Abstract
Background
Sarcopenia is a disease diagnosed in the elderly. In patients with sarcopenia, the muscle mass decreases every year. The occurrence of sarcopenia is greatly affected by extrinsic factors such as eating habits, exercise, and lifestyle. The present study aimed to determine the relationship between muscle mass traits and genes affected by epigenetic factors with three different adjustment methods using Korean Genome and Epidemiology Study (KOGES) data.
Results
We conducted a demographic study and DNA methylation profiling by three studies according to the muscle mass index (MMI) adjustment methods: appendicular skeletal muscle mass divided by body weight (MMI1); appendicular skeletal muscle mass divided by square of height (MMI2); appendicular skeletal muscle mass divided by BMI (MMI3). We analyzed differentially methylated regions (DMRs) for each group. We then restricted our subjects to be top 30% (T30) and bottom 30% (B30) based on each MMI adjustment method. Additionally, we performed enrichment analysis using PathfindR to evaluate the relationship between identified DMRs and sarcopenia. A total of 895 subjects were included in the demographic study. The values of BMI, waist, and hip showed a significant difference in all three groups. Among 446 participants, 44 subjects whose DNA methylation profiles were investigated were included for DNA methylation analysis. The results of enrichment analysis showed differences between groups. In the women group through MMI1 method, only the glutamatergic synapse pathway showed a significant result. In the men group through MMI2 method, the adherens junction pathway was the most significant. Women group through MMI2 method showed similar results, having an enriched Rap1 signaling pathway. In men group through MMI3 method, the Fc epsilon RI signaling pathway was the most enriched. Particularly, the notch signaling pathway was significantly enriched in women group through MMI3 method.
Conclusion
This study presents results about which factor should be concerned first in muscle mass index (MMI) adjustment. The present study suggested that GAB2 and JPH3 in MMI1 method, HLA-DQB1 and TBCD in MMI2 method, GAB2, NDUFB4 and ISPD in MMI3 method are potential genes that can have an impact on muscle mass. It could enable future epigenetic studies of genes based on annotation results. The present study is a nationwide study in Korea with the largest size up to date that compares adjustment indices for MMI in epigenetic research.
Similar content being viewed by others
Introduction
Although old age is not a sickness, numerous diseases and syndromes are more common in the elderly. Recently, interest in research on sarcopenia is increasing as the number of elderly people increases [1,2,3]. In Korea, research for establishing the criteria of sarcopenia and their adequacy for diagnosis has been actively underway through policy-making [4]. Sarcopenia has different adjustment indices and diagnostic criteria for each continent according to race, culture, diet, and so on [5]. The Asian Working Group for Sarcopenia (AWGS) consensus has published the diagnostic standard of sarcopenia by height-adjusted muscle mass [6]. However, Korean and Asian studies have reported sarcopenia adjustment indices using weight and body mass index (BMI). Several studies have reported different adjustment methods [7,8,9,10,11]. In addition, it has been reported that the prevalence rate of sarcopenia might vary depending on the diagnosis index [6]. Thus, further research is needed.
Sarcopenia is a disease diagnosed in the elderly. Muscle mass is known to decrease every year since the age of 30. The risk of sarcopenia is rapidly increased during middle age when exercise, hormonal changes, and digestive ability are rapidly decreasing [12,13,14]. During this period, the risk of metabolic diseases including osteoporosis and hyperlipidemia due to menopause in women is also high [15,16,17]. In addition, sarcopenia is a disease that is greatly affected by extrinsic factors such as eating habits, exercise, and lifestyle [18,19,20]. However, studies on the relationship between sarcopenia and extrinsic factors and how it varies depending on the adjustment index in East Asia, including Korea, are insufficient.
Therefore, the present study aimed to determine the relationship between muscle mass traits and epigenetic genes with three different adjustment indices (weight adjustment, square of height adjustment, and BMI adjustment) using the Korean Genome and Epidemiology Study (KOGES) data.
Materials and methods
Study subjects
Data used in this study were from the Korean Genome and Epidemiology Study (KoGES) performed by the National Research Institute of Health, Centers for Disease Control and Prevention, Ministry for Health and Welfare, Republic of Korea. The number of baseline participants was 10,030. They lived in Ansan or Ansung located in Gyeonggi Province, South Korea. In this cohort, participants aged 40 to 69 years. 9,351 subjects who had laboratory data were included. Using the bioelectrical impedance analysis (BIA) (body composition analyzer, models ZEUS 9.9, JAWON MEDICAL CO., LTD, Seoul, Korea), skeletal muscle mass was measured. We conducted three studies according to the muscle mass index (MMI) adjustment methods: appendicular skeletal muscle mass divided by body weight (MMI1); appendicular skeletal muscle mass divided by square of height (MMI2); appendicular skeletal muscle mass divided by BMI (MMI3). We then restricted our subjects to be top 30% (T30) and bottom 30% (B30) based on each MMI adjustment method.
The study was approved by the Institutional Review Board (IRB) of Korea University (Approval Number: KUIRB-2020-0191-01). All study subjects provided written informed consent.
Body composition measurement, demographic factors, and medical history
Method of body composition measurement, demographic factors, and medical history were previously described [21]. All participants attended a community clinic for clinical assessments at each follow-up visit. BMI was calculated as weight in kg divided by the square of height in meters. Weight was determined for an individual wearing light clothes without shoes (barefoot). Waist and hip circumference were also measured. The remaining survey items consisted of drinking & smoking status, level of education, and monthly income. History of hypertension, diabetes, gastritis/stomach ulcer, allergy, myocardial infarction, thyroid disorder, congestive heart failure, coronary artery disease, hyperlipidemia, asthma, chronic lung disorder, peripheral vascular disease, kidney disease, various tumors, cerebrovascular disease, head trauma, urinary tract infection, gout, degenerative arthritis, and rheumatoid arthritis was also taken.
DNA methylation profiling
The present study collected epidemiology data once every two years. DNA methylation profiles were investigated during the 4th follow-up (2009–2010) for 446 participants. In the DNA methylation study, participants were also restricted to T30 and B30. An Infinium HumanMethylation 450K beadChip (Illumina, Inc., San Diego, CA, USA) was used to obtain KoGES DNA methylation data. Quality control procedure was applied to DNA methylation data. The beta value indicating DNA methylation level was calculated as [ (Methylated reads) / (Unmethylated reads) + (Methylated reads) ]. After filtering, a total of 389,321 CpGs remained for epigenome-wide association analysis. After that, we merged CpG sites data and annotation data of Illumina Human Methylation EPIC manifest package using R. Differentially methylated regions (DMRs) were identified using a t-test performed between T30 group and B30 group using the criteria of p < 5*10^-3 and | fold change | > 0.2 to find differentially methylated CpG sites. The gene of annotation data merged with a CpG site with a significant difference in bata value was displayed in a volcano plot. Enrichment analyses of DMRs were performed using the “pathfindR” package which integrates pathway/gene set annotations from sources such as Kyoto Encylopedia of Genes and Genomes (KEGG), Reactome, BioCarta, and Gene Ontology (GO) [22].
Statistical analysis
Continuous data are reported as mean ± standard deviation and categorical data are reported as n (%). To find any significant difference in baseline characteristics or clinical factors between T30 and B30, unpaired t-test was used for continuous variables if normality assumption was met. Otherwise, Wilcoxon’s rank-sum test was performed. Differences in proportions between T30 and B30 were analyzed using Chi-squared test for categorical variables. If the assumption of Chi-squared test did not meet, Fisher’s exact test was performed.
An unpaired T-test was performed to identify genes from annotation results of methylated CpG sites between muscle mass traits groups. Methylated CpG sites were visualized by a volcano plot. Each CpG site was differentiated by the following criteria: p < 5*10^-3 and | fold change | > 0.2.
All statistical analyses were carried out using R software version 4.1.0 (R Core Team. R Foundation for Statistical Computing, Vienna, Austria, 2020). The significance level was set at p < 0.05.
Results
Demographic characteristics
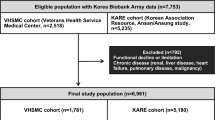
A total of 9,351 middle-aged people participated in this study. Those in B30 were considered to have insufficient muscle mass, while those in T30 were considered to have sufficient muscle mass. 7,452 people who were not included in T30 or B30 were excluded. A total of 1,004 participants with at least one missing value were also excluded. Finally, a total of 895 subjects (389 males and 506 females) were included in this study. In MMI1 method, there were 215 people with T30 and 174 people with B30 in the male group and 228 people with T30 and 278 people with B30 in the female group. In MMI2 method, there were 169 people with T30 and 220 people with B30 in the male group and 308 people with T30 and 198 people with B30 in the female group. When adjusted for BMI, there were 205 subjects with T30 and 184 subjects with B30 in the male group and 227 people with T30 and 279 people B30 in the female group.
Demographics according to weight adjustment index (MMI1)
Statistically significant differences in BMI, waist, hip, smoking status, history of gastritis/stomach ulcer, history of hyperlipidemia, history of various tumors, and sedentary life status between T30 group and B30 group were found in men. Particularly, BMI, waist, hip, smoking status, and gastritis/stomach ulcer history showed high significance. Age, BMI, waist, hip, hypertension history, diabetes history, gout history, and degenerative arthritis history showed significant differences between T30 group and B30 group were found in women. Particularly, age, BMI, waist, hip, hypertension history, gout history, and degenerative arthritis history showed high significance (Table 1).
Demographics according to the square of height adjustment index (MMI2)
Statistically significant differences in age, BMI, waist, hip, history of hyperlipidemia, and sedentary life status between T30 group and B30 group were found in men. Particularly, age, BMI, waist, and hip showed high significance with p-value < 0.005. Age, BMI, waist, hip, and hypertension history showed significant differences T30 group and B30 group in women (Table 2).
Demographics according to BMI adjustment index (MMI3)
Statistically significant differences in BMI, waist, hip, smoking status, history of gastritis/stomach ulcer, and history of hyperlipidemia between T30 group and B30 group were found for men. Particularly, BMI, waist, hip, smoking status, and history of gastritis/stomach ulcer showed high significance with p-values less than 0.005. Age, BMI, waist, hip, history of hypertension, history of diabetes, history of gout, and history of degenerative arthritis were significantly different between T30 group and B30 group in women. Particularly, age, BMI, waist, hip, hypertension history, gout history, and degenerative arthritis history showed high significance (Table 3).
Epigenome-wide association analysis of muscle status and gene from annotation results
Among 446 participants, 44 met the criteria and were included in T30 or B30 group. Finally, the analysis included these 44 subjects whose DNA methylation profiles were investigated. Based on epigenome-wide association analysis of DNA methylation, genes from annotation results of each group were certified using a volcano plot (Figs. 1 and 2). In men group through MMI1 method, five down-regulated genes (GAB2, WDR41, HCCA2, C14orf139, and RNASEN) and one up-regulated gene (GPR83) were associated with middle-age muscle loss. In women group through MMI1 method (|Log2 fold change| > 0.21 and P < 0.05), five down-regulated genes (JPH3, FSCN2, UMODL1, NPLOC4, and NDUFB4) and one up-regulated gene (CPLX2) were associated with middle-age muscle loss. In men group through MMI2 method, there was only one down-regulated gene (HLA-DQB1). However, in women group through MMI2 method, there were one down-regulated gene (TBCD) and three up-regulated genes (TRPS1, UBR2, TOX2). In MMI3 method, several muscle-loss-related genes were found in men and women: four down-regulated genes (C14orf139, HCCA2, RNASEN, GAB2) and three up-regulated genes (FAM32A, TMCO3, GPR83) in men; seven down-regulated genes (WDR41, ANKLE2, UMODL1, SHANK2, C21orf70, NPLOC4, NDUFB4) and three up-regulated genes (CPLX2, ISPD, ADARB2) in women group. All results of muscle atrophy-related genes are shown in Table 4.
Enrichment analysis of DMRs
Enrichment analysis of DMRs are presented in Fig. 3. In men group through MMI1 method (|Log2 fold change| > 0.21 and P < 0.05), Fc epsilon RI signaling pathway which is associated with inducing mast cell degranulation and plays a critical role in human airway smooth muscle cell function was the most enriched with statistical significance [23]. In women group through MMI1 method (|Log2 fold change| > 0.21 and P < 0.05), only the glutamatergic synapse pathway showed a significant result. Glutamate is known to act as a mediator between the motor nerve tip and skeletal muscle fibers [24]. In men group through MMI2 method (|Log2 fold change| > 0.07 and P < 0.05), the adherens junction pathway that join mature myocytes and give myofibrils places to attach to the membrane [25]was the most significant. Moreover, the Rap1 signaling pathway associated with skeletal muscle cell differentiation also showed significant results in the same group [26]. Women group through MMI2 method (|Log2 fold change| > 0.07 and P < 0.05), showed similar results, having an enriched Rap1 signaling pathway. In men group through MMI3 method (|Log2 fold change| > 0.21 and P < 0.05), the Fc epsilon RI signaling pathway was the most enriched. Particularly, the notch signaling pathway considered as a key player in skeletal muscle regeneration and development [27] was significantly enriched in women group through MMI3 method (|Log2 fold change| > 0.21 and P < 0.05). According to these results, changes in pathways of genes related to DMR between T30 and B30 might be due to differences in adjustment method rather than due to gender differences.
Discussion
The present epigenetic nationwide cohort study was conducted based on MMI adjusted by weight, square of height, and BMI. In the group of men adjusted by weight, there were significant differences in BMI, the circumference of waist, and the circumference of hip. These obesity-related features are associated with muscle loss [28]. Smoking also showed a significant difference in the present study. One study has reported that smoking is associated with muscle loss in men [29]. Sedentary life known to be associated with sarcopenia or muscle loss was also significantly different in the present study. In a group of women adjusted by weight, MMI values were decreased with increasing age [30]. The relationship between age and muscle loss showed relevances in many studies [31,32,33,34].In addition, there were significant differences in people with a history of high blood pressure and diabetes, both of which are known to be associated with sarcopenia or muscle loss [35,36,37,38].
Through the MMI2 method, age, BMI, waist, and hip revealed significant results in the men group. Significant results were also observed in individuals with a history of asthma illness. The decline in respiratory muscle mass, function, and power could be related to aging [39]. Several studies have consistently revealed the association between respiratory diseases such as asthma and sarcopenia or muscle loss [39, 40]. In addition, the men group through MMI2 method showed a significant difference in sedentary life. In the women group through MMI2 method, there were significant differences in BMI, the circumference of waist, and hip. However, there was no significant result according to age difference. In particular, there was a significant difference in people with a history of degenerative arthritis. As muscle wasting is a natural part of aging, sarcopenia prevalence has been recently proven in individuals with OA [41, 42]. It has been suggested that muscle wasting has a direct impact on joint stability and that loss of mobility can lead to articular cartilage degeneration [43].
In the group of men adjusted by BMI, there were significant differences in BMI, the circumference of waist and hip, and smoke. However, there was no significant difference in sedentary life. In the group of men adjusted by BMI, there were significant differences in age, BMI, waist, hip, history of hypertension, history of diabetes, and history of degenerative arthritis. Additionally, people with a history of gout showed low adjusted MMI results. According to a study by KM Beavers, reduced skeletal muscle mass is linked to increased serum uric acid levels [44]. Many studies have shown that gout might be related to muscle mass loss or sarcopenia [45, 46].
DNA methylation may play an important role in muscle loss during aging [20]. Therefore, finding the characteristics of age-related DNA methylation change might be the most promising way to find biomarkers of muscle aging [47, 48]. Several studies have determined methylation levels of certain representative genes in young and old people and found an age-related increase in methylation between NDUFB6 and COX7A1 that is important in metabolic mechanisms [49, 50].
Likewise, a gene annotation study using DMR was performed in this study. In the group of men adjusted by weight, GAB2 was a downregulated gene. According to a study by Eric Edstrom et al., Shc-GRB2-GAB (Shc, Src homology 2 domain-containing; GRB2, growth factor receptor bound protein-2; GAB, GRB2 associated binding protein) adaptor might interact with IGF-1 receptor, which activates PI3K-AKT. Through this pathway, AKT could block atrophy and stimulate myofiber hypertrophy [51]. In contrast, JPH3 was a downregulated gene in the group of women adjusted by weight. Junctophilins are major components responsible for the synthesis of JMCs (junctional membrane complexes) in skeletal and cardiac muscles [52]. Li et al. have suggested that JPH3 plays a critical function in maintaining the proper distance and crosstalk between the ER and mitochondria via the Pgc-1 pathway in beta cells [53]. Pgc-1α in nuclei is known to promote target molecule transcription in skeletal muscles [53]. Muscle atrophy-related genes in the weight adjustment group is summarized in Supplementary Table 1.
In the group of men adjusted by height squared, HLA-DQB1 was a downregulated gene. According to a study by Singh et al., HLA-DQB1 is specifically enriched in skeletal muscles and highly associated with the handgrip trait [54]. HLA-DQB1 is nominally associated with sarcopenia (EWGSOP combined definition). The influence of HLA type is more significant in women than in men [55, 56]. In the group of women adjusted by height square, TBCD was a downregulated gene and UBR2 was an upregulated gene. Carrio et al. have suggested that the TBCD gene is one gene with differentially methylated CpG (dmCpG) affected by aging. The most enriched words and pathways among genes with two or more intragenic dmCpG sites were “muscle cell” (P = 0.0004), indicating that dmCpG sites found in the elderly were related to muscle tissue functions and neuromuscular junctions [57]. The TBCD gene has the most intragenic dmCpG sites (46 distinct sites, or 13.2% of the total number of CpG sites in the gene) [58]. Additionally, Ubr2 is up-regulated in disuse atrophying skeletal muscles of mice [59]. Muscle atrophy-related genes in the square of height adjustment group are summarized in Supplementary Table 2.
The group of men adjusted by BMI also showed GAB2 as a downregulated gene. On the other hand, the group of women adjusted by BMI showed two downregulated genes (NPLOC4 and NDUFB4) and one upregulated gene (ISPD) known to be associated with muscle loss. NPLOC4 is known as a muscle degeneration-related gene because it has two SNPs (rs6565597 and rs9894429) that are related to age-related macular degeneration [60, 61]. NDUFB4 is also shown to be higher in muscles. It codes for subunits of the respiratory chain’s complex I which is related to energy metabolism.56 However, ISPD was upregulated in the low-adjusted MMI group. Marcela P. Cataldi has shown that ISPD overexpression increases functional glycosylation of α subunit of dystroglycan (F-α-DG) in skeletal muscles [62]. Because it is known that dystroglycan-null could be caused by muscle dystrophy, the muscle loss group may compensatively show ISPD upregulation. Muscle atrophy-related genes in the weight adjustment group are summarized in Supplementary Table 3. Overall molecular mechanisms related to muscle loss are presented in Fig. 4.
In conclusion, this study presents results about which factor should be concerned first in MMI adjustment. It could enable future epigenetic studies of genes based on annotation results. The present study is a nationwide study in Korea with the largest size up to date that compares adjustment methods for MMI in epigenetic research.
There are some limitations in the present study. First, compared to a large number of participants, the number of genetic tests was insufficient. In addition, because it was a cohort study, there was no restriction on the composition of participants. Tests for sarcopenia screening such as the handgrip test were not conducted. Participants were a middle-age group. There were no results from elderly participants. Second, eQTL analysis was not performed. If eQTL analysis could be added later in an extended cohort study, it will be an in-depth study on the difference between genotype and phenotype of sarcopenia. Therefore, further study including elderly participants with sufficient genetic tests is needed.
Data availability
The data presented in the study are included in the article. Further inquiries can be directed to the corresponding authors.
References
Delmonico MJ, Beck DT. The current understanding of Sarcopenia: emerging tools and interventional possibilities. Am J Lifestyle Med. 2017;11:167–81.
Chen L-K, Liu L-K, Woo J, Assantachai P, Auyeung T-W, Bahyah KS, et al. Sarcopenia in Asia: consensus report of the asian Working Group for Sarcopenia. J Am Med Dir Assoc. 2014;15:95–101.
Muscaritoli M, Anker SD, Argilés J, Aversa Z, Bauer JM, Biolo G, et al. Consensus definition of sarcopenia, cachexia and pre-cachexia: joint document elaborated by Special Interest Groups (SIG) “cachexia-anorexia in chronic wasting diseases” and “nutrition in geriatrics. Clin Nutr. 2010;29:154–9.
Park HM. Current status of Sarcopenia in Korea: a focus on korean Geripausal Women. Ann Geriatr Med Res. 2018;22:52–61.
Kruger HS, Micklesfield LK, Wright HH, Havemann-Nel L, Goedecke JH. Ethnic-specific cut-points for sarcopenia: evidence from black south african women. Eur J Clin Nutr. 2015;69:843–9.
Chen L-K, Woo J, Assantachai P, Auyeung T-W, Chou M-Y, Iijima K, et al. Asian Working Group for Sarcopenia: 2019 Consensus Update on Sarcopenia diagnosis and treatment. J Am Med Dir Assoc. 2020;21:300–307e2.
Sung MJ, Park JY, Lee HW, Kim BK, Kim DY, Ahn SH et al. Predictors of Sarcopenia in an obese Asian Population. Nutr Cancer. 2021;:1–10.
Roh E, Choi KM. Health Consequences of sarcopenic obesity: a narrative review. Front Endocrinol. 2020;11:332.
Miura H, Sakaguchi K, Ogawa W, Tamori Y. Clinical features of 65-year-old individuals in Japan diagnosed with possible sarcopenia based on the asian Working Group for Sarcopenia 2019 criteria. Geriatr Gerontol Int. 2021;21:689–96.
Ryu M, Jo J, Lee Y, Chung Y-S, Kim K-M, Baek W-C. Association of physical activity with sarcopenia and sarcopenic obesity in community-dwelling older adults: the Fourth Korea National Health and Nutrition Examination Survey. Age Ageing. 2013;42:734–40.
Kim J-H, Cho JJ, Park YS. Relationship between sarcopenic obesity and Cardiovascular Disease Risk as estimated by the Framingham risk score. J Korean Med Sci. 2015;30:264–71.
Bijlsma AY, Meskers CGM, Ling CHY, Narici M, Kurrle SE, Cameron ID, et al. Defining sarcopenia: the impact of different diagnostic criteria on the prevalence of sarcopenia in a large middle aged cohort. Age (Dordr). 2013;35:871–81.
Ryan AS, Ivey FM, Serra MC, Hartstein J, Hafer-Macko CE. Sarcopenia and physical function in Middle-Aged and older stroke survivors. Arch Phys Med Rehabil. 2017;98:495–9.
Walrand S, Guillet C, Salles J, Cano N, Boirie Y. Physiopathological mechanism of Sarcopenia. Clin Geriatr Med. 2011;27:365–85.
Fonseca MIH, da Silva IT, Ferreira SRG. Impact of menopause and diabetes on atherogenic lipid profile: is it worth to analyse lipoprotein subfractions to assess cardiovascular risk in women? Diabetol Metab Syndr. 2017;9:22.
Li Y, Zhao L, Yu D, Ding G. The prevalence and risk factors of dyslipidemia in different diabetic progression stages among middle-aged and elderly populations in China. PLoS ONE. 2018;13:e0205709.
Al-Qutob RJ, Mawajdeh SM, Khalil AA, Schmidt AB, Hannak AO, Masri BK. The magnitude of osteoporosis in middle aged women. Saudi Med J. 2001;22:1109–17.
Cruz-Jentoft AJ, Baeyens JP, Bauer JM, Boirie Y, Cederholm T, Landi F, et al. Sarcopenia: european consensus on definition and diagnosis: report of the european Working Group on Sarcopenia in Older People. Age Ageing. 2010;39:412–23.
Kim TN, Choi KM, Sarcopenia. Definition, epidemiology, and pathophysiology. J Bone Metab. 2013;20:1–10.
Gensous N, Bacalini MG, Franceschi C, Meskers CGM, Maier AB, Garagnani P. Age-Related DNA methylation changes: potential impact on skeletal muscle aging in humans. Front Physiol. 2019;10:996.
Gim J-A, Lee S, Kim SC, Baek K-W, Yoo J-I. Demographic and genome wide association analyses according to muscle Mass using data of the Korean Genome and Epidemiology Study. J Korean Med Sci. 2022;37:e346.
Ulgen E, Ozisik O, Sezerman OU. pathfindR: an R Package for Comprehensive Identification of Enriched Pathways in Omics Data through active subnetworks. Front Genet. 2019;10:858.
Gounni AS, Wellemans V, Yang J, Bellesort F, Kassiri K, Gangloff S, et al. Human airway smooth muscle cells express the high affinity receptor for IgE (fc epsilon RI): a critical role of fc epsilon RI in human airway smooth muscle cell function. J Immunol. 2005;175:2613–21.
Colombo MN, Francolini M. Glutamate at the Vertebrate Neuromuscular Junction: from modulation to neurotransmission. Cells. 2019;8:996.
Ong LL, Kim N, Mima T, Cohen-Gould L, Mikawa T. Trabecular myocytes of the embryonic heart require N-cadherin for migratory unit identity. Dev Biol. 1998;193:1–9.
Pizon V, Cifuentes-Diaz C, Mège RM, Baldacci G, Rieger F. Expression and localization of RAP1 proteins during myogenic differentiation. Eur J Cell Biol. 1996;69:224–35.
Buas M, Kadesch T. Regulation of skeletal myogenesis by Notch. Exp Cell Res. 2010;316:3028–33.
Wannamethee SG, Atkins JL. Muscle loss and obesity: the health implications of sarcopenia and sarcopenic obesity. Proceedings of the Nutrition Society. 2015;74:405–12.
Jo Y, Linton JA, Choi J, Moon J, Kim J, Lee J, et al. Association between cigarette smoking and Sarcopenia according to obesity in the Middle-Aged and Elderly Korean Population: the Korea National Health and Nutrition Examination Survey (2008–2011). Korean J Fam Med. 2019;40:87–92.
Smith L, Tully M, Jacob L, Blackburn N, Adlakha D, Caserotti P, et al. The Association between Sedentary Behavior and Sarcopenia among adults aged ≥ 65 years in low- and Middle-Income Countries. Int J Environ Res Public Health. 2020;17:1708.
Kirwan R, McCullough D, Butler T, Perez de Heredia F, Davies IG, Stewart C. Sarcopenia during COVID-19 lockdown restrictions: long-term health effects of short-term muscle loss. GeroScience. 2020;42:1547–78.
Wilkinson DJ, Piasecki M, Atherton PJ. The age-related loss of skeletal muscle mass and function: measurement and physiology of muscle fibre atrophy and muscle fibre loss in humans. Ageing Res Rev. 2018;47:123–32.
Volpi E, Nazemi R, Fujita S. Muscle tissue changes with aging. Curr Opin Clin Nutr Metab Care. 2004;7:405–10.
Seene T, Kaasik P. Muscle weakness in the elderly: role of sarcopenia, dynapenia, and possibilities for rehabilitation. Eur Rev Aging Phys Act. 2012;9:109–17.
Trierweiler H, Kisielewicz G, Hoffmann Jonasson T, Rasmussen Petterle R, Aguiar Moreira C. Zeghbi Cochenski Borba V. Sarcopenia: a chronic complication of type 2 diabetes mellitus. Diabetol Metab Syndr. 2018;10:25.
Kalyani RR, Corriere M, Ferrucci L. Age-related and disease-related muscle loss: the effect of diabetes, obesity, and other diseases. Lancet Diabetes Endocrinol. 2014;2:819–29.
Srikanthan P, Hevener AL, Karlamangla AS. Sarcopenia exacerbates obesity-associated insulin resistance and dysglycemia: findings from the National Health and Nutrition Examination Survey III. PLoS ONE. 2010;5:e10805.
Shu J, Matarese A, Santulli G. Diabetes, body fat, skeletal muscle, and hypertension: the ominous chiasmus? J Clin Hypertens (Greenwich). 2018;21:239–42.
Nagano A, Wakabayashi H, Maeda K, Kokura Y, Miyazaki S, Mori T et al. Respiratory Sarcopenia and Sarcopenic Respiratory disability: concepts, diagnosis, and treatment. J Nutr Health Aging. 2021;:1–9.
Benz IGE, Trajanoska K, Schoufour J, Lahousse L, Roos ED, Terzikhan N, et al. Sarcopenia in elderly population with chronic respiratory diseases: a population-based study. Eur Respir J. 2018;52:62.
Larsson L, Degens H, Li M, Salviati L, Lee YI, Thompson W, et al. Sarcopenia: aging-related loss of muscle Mass and function. Physiol Rev. 2019;99:427–511.
Jin WS, Choi EJ, Lee SY, Bae EJ, Lee T-H, Park J. Relationships among obesity, Sarcopenia, and Osteoarthritis in the Elderly. J Obes Metab Syndr. 2017;26:36–44.
Shorter E, Sannicandro AJ, Poulet B, Goljanek-Whysall K. Skeletal muscle wasting and its relationship with osteoarthritis: a Mini-Review of Mechanisms and current interventions. Curr Rheumatol Rep. 2019;21:40.
Beavers KM, Beavers DP, Serra MC, Bowden RG, Wilson RL. Low relative skeletal muscle mass indicative of sarcopenia is associated with elevations in serum uric acid levels: findings from NHANES III. J Nutr Health Aging. 2009;13:177–82.
Lee J, Hong YS, Park S-H, Kang KY. High serum uric acid level is associated with greater handgrip strength in the aged population. Arthritis Res Therapy. 2019;21:73.
Floriano JP, Nahas PC, de Branco FMS, dos Reis AS, Rossato LT, Santos HO, et al. Serum uric acid is positively Associated with muscle Mass and Strength, but not with functional capacity, in kidney transplant patients. Nutrients. 2020;12:2390.
Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:3156.
Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–84.
Ling C, Poulsen P, Simonsson S, Rönn T, Holmkvist J, Almgren P, et al. Genetic and epigenetic factors are associated with expression of respiratory chain component NDUFB6 in human skeletal muscle. J Clin Invest. 2007;117:3427–35.
Rönn T, Poulsen P, Hansson O, Holmkvist J, Almgren P, Nilsson P, et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia. 2008;51:1159.
Edström E, Altun M, Hägglund M, Ulfhake B. Atrogin-1/MAFbx and MuRF1 are downregulated in aging-related loss of skeletal muscle. J Gerontol A Biol Sci Med Sci. 2006;61:663–74.
Piggott CA, Jin Y, Junctophilins. Key membrane tethers in muscles and neurons. Front Mol Neurosci. 2021;14:141.
Li L, Pan Z-F, Huang X, Wu B-W, Li T, Kang M-X, et al. Junctophilin 3 expresses in pancreatic beta cells and is required for glucose-stimulated insulin secretion. Cell Death Dis. 2016;7:e2275.
Singh AN, Gasman B. Disentangling the Genetics of Sarcopenia: prioritization of NUDT3 and KLF5 as genes for lean mass and HLA-DQB1-AS1 for hand grip strength based on associated SNPs. 2021.
Jones G, Trajanoska K, Santanasto AJ, Stringa N, Kuo C-L, Atkins JL, et al. Genome-wide meta-analysis of muscle weakness identifies 15 susceptibility loci in older men and women. Nat Commun. 2021;12:654.
Jones G, Pilling LC, Kuo C-L, Kuchel G, Ferrucci L, Melzer D. Sarcopenia and Variation in the human leukocyte Antigen Complex. J Gerontol A Biol Sci Med Sci. 2020;75:301–8.
Carrió E, Suelves M. DNA methylation dynamics in muscle development and disease. Front Aging Neurosci. 2015;7:19.
Zykovich A, Hubbard A, Flynn JM, Tarnopolsky M, Fraga MF, Kerksick C, et al. Genome-wide DNA methylation changes with age in disease-free human skeletal muscle. Aging Cell. 2014;13:360–6.
Hockerman GH, Dethrow NM, Hameed S, Doran M, Jaeger C, Wang W-H, et al. The Ubr2 gene is expressed in skeletal muscle atrophying as a result of hind limb suspension, but not Merg1a expression alone. Eur J Transl Myol. 2014;24:3319.
Fritsche LG, Igl W, Bailey JNC, Grassmann F, Sengupta S, Bragg-Gresham JL, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–43.
Kobilo T, Guerrieri D, Zhang Y, Collica SC, Becker KG, van Praag H. AMPK agonist AICAR improves cognition and motor coordination in young and aged mice. Learn Mem. 2014;21:119–26.
Cataldi MP, Blaeser A, Lu P, Leroy V, Lu QL. ISPD overexpression enhances Ribitol-Induced glycosylation of α-Dystroglycan in Dystrophic FKRP Mutant mice. Mol Ther Methods Clin Dev. 2019;17:271–80.
Acknowledgements
This work was supported by biomedical research instiute fund (GNUHBRIF-2019-0013) from the Gyeongsang National University Hospital.
Funding
This study was conducted with the research design support from the Rural Development Administration (PJ014155052019). The funding body played no role in the design of the study and collection, analysis, interpretation of data, and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
JA.G, SY.L, and JI.Y made contributions to the study’s conception and design. KW.B, and SH. S collected and processed the data. SY.L and SC.K analyzed and interpreted the data. SY.L wrote the manuscript. JA.G and JI.Y revised the manuscript. All authors contributed to the article and approved the submitted manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and study was approved by the Institutional Review Board (IRB) of Korea University (Approval Number: KUIRB-2020-0191-01). All study subjects provided written informed consent.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gim, JA., Lee, SY., Kim, S.C. et al. Relationship between DNA methylation changes and skeletal muscle mass. BMC Genom Data 24, 48 (2023). https://doi.org/10.1186/s12863-023-01152-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-023-01152-3