Fst, 14 SNPs were observed under selection in C3AR1, LTBR, SOCS2, CSF3, NOS2, STAT5B, TGFB2, IL2RA, and TLR7 genes (Figure 2, Additional file 3: Table S2). Using this approach, Fst values greater than 0.20 were observed in the sheep populations and within OAR 3, 11, 12, 13 and X. The sign of alpha was always positive which indicates that, in all cases, directional selection was present in the SNPs under selection.
Manhattan plots of the Bayesian Fst values per SNP from targeted genomic regions in sheep. The SNP data is ordered based on chromosomal location (x-axis). The plots includes: Katahdin and St. Croix vs Dorper (KS vs D, pooled resistant breeds vs susceptible breed), Katahdin vs Dorper (K vs D, resistant vs susceptible breed), St. Croix vs Dorper (S vs D, resistant vs susceptible breed) and St. Croix vs Katahdin (S vs K, resistant vs resistant breed) analyses. The genes with SNPs under selection in each analysis are presented in a box on the right side of Manhattan plots
Shared signatures of selection between both approaches were identified in C3AR1, SOCS2, NOS2, CSF3, TGFB2, IL2RA and STAT5B genes (Table 1 and Figure 3). For Katahdin and St. Croix vs Dorper analysis (Table 1), OAR 11 and 12 and 13 contained extreme Fst values in CSF3, NOS2, TGFB2 and IL2RA genes. The NOS2 and IL2RA genes were the genomic regions with most loci under selection. The SNPs (OAR11:18963484 and OAR11:18963494) within exon 7 and 16 of NOS2 gene (A/G and T/C) are synonymous mutations.
For the Katahdin vs Dorper analysis (Table 1), 8 SNPs under selection were observed in SOCS2, NOS2, TGFB2 and IL2RA genes within OAR3, 11, 12 and 13, respectively. The SNPs in NOS2, TGFB2, and IL2RA genes were also observed with high genetic differentiation in the Katahdin and St. Croix vs Dorper analysis. The OAR3:129558034 (SOCS2), OAR11:18963484 (NOS2), and OAR11:18963494 (NOS2) are synonymous mutations.
For the St. Croix vs Dorper analysis (Table 1), 5 SNPs within OAR 3, 11, 12 and 13 were observed under selection in untranslated and intronic regions. The genes showing high genetic differentiation were C3AR1, CSF3, STAT5B, TGFB2, and IL2RA. The highest Fst value (0.34 for BayeScan and 0.48 for R software) was found in a SNP (OAR3:206099209) located in the 5’UTR region of CSF3gene. The SNPs (OAR11:39857496, OAR12:19965761, and OAR13:10442920) in CSF3, TGFB2 and IL2RA genes were also observed under selection in the Katahdin and St. Croix vs Dorper analysis.
Signatures of selection using Fst in goats
For goats, genes within CHR 1, 3, 8, 16, 19 and 22 contained 13 SNPs under selection using the frequentist Fst (Additional file 4: Table S3, Additional file 1: Fig. S1). Using this approach, genes with loci under selection in goat populations were IL12A, TLR4, TGFB2, ITGA9, CD86, CD1D, NOS2, and IL13RA1. Signatures of selection detected with the Bayesian Fst were identified in 11 SNPs harboring IL12A, TLR4, IL33, TGFB2, ITGA9, CD86, CD1D, and IL13RA1 genes (Additional file 5: Table S4, Figure 4). All the SNPs were under directional selection and located in exonic, intronic and untranslated regions (Figure 4).
Manhattan plots of the Bayesian Fst values per SNP from targeted genomic regions in goats. The SNP data is ordered based on chromosomal location (x-axis). The plots includes: Kiko and Spanish vs Boer (KS vs B, pooled resistant breeds vs susceptible breed), Kiko vs Boer (K vs B, resistant vs susceptible breed), Spanish vs Boer (S vs B, resistant vs susceptible breed) and Spanish vs Kiko (S vs K, resistant vs resistant breed) analyses. The genes with SNPs under selection in each analysis are presented in a box on the right side of Manhattan plots
Information regarding shared SNPs under selection in goats using both frequentist and Bayesian approaches is presented in Table 2 and Figure 3. For the Kiko and Spanish vs Boer analysis (Table 2), IL12A, TLR4 and ITGA9 genes had 4 SNPs under selection. The highest Fst value was observed in the CHR8:106725462 (TLR4). The CHR8:106725462 and CHR8:106725265 in exon 3 of TLR4 gene are synonymous mutations.
For the Kiko vs Boer analysis (Table 2), high genetic differentiation was observed in a SNP (CHR1:66217253) located in an intronic region of CD86 gene. For the Spanish vs Boer analysis (Table 2), CD1D, TGFB2, ITGA9 and IL13RA1 genes contained 4 SNPs under selection. The SNP (T/G) observed under selection in exon 2 of CD1D gene generates a synonymous mutation. Loci under selection in TGFB2, ITGA9 and IL13RA1 genes were identified in the untranslated regions.
Finally, for the Spanish vs Kiko analysis (Table 2), 5 SNPs within TLR4, and ITGA9 genes were observed under selection. The majority of the SNPs with divergent allelic frequencies were located in TLR4 gene and within exon 3 and 4. Two (CHR8:106725462 and CHR8:106725265) out of 4 of the SNPs in TLR4 gene were also observed under selection in the Kiko and Spanish vs Boer analysis. The CHR22:11106216 was also under directional selection in the Kiko and Spanish vs Boer analysis, and in the Spanish vs Boer analysis.
Genomic regions under selection in both species
After examination of the Fst results per species, several loci in TGFB2, NOS2, and TLR4 genes were observed under selection in both species and are presented in Table 3 and Figure 3.
Discussion
Domestication, breed formation, and selective breeding leave detectable patterns within the genome of livestock species such as sheep and goats. Identification of these genomic patterns in the DNA sequence could help to identify of genes controlling resistance to H. contortus or other gastrointestinal parasites. Several studies have attempted to identify the genetic variation controlling gastrointestinal parasite resistance in sheep and goats by using SNP markers and Genome Wide Association Studies (GWAS) but few research studies has been devoted to identify signatures of selection for GNI resistance in these species [27,28,29]. Signatures of selection for resistance to GNI have not been identified in goats, and for sheep, only Perendale and Romney breeds have been evaluated [24]. In the present study, we unravel signatures of selection using a targeted sequencing approach in three different breeds of sheep and goats. The SNPs potentially under selection identified in this study spanned a myriad of candidate genes related to immune response and cellular mechanisms against H. contortus.
In our study, all the signatures of selection identified in sheep and goats were under directional selection. Directional selection is one of the primary cause of phenotypic diversification and has been used to increase the frequency of favorable additive alleles [30]. This selection process has not exhausted the genetic variation for most economically important traits in livestock [31]. Our results suggest that some of the SNPs in genes related to resistance to GNI are under directional selection. This could be possible due to selection for resistance to GNI is focused on resistant individuals, and susceptible animals are usually removed from the flock or not used in the mating process.
Signatures of selection in sheep populations
For Katahdin vs Dorper, and Katahdin and St. Croix vs Dorper analyses, loci within SOCS2, NOS2, TGFB2 and IL2RA genes were observed under selection. The SOCS2 gene has been previously associated with FEC in Dorper x Red Maasai sheep using GWAS and expression of this candidate gene was observed in abomasal tissue, mesenteric lymph nodes, and Peyer’s patches from ewes and lambs [27]. Thus, it is possible that SOCS2 gene could be used as candidate gene for future studies to validate previous and current results in Dorper and Dorper × Red Maasai sheep.
The SOCS2 gene is a broad key regulator of cytokine responses, including IL2, IL3, IL4, IFN-γ, CSF, and Jak-STAT signaling pathways in bone marrow and T cells [32]. Studies on mice infected with Tripanozoma cruzi have shown that expression of SOCS2 facilitates inflammatory and immune responses to prevent myocardial dysfunction but increases parasitemia [33]. On the contrary, SOCS2−/− mice infected with Schistosoma mansoni expressed increased Th2 response with higher IgE and eosinophil production than SOCS2+/+ mice [34]. Also, SOCS2−/− mice have shown increased body weight and gigantism possibly due to downregulation of growth hormone and insulin-like growth factor-I (IGF-I) signaling [35]. In Scottish Blackface sheep infected with Teladorsagia circumcincta, SOCS2 gene was found differentially expressed between resistant and control animals [36].
Nitrogen oxygen synthase 2 or NOS2 is a key molecule involved in Th1 response. It participates in the production of nitric oxide to kill invading microbes in phagocytes during classical macrophage activation by IFN-γ and TNF-α. Differential expression of this gene has been observed in the abomasum of Merino sheep during H. contortus challenge [37]. In that study, mRNA expression NOS2 gene was downregulated in susceptible individuals. While there is a proposed interplay between Th1, Th2, and Treg responses during GNI [38], susceptibility to these infections has been related to Th1 and Th17 responses, and Th2 has been associated with resistance to helminth infections in sheep [39].
TGFB2 protein has been reported as an anti-inflammatory cytokine, and was observed in high concentration in the gut mucosa of sheep after H. contortus infection [40]. In pigs, PAS1, a product of Ascaris suum, induces IL-10 and TGFB2 production in macrophages and has been related to loss of pro-inflammatory cytokine production [41]. In humans and animal models, it has been shown that inhibition of T-cell proliferation might be triggered by an increase of IL-10 and TGFB production in antigen presenting cells or T-cells as a result of down-modulatory molecules that are released by the parasites to enhance survival [42]. Thus, parasites are prone to use IL-10 and TGFB to downregulate host immune response.
IL2RA protein is mainly expressed in CD4+ Treg cells and it constitutes one of the three subunits of the IL2R. In humans, induction of Treg cells increases during natural and long term gastrointestinal nematode infections [43, 44]. In sheep, expression of the mRNA of IL2RA gene in the abomasum has been related to subsequent H. contortus infections in resistant sheep while its expression in the jejunal mucosa has been linked to susceptibility of Trichostrongylus columbriformis [37]. Thus, differential expression between susceptible and resistant individuals could depend on the stage of the host immune response, the infection period, as well as the nematode species.
For St. Croix vs Dorper, and Katahdin and St. Croix vs Dorper analyses, SNPs in C3AR1, CSF3, STAT5B, TGFB2 and IL2RA genes were found to be under selection. C3AR1 protein plays an important role during innate immune responses. It is part of the complement cascade. Reduced T cell responses has been observed when host animals lack of C3AR [45]. Recent work in mice, using bone marrow transplant and RNA Seq analysis, identified that signaling by C3AR mediates macrophage recruitment after induced injury with cardiotoxin and muscle regeneration [46]. The exact role of C3AR1 has not been evaluated in sheep during gastrointestinal nematode infections, but some studies suggest that the complement activation is one of the first mechanisms of protection against helminth infections [47] and classical and alternative complement pathways can be activated in resistant sheep to H. contortus [48].
The STAT5 gene can be activated by many cytokines such as GM-CSF and thymic stromal lymphopoietin (TSLP) in the dendritic cells. Activation of STAT5 by TSLP has been shown to trigger Th2 responses at barrier surfaces [49]. Also, STAT5 signaling has been related to many biological processes, such as TCR signaling and basal proliferation of naïve CD4+ T cells [50]. Moreover, STAT5B mediates the signal transduction of IL2, IL4, CSF1, and different growth hormones. Thus, it is possible that STAT5B gene is responsible for many cellular functions during H. contortus exposure and further analysis is required to confirm our results.
Signatures of selection in goat populations
For many years, there has been a debate about the immune mechanisms controlling H. contortus infections in sheep and goats. The same helminth species can parasitize both species but previous studies suggest higher levels of infection in goats [51].
For Kiko and Spanish vs Boer, genes with genetic differentiation were IL12A, TLR4 and ITGA9. IL12 protein is a major cytokine that controls the maturation of CD4+ T cells into Th1 cells and promotes IFNG production in response to intracellular parasites. IL12 protein is composed of an alpha chain (p35 subunit) and a beta chain (p40 subunit) linked by a disulfide bond [52]. In Nelore cattle, some studies have suggested that susceptibility to gastrointestinal parasites is associated with an increase of Th1 response with high elevated worm burden and elevated IFNG and IL12 production [53].
Toll-like receptors (TLR) are vital for the detection of invading pathogens and are commonly expressed in antigen presenting cells and other immune cells [54]. In resistant sheep infected with H. contortus and T. colubriformis, upregulation of several TLR genes, including TLR4, was observed in the abomasum. In the same study, susceptible individuals presented lower expression of this gene [37]. Contrary to sheep, susceptible Angus yearlings infected with Ostertagia, Cooperia and Nematodirus spp., TLR4 showed higher expression in the mesenteric lymph nodes [55]. In goats, increased expression of TLR4 gene in blood has been observed during inclusion of Sericea lespedeza in the diet [56]. This observation could be related to biologically active tannin fractions from plants containing tannins such as S. lespedeza. Several studies have shown plant tannins are able to modulate the innate immune response and act as γ-T cell agonists [56, 57].
The ITGA9 gene encodes an alpha integrin that compose the integral membrane glycoproteins that mediates cell-cell and cell matrix adhesion. In resistant Merino sheep infected with H. contortus, transcriptome analysis results revealed ITGA9 gene as part of an enriched gene set related to the extracellular matrix receptor interaction pathway [58]. The exact role of ITGA9 gene in goats is unknown, but further analysis could help to understand possible mechanisms of protection against H. contortus and other gastrointestinal parasites.
For Kiko vs Boer, a SNP in CD86 gene was observed under selection. This gene encodes a membrane bound protein in antigen presenting cells that binds CD28 and CTLA-4 proteins localized in the T cell membrane. Binding with C28 leads to T cell proliferation and cytokine production, while binding with CTLA-4 negatively regulates T cell response [59]. Thus, it is possible that CD86 controls some T cell mechanisms in goats.
For Spanish vs Boer analysis, signatures of selection were identified in CD1D, TGFB2, ITGA9 and IL13RA1 genes. CD1D is a major histocompatibility complex class I related protein that regulates presentation of glycolipids antigens to natural killer T cells [60]. In resistant cattle naturally exposed to gastrointestinal parasites, CD1D was upregulated in the mesenteric lymph nodes [55]. In goats, the role of CD1D has not been studied but it is possible that this gene could play an important role during presentation of glycoproteins from H. contortus to T cells.
As observed in sheep, TGFB2 could be used by H. contortus to promote infection. In tropical cattle, susceptibility to Theileria annulata has been associated with TGFB2 induction and increased TGF-b2 production by Theileria-infected macrophages promote invasiveness [61]. In sheep, TGFB-like molecules have been identified in larvae from H. contortus and T. circumcincta [62]. Thus, activation of TGFB and TGFB-like molecules from gastrointestinal parasites could control downregulation of the immune response. The exact role of TGFB2 in goats is unclear and more research is needed to understand its function during H. contortus infections.
IL13RA1 subunit together with IL4RA can form a functional receptor for IL13 [63]. In goats, no evidence of IL13RA1 function has been reported but in Hereford Shorthorn cattle infected with Boophilus microplus, results showed that IL13RA1 precursor was differentially expressed [64].
Finally for Spanish vs Kiko, TLR4 and ITGA9 had SNPs under selection. For this analysis, the same SNP identified under selection in ITGA9 gene in Kiko and Spanish vs Boer analysis was observed. For TLR4, 2 more SNPs in exon 4 were identified under selection. Thus, it seems that for goats, TLR4 and ITGA9 genes could play important roles during H. contortus infection.
Common signatures of selection in sheep and goats
During the last two decades, results have shown differences in feeding behavior and gastrointestinal nematode parasitism between sheep and goats. Feeding behavior is one important aspect that differentiates sheep and goats. Sheep are typically raised in grazing systems with parasites commonly found within the pastures, and have to counteract the negative effects of GNI by developing an effective immune response. Goats are common browsers which allows them to rely less on immune response mechanisms [65].
For many years, there has been a question of the importance of immune effector molecules and the mucosal response in goats during GNI. Our findings suggest a possible interplay between Th1 and Th2 responses with conserved breed specific mechanisms. For both species, our results suggest a possible interplay between Th1 and Th2 response during GNI, as previously described by Hassan et al. [39] and Pernthaner et al. [66].
One of the most interesting findings from this study is the identification of shared immune response mechanisms between sheep and goats (Fig. 3). It is possible that some immune response mechanisms are shared between both species and induce an effective immune response against H. contortus. The NOS2, TGFB2, and TLR4 genes, observed under selection in both species, are key modulators of Th1 and Th2 responses, and active players of antigen recognition. Several studies in cashmere and dairy goats have evaluated the responsiveness of resistant animals to GNI and have found a negative correlated response between worm counts and eosinophil, mast cell, and globule leucocyte counts [67,68,69]. In sheep, similar cellular immune response has been associated to GNI [70, 71]. Thus, it is possible that some mechanisms of immune protection are shared between these species but more studies are required to understand these events during H. contortus infection.
The conserved mechanisms of protective response against H. contortus are most likely to be useful targets in the development of alternative nematode control strategies in both species, as they can be widely applied in production systems. For this reason our future efforts will focus on validation of the results observed in the present study to unravel genetic mechanisms used for controlling H. contortus or other GNI between sheep and goats.
Conclusion
Results from this study support the hypothesis that resistance to gastrointestinal parasites such as H. contortus is likely to be controlled by many loci. Different immune response mechanisms between sheep and goats are used to control H. contortus but some aspects are shared in both species. Shared mechanisms of immune protection could be useful targets in breeding programs aimed to produce resistant animals and future research is necessary to validate our findings.
Methods
Animal populations
The research protocol for the present study was approved by the Langston University Animal Care and Use Committee. Sire candidates were randomly selected in the first year from five commercial farms in Arkansas (CWC Farm), Kansas (Hogan Ranch), and Missouri (CMI Dorpers, Thousand Oak Ranch and Persimmon Creek Ranch) and American Institute for Goat Research at Langston University, Oklahoma and transferred to Langston University for a central sire performance test with an artificial H. contortus infection described later. In the second and third years, young male offspring of resistant or moderate resistant breeding groups to GIN were tested with the artificial H. contortus infection. Sheep and goats were grouped per breed and species in adjacent pens with automated feeders allowing free-choice access to a 15% crude protein diet at Langston University. Overall, 145 offspring sheep from Dorper (n = 48), Katahdin (n = 57), and St. Croix (n = 40) breeds and 144 offspring goats from Boer (n = 52), Kiko (n = 44) and Spanish (n = 48) breeds were used in this study.
Deworming and H. contortus artificial infection methods are described in a previous publication [25]. Briefly, sheep and goats were treated with albendazole (Valbazen®; 10 and 20 mg per kg of body weight, respectively) and levamisole (Prohibit®; 12 and 18 mg per kg of body weight, respectively) during 2 weeks of adaptation. Then, animals were screened for FEC reduction (< 100 epg) and received an oral dose of 10,000 L3 larvae of H. contortus. FEC was recorded at 28, 35 and 42 days post-infection. Animals returned to commercial farms at the end of the study.
Genotyping and data quality control
Blood samples from sheep and goats were collected by puncture of the jugular vein using vacutainer tubes with anticoagulant EDTA. Subsequently, genomic DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) according the manufacturer’s instructions and stored at − 20 °C. The DNA yield was calculated from a spectrophotometric measurement at 260 nm (NanoDrop-1000, Thermo Scientific), and the purity was assessed using a ratio 260/280 nm.
Two hundred and fifty ng/μL of genomic DNA was genotyped using Capture-Sequencing by RAPiD Genomics (Gainesville, Florida) to target 100 genes related to the immune response against H. contortus or other GNI. The candidate gene panel was selected for targeted sequencing based on results from previous studies in sheep [55, 72,73,74,75,76] and goats [77]. In addition, genes related to the immune response against H. contortus and other GNI were considered as candidates for targeted sequencing (Additional file 6: Table S5).
The Oar_v4.0 reference genome available at the National Center for Biotechnology Information (NCBI) genome browser was used to design biotinylated 120-mer probes that captured sequences at each target locus. For library preparation, Nextera tagmentation protocol from Illumina was used. Then, biotin-labeled probes hybridized denatured libraries and streptavidin-coated beads were used to capture the probe-library complex. Streptavidin-coated beads were magnetically pulled down and DNA fragments were eluted. Libraries were captured by complimentary surface bound oligos and library amplification was performed using bridge amplification according to Illumina’s guidelines. The probe set used for sequence capture contained 5000 probes representing 100 genes. Target enriched libraries were sequenced using the Illumina HiSeq 3000 PE100 platform to generate 2 × 101 bp paired-end reads.
Data was demultiplexed using bcl2fastq conversion software from Illumina, cleaned, and trimmed. The 3′ ends were trimmed and low quality bases with < 20 Phred quality score reads were removed. Clean reads were mapped to the sheep (Oar_v4.0) and goat (ASM170441v1) reference genomes with MOSAIK software [78]. Freebayes was used for identification of SNPs and VCFtools [79] was used to generate VCF files. Samples were filtered based on maximum missing count [3], minimum number of alleles [2], mean read depth (750), call rate (< 95%) and MAF (≤ 0.01). Thus, SNPs with a call rate < 95% and MAF ≤ 0.01 were removed.
Principal component analysis plots were generated to illustrate population structure using JMP Genomics 9 software from SAS (SAS Institute Inc., Cary, NC). Individuals included in the principal component analysis and further Fst analysis were selected based on the identity by state threshold of ≤0.5. For these analyses, one hundred and twenty sheep from Dorper (n = 40), Katahdin (n = 40), and St. Croix (n = 40) breeds and 129 goats from Boer (n = 43), Kiko (n = 43) and Spanish (n = 43) breeds were used.
Identification of signatures of selection using allele frequencies
Signatures of selection were identified using Fst statistic at each SNP using frequentist and Bayesian approaches which are focused on the identification of differences in allele frequencies between subpopulations. To identify genetic divergence between subpopulations, analyses between resistant and susceptible breeds within species were carried out. St. Croix and Katahdin were considered resistant sheep breeds and were compared against Dorper which was considered the susceptible sheep breed. Analyses were performed as follow: Katahdin and St. Croix vs Dorper (KS vs D, pooled resistant breeds vs susceptible breed), Katahdin vs Dorper (K vs D, resistant vs susceptible breed), and St. Croix vs Dorper breeds (S vs D, resistant vs susceptible breed). Similarly, for goats, Spanish and Kiko breeds were classified as resistant and compared against the susceptible Boer breed. Identification of signatures of selection in goats between resistant and susceptible breeds were performed as follows: Kiko and Spanish vs Boer (KS vs B, pooled resistant breeds vs susceptible breed), Kiko vs Boer (K vs B, resistant vs susceptible breed), and Spanish vs Boer (S vs B, resistant vs susceptible).
To identify any signatures of selection different in the two resistant breeds, an additional analysis was performed per species by comparing the St. Croix against Katahdin (S vs K, resistant vs resistant breed) for sheep, and Spanish against Kiko (S vs K, resistant vs resistant breed) for goats.
For the frequentist Fst, calculation of average allele frequency across breeds, estimation of total variance, and deviation of each population from mean and Fst computation were performed using the R software and the R codes from Gondro et al. [22]. The formula used to estimate Fst values was the following:
where σ2subpopulation is the variance of the deviation of each population from mean and σ2total is the total variance. Estimates corresponding to the 1% extreme Fst values were used to define a significance threshold and identify regions under selection.
Bayesian Fst was estimated using BayeScan software. In this approach, a Bayesian likelihood method implemented via reversible jump Markov Chain Monte Carlo (MCMC) was used which assumes that allele frequencies follow a Dirichlet distribution [80]. The main advantage of this approach is that Fst statistic is modelled using logistic regression methods by decomposing locus–population Fst coefficients into a population-specific component (beta), shared by all loci and a locus-specific component (alpha) shared by all the populations. Then, departure from neutrality at a given locus is assumed when the locus-specific component is necessary to explain the observed pattern of diversity (alpha significantly different from 0). Diversifying selection can be assumed if positive values of alpha are observed, whereas negative alpha values suggest balancing or purifying selection. Consequently, two alternative models are generated for each locus, including or not the alpha component to model selection. BayeScan software uses a reversible-jump MCMC algorithm to estimate the posterior probability of these models [80,81,82]. For the Markov chain Monte Carlo algorithm implemented in BayeScan, 20 pilot runs of 5000 iterations were used to adjust the proposal distribution to have acceptance rates between 0.25 and 0.45 for the runs. Then, a burn-in of 10,000 iterations followed by 100,000 iterations were used for estimation [80,81,82].
Candidate SNPs under selection were located using the sheep (Oar_v4.0) and goat (ASM170441v1) reference genomes from NCBI. The Online Mendelian Inheritance in Man website and scientific literature were used to determine gene function.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available on NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject), accession number PRJEB32310 (sheep) and PRJEB32312 (goat).
Abbreviations
- CHR:
-
Chromosome number in goat
- FEC:
-
Fecal egg count
- GIN:
-
Gastrointestinal nematodes
- OAR:
-
Chromosome number in sheep
References
Terrill TH, Miller JE, Burke JM, Mosjidis JA, Kaplan RM. Experiences with integrated concepts for the control of Haemonchus contortus in sheep and goats in the United States. Vet Parasitol. 2012;186(1–2):28–37. https://doi.org/10.1016/j.vetpar.2011.11.043.
Miller JE, Bahirathan M, Lemarie SL, Hembry FG, Kearney MT, Barras SR. Epidemiology of gastrointestinal nematode parasitism in Suffolk and Gulf Coast native sheep with special emphasis on relative susceptibility to Haemonchus contortus infection. Vet Parasitol. 1998;74(1):55–74. https://doi.org/10.1016/S0304-4017(97)00094-0.
Uhlinger C, Fleming S, Moncol D. Survey for drug-resistant gastrointestinal nematodes in 13 commercial sheep flocks. JAVMA. 1992;201:77–80.
Mortensen LL, Williamson LH, Terrill TH, Kircher RA, Larsen M, Kaplan RM. Evaluation of prevalence and clinical implications of anthelmintic resistance in gastrointestinal nematodes in goats. JAVMA. 2003;223:495–500.
Andersson L. How selective sweeps in domestic animals provide new insight into biological mechanisms. J Intern Med. 2012. https://doi.org/10.1111/j.1365-2796.2011.02450.x.
Zvinorova PI, Halimani TE, Muchadeyi FC, Matika O, Riggio V, Dzama K. Breeding for resistance to gastrointestinal nematodes - the potential in low-input/output small ruminant production systems. Vet Parasitol. 2016. https://doi.org/10.1016/j.vetpar.2016.05.015.
Baker RLB, Reynolds L, Kassi AL, Rege JEO, Bekelye T, Mukassa-Mugerwa E, Rey B. Prospects for breeding for resistance to endoparasites in small ruminants in Africa- a new ILCA research programme. Small Ruminant Research and Development in Africa. Proceedings of the Second Biennial Conference of the African Small Ruminant Research Network. 1994; 223. ILRI (aka ILCA and ILRAD).
Baker RL. Genetic resistance to endoparasites in sheep and goats. A review of genetic resistance to gastrointestinal nematode parasites in sheep and goats in the tropics and evidence for resistance in some sheep and goat breeds in sub-humid costal Kenya. Anim. Genet Resour Inf. 1998;24:13–30.
Beraldi D, McRae AF, Gratten J, Pilkington JG, Slate J, Visscher PM, Pemberton JM. Quantitative trait loci (QTL) mapping of resistance to strongyles and coccidia in the free-living Soay sheep (Ovis aries). Int J Parasitol. 2007;37:121–9.
Gauly M, Kraus M, Vervelde L, Van Leeuwen MAW, Erhardt G. Estimating genetic differences in natural resistance in Rhön and Merinoland sheep following experimental Haemonchus contortus infection. Vet Parasitol. 2002;106:55–67.
Mandonnet N, Aumont G, Fleury J, Arquet R, Varo H, Gruner L, Bouix J, Khang JV. Assessment of genetic variability of resistance to gastrointestinal nematode parasites in creole goats in the humid tropics. J Anim Sci. 2001;79:1706–12.
Mandonnet N, Menendez-Buxadera A, Arquet R, Mahieu M, Bachand M, Aumont G. Genetic variability in resistance to gastro-intestinal strongyles during early lactation in creole goats. Anim Sci. 2006;82:283–7.
Pollot GE, Karlsson LJE, Eady S, Greeff JC. Genetic parameters for indicators of host resistance to parasites from weaning to hogget age in merino sheep. J Anim Sci. 2004;82:2852–64.
Vagenas D, Jackson F, Russel AJF, Merchant M, Wright A, Bishop SC. Genetic control of resistance to gastro-intestinal parasites in crossbred cashmere-producing goats: responses to selection, genetic parameters and relationships with production traits. Anim Sci. 2002;74:199–208.
Vanimisetti HB, Andrew SL, Zajac AM, Notter DR. Inheritance of fecal egg count and packed cell volume and their relationship with production traits in sheep infected with Haemonchus contortus. J Anim Sci. 2004;82:1602–11.
Stear MJ, Boag B, Cattadori I, Murphy L. Genetic variation in resistance to mixed, predominantly Teladorsagia circumcincta nematode infections of sheep: from heritabilities to gene identification. Parasite Immunol. 2009;31:274–82.
Bisset SA, Morris CA, McEwan JC, Vlassoff A. Breeding sheep in New Zealand that are less reliant on anthelmintics to maintain health and productivity. NZ Vet J. 2001;49.
Eady SJ, Woolaston RR, Mortimer SI, Lewer RP, Raadsma HW. Resistance to nematode parasites in Merino sheep: sources of genetic variation. Aust J Agric Res. 1996:47.
McEwan JC, Dodds KG, Greer GJ, Bain WE, Duncan SJ, Wheeler R, et al. Genetic estimates for parasite resistance traits in sheep and their correlations with production traits. NZ J Zool. 1995;22:177.
Chessa B, Pereira F, Arnaud F, Amorim A, Goyache F, Mainland I, et al. Revealing the history of sheep domestication using retrovirus integrations. Science. 2009. https://doi.org/10.1126/science.1170587.
Qanbari S, Pausch H, Jansen S, Somel M, Strom TM, Fries R, et al. Classic selective sweeps revealed by massive sequencing in cattle. PLoS Genet. 2014;10(2):e1004148. https://doi.org/10.1371/journal.pgen.1004148.
Gondro C, van der Werf J, Hayes B. In: genome-wide association studies and genomic prediction. LLC: Methods in Molecular Biology. Springer Science Business Media; 2013. p. 423–36. https://doi.org/10.1007/978-1-62703-447-0.
Purfield DC, McParland S, Wall E, Berry DP. The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. PLoS One. 2017; doi.org:https://doi.org/10.1371/journal.pone.0176780.
McRae KM, McEwan JC, Dodds KG, Gemmell NJ. Signatures of selection in sheep bred for resistance or susceptibility to gastrointestinal nematodes. BMC Genomics. 2014. https://doi.org/10.1186/1471-2164-15-637.
Onzima RB, Upadhyay MR, Doekes HP, et al. Genome-wide characterization of selection signatures and runs of Homozygosity in Ugandan goat breeds. Front Genet. 2018;9:318. https://doi.org/10.3389/fgene.2018.00318.
Estrada-Reyes, Z. M., Tsukahara, Y., Goetsch, A. L., Gipson, T. A., Sahlu, T., Puchala, R., et al. Effect of Ovar-DRA and Ovar-DRB1 genotype in small ruminants with haemonchosis. Parasite Immunol. 2018; doi.org:https://doi.org/10.1111/pim.12534.
Kemper KE, Emery DL, Bishop SC, Oddy H, Hayes BJ, Dominik S, et al. The distribution of SNP marker effects for faecal worm egg count in sheep, and the feasibility of using these markers to predict genetic merit for resistance to worm infections. Genet Res. 2011. https://doi.org/10.1017/S0016672311000097.
Benavides MV, Sonstegard TS, Kemp S, Mugambi JM, Gibson JP, Baker RL, et al. Identification of novel loci associated with gastrointestinal parasite resistance in a red Maasai × Dorper backcross population. PLoS One. 2015. https://doi.org/10.1371/journal.pone.0122797.
Silva FF. Bambou JC. Barbier C, Fleury J, Machado T, et al. Genome wide association study reveals new candidate genes for resistance to nematodes in Creole goat. Small Rumin. Res: Oliveira JA; 2018. https://doi.org/10.1016/j.smallrumres.2018.06.004.
Rieseberg LH, Widmer A, Arntz AM, Burke JM. Directional selection is the primary cause of phenotypic diversification 2002; 99(19): 12242–12245.
Hill WG, Kirkpatrick M. What animal breeding has taught us about evolution? 2010; 41: 1–19. In Futuyma DJ, Shafer HB, Simberloff D. Eds. Annual review of ecology, evolution, and systematics. Annual reviews, Palo Alto.
Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, et al. A family of cytokine-inducible inhibitors of signaling. Nature. 1997. https://doi.org/10.1038/43206.
Esper L, Roman-Campos D, Lara A, Brant F, Castro LL, Barroso A, et al. Role of SOCS2 in Modulating Heart Damage and Function in a Murine Model of Acute Chagas Disease. Am. J. of Pathol. 2012; doi.org:https://doi.org/10.1016/j.ajpath.2012.03.042.
Knosp CA, Carroll HP, Elliott J, Saunders SP, Nel HJ, Amu S, et al. SOCS2 regulates T helper type 2 differentiation and the generation of type 2 allergic responses. J. Exp. Med. 2011; doi: https://doi.org/10.1084/jem.20101167 PMID: 21646394.
Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA, et al. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature. 2000; doi.org:https://doi.org/10.1038/35016611.
Gossner A, Wilkie H, Joshi A, Hopkins J. Exploring the abomasal lymph node transcriptome for genes associated with resistance to the sheep nematode Teladorsagia circumcincta. Vet. Res. 2013; doi: https://doi.org/10.1186/1297-9716-44-68 PMID: 23927007.
Ingham A, Reverter A, Windon R, Hunt P, Menzies M. Gastrointestinal nematode challenge induces some conserved gene expression changes in the gut mucosa of genetically resistant sheep. Int J Parasitol. 2008. https://doi.org/10.1016/j.ijpara.2007.07.012.
Maizels RM, McSorley HJ. Regulation of the host immune system by helminth parasites. J Allergy Clin Immunol. 2016. https://doi.org/10.1016/j.jaci.2016.07.007.
Hassan M, Hanrahan JP, Good B, Mulcahy G, Sweeney T. A differential interplay between the expression of Th1/Th2/Treg related cytokine genes in Teladorsagia circumcincta infected DRB1*1101 carrier lambs. Vet Res. 2011. https://doi.org/10.1186/1297-9716-42-45.
Robinson N, Pleasance J, Piedrafita D, Meeusen EN. The kinetics of local cytokine and galectin expression after challenge infection with the gastrointestinal nematode Haemonchus contortus. Int J Parasitol. 2011. https://doi.org/10.1016/j.ijpara.2010.11.006.
Oshiro TM, Macedo MS, Macedo-Soares MF. Anti-inflammatory activity of PAS-1, a protein component of Ascaris suum. Inflamm Res. 2005. https://doi.org/10.1007/s00011-004-1316-7.
Maizels RM, Yazdanbakhsh M. Immune Regulation by helminth parasites: cellular and molecular mechanisms. Nat Rev Immunol. 2003; doi.org:https://doi.org/10.1038/nri1183.
Finlay CM, Walsh KP, Mills KH. Induction of regulatory cells by helminth parasites: exploitation for the treatment of inflammatory diseases. Immunol Rev. 2014;259(1):206–30.
Smits HH, Yazdanbakhsh M. Chronic helminth infections modulate allergen-specific immune responses: protection against development of allergic disorders? Ann Med. 2007;39(6):428–39.
Li E, Tako EA, Singer SM. Complement activation by Giardia duodenalis parasites through the lectin pathway contributes to mast cell responses and parasite control. Infect Immun. 2016;84(4):1092–9. https://doi.org/10.1128/IAI.00074-16.Editor.
Zhang C, Wang C, Li Y, Takashi M, Liu C, Cui W, et al. Complement C3a signaling facilitates skeletal muscle regeneration by regulating monocyte function and trafficking. Nat Commun. https://doi.org/10.1038/s41467-017-01526-z.
Li RW, Hou Y, Li C, Gasbarre LC. Localized complement activation in the development of protective immunity against Ostertagia ostertagi infections in cattle. Vet Parasitol. 2010;174:247–56.
Guo Z, González JF, Hernandez JN, McNeilly TN, Corripio-Miyar Y, Frew D, et al. Possible mechanisms of host resistance to Haemonchus contortus infection in sheep breeds native to the Canary Islands. Sci Rep. 2016;6:1–14. https://doi.org/10.1038/srep26200.
Bell BD, Kitajima M, Larson RP, Stoklasek TA, Dang K, Sakamoto K, et al. The transcription factor STAT5 is critical in dendritic cells for the development of TH2 but not TH1 responses. Nat Immunol. 2013;14(4):364–71. https://doi.org/10.1038/ni.2541.
Taylor DK, Walsh PT, LaRosa DF, Zhang J, Burchil MA, Farrar MA, Turka LA. Constitutive activation of STAT5 supersedes the requirement for cytokine and TCR engagement of cd4+ t cells in steady-state homeostasis. J Immunol. 2006. https://doi.org/10.4049/jimmunol.177.4.2216.
Jallow OA, McGregor BA, Anderson N, Holmes JHG. Intake of trichostrongylid larvae by goats and sheep grazing together. Aust. Vet. J. 1994; doi.org:https://doi.org/10.1111/j.1751-0813.1994.tb00927.x.
Trinchieri G. Interleukin-12 : A proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu Rev Immunol. 1995;13:251–76.
Bricarello PA, Zaros LG, Coutinho LL, Rocha RA, Silva MB, Kooyman FNJ, et al. Immunological responses and cytokine gene expression analysis to Cooperia punctata infections in resistant and susceptible Nelore cattle. Vet Parasitol. 2008;155(1–2):95–103. https://doi.org/10.1016/j.vetpar.2008.03.016.
O’ Neill LAJ. How Toll-like receptors signal : what we know and what we don’t know. Curr Opin Immunol. 2006; 3–9. doi:https://doi.org/10.1016/j.coi.2005.11.012.
Araujo RN, Padilha T, Zarlenga D, Sonstegard T, Connor EE, Van Tassel C, Lima WS, Nascimento E, Gasbarre LC. Use of a candidate gene array to delineate gene expression patterns in cattle selected for resistance or susceptibility to intestinal nematodes. Vet Parasitol. 2009;162(1–2):106–15. https://doi.org/10.1016/j.vetpar.2008.12.017.
Worku M, Abdalla A, Adjei-Fremah S, Ismail H. The impact of diet on expression of genes involved in innate immunity in goat blood. J Agric Sci. 2016;8(3):1. https://doi.org/10.5539/jas.v8n3p1.
Holderness J, Hedges JF, Daughenbaugh K, Kimmel E, Graff J, Freedman B, Jutila MA. Response of γδ T cells to plant-derived tannins. Crit Rev Immunol. 2008;28(5):377–402.
Zhang R, Liu F, Hunt P, et al. Transcriptome analysis unraveled potential mechanisms of resistance to Haemonchus contortus infection in merino sheep populations bred for parasite resistance. Vet Res. 2019:1–13. https://doi.org/10.1186/s13567-019-0622-6.
Zheng Y, Manzotti CN, Liu M, Burke F, Mead KI, Sansom DM. CD86 and CD80 differentially modulate the suppressive function of human regulatory T cells. J Immunol. 2004;172(5):2778–84. https://doi.org/10.4049/jimmunol.172.5.2778.
Brozovic S, Nagaishi T, Yoshida M, Betz S, Salas A, Chen D, et al. CD1d function is regulated by microsomal triglyceride transfer protein. Nat Med. 2004;10(5):535–9. https://doi.org/10.1038/nm1043.
Chaussepied M, Janski N, Baumgartner M, Lizundia R, Jensen K, Weir W, Shiels BR, Weitzman JB, Glass EJ, Werling D, Langsley G. TGF-b2 induction regulates invasiveness of Theileria-transformed leukocytes and disease susceptibility. PLoS Pathog. 2010;18;6(11):e1001197. https://doi.org/10.1371/journal.ppat.1001197.
McSorley HJ, Grainger JR, Harcus Y, Murray J, Nisbet AJ, Knox DP, Maizels RM. daf-7-related TGF-beta homologues from Trichostrongyloid nematodes show contrasting life-cycle expression patterns. Parasitology. 2010;137:159–71.
Aman MJ, Tayebi N, Obiri NI, Puri RK, Modi WS, Leonard WJ. cDNA cloning and characterization of the human interleukin 13 receptor α chain. J Biol Chem. 1996;271(46):29265–70.
Wang YHA, Reverter AA, Kemp DA, McWilliam SM, Ingham A, Davis CA, et al. Gene expression profiling of Hereford shorthorn cattle following challenge with Boophilus microplus tick larvae. Aust J Exp Agric. 2007:1397–407.
Hoste H, Torres-Acosta JFJ, Aguilar-Caballero AJ. Nutrition-parasite interactions in goats: is immunoregulation involved in the control of gastrointestinal nematodes? Parasite Immunol. 2008;30:79–88. https://doi.org/10.1111/j.1365-3024.2007.00987.x.
Pernthaner A, Cole SA, Morrison L, Hein WR. Increased expression of interleukin-5 (IL-5), IL-13, and tumor necrosis factor alpha genes in intestinal lymph cells of sheep selected for enhanced resistance to nematodes during infection with Trichostrongylus colubriformis. Infect Immunol. 2005. https://doi.org/10.1128/IAI.73.4.2175-2183.2005.
Patterson DM, Jackson F, Huntley JF, Stevenson LM, Jones DG, Jackson E, et al. Studies on caprine responsiveness to nematodiasis: segregation of male goats into responders and non-responders. Int J Parasitol. 1996a. https://doi.org/10.1016/0020-7519(95)00121-2.
Patterson DM, Jackson F, Huntley JF, Stevenson LM, Jones DG, Jackson E, et al. The response of breeding does to nematodiasis: segregation into ‘responders’ and ‘non-responders’. Int J Parasitol 1996b; doi: https://doi.org/10.1016/S0020-7519(96)00125-7.
Etter E, Hoste H, Chartier C, et al. Susceptibility of high and low producer dairy goats to single experimental infection with Trichostrongylus colubriformis. Parasitol Res 2000; doi.org:https://doi.org/10.1007/s004360000280.
Meeusen EN, Balic A, Bowles VM. Cells, cytokines and other molecules associated with rejection of gastrointestinal nematode parasites. Vet Immunol Immunopathol. 2005. https://doi.org/10.1016/j.vetimm.2005.07.002.
Alba-Hurtado F, Muñoz-Guzmán MA. Immune responses associated with resistance to haemonchosis in sheep. Biomed Res Int. 2013. https://doi.org/10.1155/2013/162158.
MacKinnon KM, Burton JL, Zajac AM, Notter DR. Microarray analysis reveals difference in gene expression profiles of hair and wool sheep infected with Haemonchus contortus. Vet Immunol Immunopathol. 2009;130(3–4):210–20. https://doi.org/10.1016/j.vetimm.2009.02.013.
McRae KM, Stear MJ, Good B, Keane OM. The host immune response to gastrointestinal nematode infection in sheep. Parasite Immunol. 2015;10:605–13. https://doi.org/10.1111/pim.12290.
Sweeney T, Hanrahan JP, Ryan MT, Good B. Immunogenomics of gastrointestinal nematode infection in ruminants: breeding for resistance to produce food sustainably and safely. Parasite Immunol. 2016;38(9):569–86. https://doi.org/10.1111/pim.12347.
Venturina VM, Gossner AG, Hopkins J. The immunology and genetics of resistance of sheep to Teladorsagia circumcincta. Vet Res Commun. 2013;37(2):171–81. https://doi.org/10.1007/s11259-013-9559-9.
Benavides MV, Sonstegard TS, Van Tassell C. Genomic regions associated with sheep resistance to gastrointestinal nematodes. Trends Parasitol. 2016;32(6):470–80. https://doi.org/10.1016/j.pt.2016.03.007.
Bhuiyan, A.A., Li, J., Wu, Z., Ni, Pan., Adetula, A.A., Wang, H., Zhang, C., Tang, X., Bhuyan, A.A., Zhao, S. and Du, X. 2017. Exploring the genetic resistance to gastrointestinal nematodes infection in goat using RNA-sequencing. Int J Mol Sci 18(4). doi:https://doi.org/10.3390/ijms18040751.
Lee W, Stromberg MP, Ward A, Stewart C, Garrison EP, Marth GT. MOSAIK : A Hash-Based Algorithm for Accurate Next- Generation Sequencing Short-Read Mapping. 2014; 9(3). doi:https://doi.org/10.1371/journal.pone.0090581.
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, Depristo MA, Handsaker RE, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8. https://doi.org/10.1093/bioinformatics/btr330.
Foll M, Gaggiotti OE. A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–93.
Foll M, Fischer MC, Heckel G, Excoffier L. Estimating population structure from AFLP amplification intensity. Mol Ecol. 2010;19:4638–47.
Fischer MC, Foll M, Excoffier L, Heckel G. Enhanced AFLP genome scans detect local adaptation in high-altitude populations of a small rodent (Microtus arvalis). Mol Ecol. 2011;20:1450–62.
Acknowledgements
We gratefully acknowledge Dr. Francisco Peñagaricano, Assistant Professor at the University of Florida and Dr. Rostam Abdollahi-Arpanahi, Postdoctoral Researcher at the University of Florida, USA, and Assistant Professor at the University of Tehran, Iran, for their guidance.
Funding
This study was supported by UF Agricultural Station and the National Institute of Food and Agriculture, U.S. Department of Agriculture, under award No. 003673. The project was supported by the USDA National Institute of Food and Agriculture 1890 Institution Capacity Building Grant Program, Project OKLXSAHLU12, Accession Number 230541. Z.M. Estrada- Reyes was partially supported by the Consejo Nacional de Ciencias y Tecnología, CONACYT. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. ZMER conducted all analyses of the samples evaluates and drafted the manuscript. RGM conceived and assisted with the analysis and manuscript. RRA assisted with the data analysis. YT, ALG, TAG, TS, RP, ZW and SPH assisted with the manuscript, collected the FEC data, and selected the animals with low FEC.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The research protocol for the present study was approved by the Langston University Animal Care and Use Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Manhattan plots of the frequentist Fst values per SNP from targeted genomic regions in sheep and goats. The SNP data is ordered based on chromosomal location (x-axis). For sheep, plots includes: Katahdin and St. Croix vs Dorper (KS vs D, pooled resistant breeds vs susceptible breed), Katahdin vs Dorper (K vs D, resistant vs susceptible breed), St. Croix vs Dorper (S vs D, resistant vs susceptible breed) and St. Croix vs Katahdin (S vs K, resistant vs resistant breed) analyses. For goats, plots includes: Kiko and Spanish vs Boer (KS vs B, pooled resistant breeds vs susceptible breed), Kiko vs Boer (K vs B, resistant vs susceptible breed), Spanish vs Boer (S vs B, resistant vs susceptible breed) and Spanish vs Kiko (S vs K, resistant vs resistant breed) analyses. The genes with SNPs under selection in each analysis are presented in a box on the right side of Manhattan plots.
Additional file 2: Table S1.
Signatures of selection identified between resistant (Katahdin or St. Croix) and susceptible (Dorper) sheep breeds using frequentist Fst. Breeds compared (comparison), gene name, gene region, SNP name (chromosome and position), SNP, mutation type (synonymous or missense), and Fst value for the SNPs under selection.
Additional file 3: Table S2.
Signatures of selection identified between resistant (Katahdin or St. Croix) and susceptible (Dorper) sheep breeds using Bayesian Fst. Breeds compared (comparison), gene name, gene region, SNP name (chromosome and position), SNP, mutation type (synonymous or missense), and Fst value for the SNPs under selection.
Additional file 4: Table S3.
Signatures of selection identified between resistant (Kiko or Spanish) and susceptible (Boer) goat breeds using frequentist Fst. Breeds compared (comparison), gene name, gene region, SNP name (chromosome and position), SNP, mutation type (synonymous or missense), and Fst value for the SNPs under selection.
Additional file 5: Table S4.
Signatures of selection identified between resistant (Kiko or Spanish) and susceptible (Boer) goat breeds using Bayesian Fst. Breeds compared (comparison), gene name, gene region, SNP name (chromosome and position), SNP, mutation type (synonymous or missense), and Fst value for the SNPs under selection.
Additional file 6: Table S5.
Gene table
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Estrada-Reyes, Z.M., Tsukahara, Y., Amadeu, R.R. et al. Signatures of selection for resistance to Haemonchus contortus in sheep and goats. BMC Genomics 20, 735 (2019). https://doi.org/10.1186/s12864-019-6150-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6150-y