Abstract
Background
The Neacomys genus is predominantly found in the Amazon region, and belongs to the most diverse tribe of the Sigmodontinae subfamily (Rodentia, Cricetidae, Oryzomyini). The systematics of this genus and questions about its diversity and range have been investigated by morphological, molecular (Cytb and COI sequences) and karyotype analysis (classic cytogenetics and chromosome painting), which have revealed candidate species and new distribution areas. Here we analyzed four species of Neacomys by chromosome painting with Hylaeamys megacephalus (HME) whole-chromosome probes, and compared the results with two previously studied Neacomys species and with other taxa from Oryzomyini and Akodontini tribes that have been hybridized with HME probes. Maximum Parsimony (MP) analyses were performed with the PAUP and T.N.T. software packages, using a non-additive (unordered) multi-state character matrix, based on chromosomal morphology, number and syntenic blocks. We also compared the chromosomal phylogeny obtained in this study with molecular topologies (Cytb and COI) that included eastern Amazonian species of Neacomys, to define the phylogenetic relationships of these taxa.
Results
The comparative chromosome painting analysis of the seven karyotypes of the six species of Neacomys shows that their diversity is due to 17 fusion/fission events and one translocation, pericentric inversions in four syntenic blocks, and constitutive heterochromatin (CH) amplification/deletion of six syntenic autosomal blocks plus the X chromosome. The chromosomal phylogeny is consistent with the molecular relationships of species of Neacomys. We describe new karyotypes and expand the distribution area for species from eastern Amazonia and detect complex rearrangements by chromosome painting among the karyotypes.
Conclusions
Our phylogeny reflects the molecular relationships of the Akodontini and Oryzomyini taxa and supports the monophyly of Neacomys. This work presents new insights about the chromosomal evolution of this group, and we conclude that the karyotypic divergence is in accord with phylogenetic relationships.
Similar content being viewed by others
Background
The genus Neacomys Thomas, 1900 comprises 12 recognized species distributed from Panama to central Brazil and Bolivia; these rodents are mainly found in the Amazon region, and belong to the most diverse tribe of the Sigmodontinae subfamily (Rodentia, Cricetidae, Oryzomyini) [1,2,3,4,5,6,7,8,9]. Recently, morphologic, molecular and cytogenetic analyses have revealed candidate species along with, new distributions and karyotypes; this work has reshaped the geographic boundaries, raised questions regarding the taxonomic status, suggested the occurrence of species complexes in some lineages (e.g., N. minutus) and reinforced the view that this genus is more diverse than was previously believed [2, 5,6,7,8,9,10].
Cytogenetic studies on Neacomys show a range of diploid number (2n) from 28 to 64, and an autosomal fundamental number (FN) from 36 to 70 (Table 1), but in most species the diploid and fundamental number are the only available information, and the results of other cytogenetic techniques are not reported. Recent studies using classic banding, FISH with telomeric and rDNA 18S probes define five new karyotypes [5], two for N. paracou (2n = 56, FN = 62; 66), one for N. dubosti (2n = 64/FN = 68) and two for an undescribed species, Neacomys sp. (2n = 58/FN = 64; 70). A third karyotype was described for this Neacomys sp., which has the same 2n (58), but a different FN (66) [8].
Two additional undescribed species identified by karyotypic and molecular data (Cytb and COI sequences): Neacomys sp. A (2n = 58/FN = 68) and Neacomys sp. B (2n = 54/FN = 66) [7], were analyzed using whole-chromosome probes from Hylaeamys megacephalus (HME) [11].
The authors linked the findings in these two species with data on three other species mentioned above [5], and defined four synapomorphies for the genus (chromosomal associations HME 6a/21, (9,10)/7b/(9,10), 12/(16,17), and 20/(13,22)/4), that agreed with the chromosomal evolutionary pattern previously described for eastern Amazonian species [5], with fusion/fission, pericentric inversions and amplification/deletion of constitutive heterochromatin events as the main cause of the karyotypic diversity.
The Neacomys genus was always recovered as a monophyletic group by the use of different markers (morphological and/or molecular - Cytb, COI, IRBP) and approaches (Maximum Parsimony, Maximum Likelihood and Bayesian Inference), with values above 74% of support and variation according to the analysis and set of data, but most of the investigations were performed in western Brazilian Amazonian samples [2, 5,6,7, 9, 12,13,14].
A molecular analysis of nine species from eastern Amazonia was performed [7], including the three undescribed taxa mentioned above: Neacomys sp. [5], Neacomys sp. A and Neacomys sp. B [7]. The resulting phylogeny demonstrated that, despite the new species, the main relationships among the species remain the same: N. paracou stands as a sister to the other species of the genus, followed by N. spinosus and two clades – one comprising N. dubosti and species from Eastern Amazonia (Neacomys sp. A, Neacomys sp. B and Neacomys sp.) and the other formed by N. minutus, N. musseri and N. guianae.
In an effort to understand the direction of chromosome change, Da Silva et al. [5] plotted the 2n and FN data on a molecular phylogeny (Cytb) and discussed the chromosomal evolution of the genus. The authors observed that the clades were structured with distinct chromosomal evolutionary patterns, and postulated that three major and independent events had occurred: two increases and one reduction from the hypothetical ancestral 2n (56). They also proposed that Robertsonian translocations were the main cause of the 2n reductions observed in N. minutus (2n = 35–36/FN = 40) and N. musseri (2n = 34/FN = 64–68) [2], but the differences in FN were caused by centromeric repositioning. The same events probably occurred during the karyotypic evolution of N. rosalindae (2n = 48/FN = 50) and N. macedoruizi (2n = 28/FN = 36) [9]. The independent occurrence of both increases and decreases of 2n during the chromosomal evolution of Neacomys (see [5]) is unusual, since a reduction of 2n is the evolutionary trend seen in other taxa (e.g., Akodon) [15].
Comparative chromosome painting for the reconstruction of phylogenies is another approach that has contributed to the knowledge on systematics and karyotypic evolution of several groups of mammals [16], including bats [17,18,19,20] and rodents from the Akodon genus [21].
Given the high chromosomal variation, the presence of distinct evolutionary pathways and the phylogenetic patterns found in Neacomys, the aim of this study is to use chromosome painting with HME probes [11] to examine the chromosomal events leading to the karyotypic diversity of five eastern Amazonian taxa of Neacomys: Neacomys sp. C, Neacomys sp. D, Neacomys sp. E, N. paracou, and N. amoenus (Fig. 1); comparing them with other taxa previously hybridized with HME probes [7, 11, 22,23,24], and investigate if the karyotypic divergence followed the phylogenetic relationships. This will allow the reconstruction of chromosomal homologies for comparison with the molecular phylogenies that include eastern Amazonian species of Neacomys [7].
Map showing collection points of samples of Neacomys. Diploid number (2n) and fundamental number (FN) are shown based on Table 1. The numbers refer to localities mentioned in Table 1. (1) Marabá – PA; (2) Afuá, Marajó Island – PA; (3) Chaves, Marajó Island – PA; (4) Mazagão – AP; (5) Santa Bárbara – PA; (6) Alta Floresta – MT; (7) Itaituba – PA; (8) Jacareacanga – PA; (9) Juruti – PA; (10) Itaituba – PA. Brazilian states are Amapá (AP), Mato Grosso (MT) and Pará (PA). This map was made using QUANTUM-GIS (QGIS) program version 2.10.1. Database was obtained from DIVA and REDLIST
Results
Classic and molecular cytogenetics
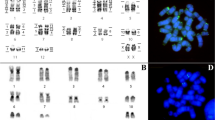
The chromosome painting analysis performed with HME probes on species of Neacomys showed that centromeric (*) and heterochromatic regions do not present hybridization signals. The results are detailed in Table 2 and Figs. 2 and 3.
G-banding with homologies revealed by Hylaeamys megacephalus (HME) probes [11]. a Neacomys sp. C (NSP-C, 2n = 58/FN = 64); b Neacomys sp. D (NSP-D, 2n = 58/FN = 70); c N. paracou (NPA, 2n = 56/FN = 64); d Neacomys sp. E (NSP-E, 2n = 62/FN = 60); e N. amoenus (NAM, 2n = 64/FN = 68). (*) Indicates centromere. (H) Indicates large block of constitutive heterochromatin
Chromosomal associations in species of Neacomys. Numbers on right correspond to homologies revealed by Hylaeamys megacephalus (HME) probes [11]. Abbreviations and numbers below correspond to the chromosomal pair from each species that show the respective chromosomal association. (*) Indicates centromere. Legend: Neacomys sp. C (NSP-C), Neacomys sp. D (NSP-D), N. paracou (NPA), Neacomys sp. E (NSP-E), and N. amoenus (NAM)
Neacomys sp. C (NSP-C, 2n = 58/FN = 64) and Neacomys sp. D (NSP-D, 2n = 58/FN = 70)
Neacomys sp. C karyotype (NSP-C, 2n = 58/FN = 64, Fig. 2a) was previously published with classic banding [5], for specimens collected in Marabá (PA) identified as “Neacomys sp.”. Here we performed chromosome painting with HME probes in this karyotype.
The Neacomys sp. D karyotype (NSP-D, 2n = 58/FN = 70, Fig. 2b) was previously published with classic banding [5], for specimens collected in Chaves town, Marajo Island (PA). Here, we performed chromosome painting with HME probes in this karyotype. We collected an additional sample from Afuá, Marajó Island, which has constitutive heterochromatin (CH) at the centromeric region of almost all autosomes, with large CH blocks in five meta/submetacentric pairs and the X chromosome. The Y chromosome is almost entirely heterochromatic (Additional file 1: Figure S1a). So, the sample of Neacomys sp. D from Afuá shows the same karyotype of Neacomys sp. D from Chaves, and they presented no distinction on the chromosome painting analysis. The divergence between the FN of NSP-C and NSP-D is due to the presence of large CH blocks in the short arms of NSP-D 22, 23, and 26 (metacentrics, Additional file 1: Figure S1a), whereas these blocks are absent in NSP-C 22, 24, and 23 (acrocentrics), respectively.
Twelve of the 24 chromosomal probes show conserved synteny (11 autosomal probes plus the X chromosome), in which five (HME 2, 3, 15, 24 and 25) hybridize to whole chromosomes of NSP-C (2, 3, 17, 11 and 27, respectively) and NSP-D (2, 3, 17, 11 and 27, respectively) and six (HME 4, 12, 18, 20, 21 and 26) hybridize to parts of other chromosomes in NSP-C (1q distal, 4q proximal, 15q proximal, 1q proximal, 5q distal and 28q, respectively) and NSP-D (1q distal, 4q proximal, 15q proximal, 1q proximal, 5q distal and 28q, respectively). The other 12 autosomal probes show more than one signal in NSP-C and NSP-D, with ten (HME 1, 6, 7, 8, (9,10), 11, 14, (16,17), 19 and 23) hybridizing to two chromosomes each; HME 5 and (13,22) show signals in three chromosomes each. In NSP-D the short arms of pairs 22, 23, and 26 did not hybridize to any HME probes because they are heterochromatic (Additional file 1: Figure S1a). Seven NSP-C and NSP-D pairs show association of syntenic blocks to multiple HME probes (Fig. 3, Table 2).
Neacomys paracou (NPA, 2n = 56/FN = 64)
Neacomys paracou karyotype has 2n = 56/FN = 64 with autosomes comprising 22 acrocentric pairs and five meta/submetacentric pairs; the X chromosome is a middle-sized acrocentric and the Y chromosome is a small-sized acrocentric (Fig. 2c; Additional file 1: Figure S1b). The CH is distributed at the centromeric region of all autosomes and the X chromosome. The Y chromosome is almost entirely heterochromatic (Additional file 1: Figure S1b).
Fourteen of the 24 chromosomal probes show conserved synteny (13 autosomal probes plus the X chromosome), in which eight (HME 2, 3, 15, 18, 23, 24, 25 and 26) hybridize to whole chromosomes of NPA (2, 3, 15, 17, 23, 11, 27 and 22, respectively) and five (HME 4, 12, 19, 20 and 21) hybridize to parts of other chromosomes (NPA 1q distal, 4q proximal, 24p distal e 24q, 1q proximal and 5q distal, respectively). The other ten autosomal probes show more than one signal in NPA, with nine (HME 1, 5, 6, 7, 8, (9,10), 11, 14 and (16,17)) hybridizing to two chromosomes each; HME (13,22) gives signals in three chromosomes. Six NPA pairs show association of syntenic blocks to multiple HME probes (Fig. 3, Table 2).
Neacomys sp. E (NSP-E, 2n = 62/FN = 60)
Neacomys sp. E has 2n = 62/FN = 60 with autosomes comprising 30 acrocentric pairs; the X chromosome is a middle-sized acrocentric and the Y chromosome is a small-sized acrocentric (Fig. 2d). The CH is distributed at the centromeric region of all autosomes and the X chromosome. The Y chromosome is almost entirely heterochromatic (Additional file 1: Figure S1c).
Thirteen of the 24 chromosomal probes show conserved synteny (12 autosomal probes plus the X chromosome), in which eight (HME 2, 3, 15, 18, 19, 24, 25 and 26) hybridize to whole chromosomes of NSP-E (2, 3, 15, 27, 18, 11, 29 and 30, respectively) and four (HME 4, 12, 20 and 21) hybridize to parts of other chromosomes (NSP-E 1q distal, 4q proximal, 1q proximal and 5q distal, respectively). The other 11 autosomal probes give more than one signal in NSP-E, with ten (HME 1, 5, 6, 7, 8, (9,10), 11, 14, (16,17) and 23) hybridizing to two chromosomes each; HME (13,22) shows signals in three chromosomes. Four NSP-E pairs show association of syntenic blocks to multiple HME probes (Fig. 3, Table 2).
Neacomys amoenus (NAM, 2n = 64/FN = 68)
The karyotype of Neacomys amoenus has 2n = 64/FN = 68 with autosomes comprising 28 acrocentric pairs and three small metacentric pairs; the X chromosome is a middle-sized submetacentric and the Y chromosome is a small-sized acrocentric (Fig. 2e). CH is distributed at the centromeric region of all autosomes. The X chromosome has a large CH block in the short arm and the Y chromosome is almost entirely heterochromatic (Additional file 1: Figure S1d).
Thirteen of the 24 chromosomal probes show conserved synteny (12 autosomal probes plus the X chromosome), in which eight (HME 2, 3, 15, 18, 19, 24, 25 and 26) hybridize to whole chromosomes of NAM (2, 3, 15, 25, 17, 11, 27 and 30, respectively) and four (HME 4, 12, 20 and 21) hybridize to parts of other chromosomes (NAM 1q distal, 4q proximal, 1q proximal and 5q distal, respectively). The other 11 autosomal probes show more than one signal in NAM, with nine (HME 1, 6, 7, 8, (9,10), 11, 14, (16,17) and 23) hybridizing to two chromosomes each; HME 5 and (13,22) show signals in three chromosomes each. Four NAM pairs show association of syntenic blocks to multiple HME probes (Fig. 3, Table 2).
Chromosomal phylogeny
The comparative analysis of 15 karyotypes from 14 species hybridized with HME whole-chromosome probes [10] generated 56 non-additive multi-state chromosomal characters (Additional file 2: Table S1), which were converted into a matrix (Additional file 3: Table S2) and submitted to parsimony analysis. Characters and character states are detailed in the Additional file 5.
Both heuristic (PAUP) and exhaustive (T.N.T.) analysis recovered the same phylogenetic relationships with distinct support values (bootstrap; Fig. 4). From PAUP analysis the most parsimonious tree was obtained with consistency index 0.691, retention index 0.764, and homoplasy index 0.308; from T.N.T. analysis the implemented algorithm was FUSE and retrieved a best score of 94 in a single tree from a total of 95, 636, 532 rearrangements. The Sigmodontinae subfamily (Fig. 4, node A) showed an initial branching in two reciprocally monophyletic clades, representing the tribes Akodontini (Fig. 4, node B) and Oryzomyini (Fig. 4, node C), both with good reliability and synapomorphies reinforcing the branches. The monophyly of Neacomys was confirmed with bootstrap values of 84% for PAUP analysis and 75% for T.N.T. analysis, and four chromosome signatures (Fig. 4, node D). We recovered a polytomy with no definition among the branches that lead to NSP-B, NSP-E, and the clade composed of NPA, NSP-A, NSP-C and NSP-D. Results are discussed using data retrieved by T.N.T.
Most parsimonious tree based on matrix of chromosomal characters in TNI, AMO, ASP, NLA (outgroup), HME, CLA, OCA-PA, OCA-RJ, NSP-A, NSP-B, NSP-C, NSP-D, NPA, NSP-E and NAM karyotypes. Karyotype abbreviations as in Table 2. Numbers above branches are maximum parsimony bootstrap values analyzed on PAUP (left) and on T.N.T. (right); below are the chromosomal signatures for each node: Node A (Sigmodontinae subfamily), node B (Akodontini tribe), node C (Oryzomyini tribe), and node D (Neacomys genus). The analysis was carried out using PAUP and T.N.T. software packages. Only values above 50% are shown. Inside the box is the ideogram of HME karyotype, as assessed based on HME probes
Discussion
New cytogenetic and distribution data for Neacomys
Neacomys sp. C and D were first described as “Neacomys sp.”, for Marabá and Chaves (Marajó Island) populations, respectively [5]. The samples of Neacomys sp. D from Afuá, Marajó Island (Fig. 1, locality 2) present the same karyotype as samples from the Chaves population (2n = 58/FN = 70) [5]. We named the specimens from Marabá (2n = 58/FN = 64) as Neacomys sp. C, and those from Afuá/Chaves as Neacomys sp. D, to differentiate their karyotypes. The two karyotypes are similar to Neacomys sp. A (2n = 58/FN = 68) [7], since they share the same diploid number (2n = 58), with variations in the FN. However, our comparative analyses reveal the differences in sections below (see Complex chromosomal evolution in Neacomys, and on Additional file 4: Table S3 and Additional file 6: Figure S2).
Neacomys from Vila Rica (Mato Grosso state, Brazil) has a new cytotype (2n = 58/FN = 66) [8]. The molecular phylogeny (Cytb sequences) [8] showed that the Vila Rica samples belonged to the “Marabá clade”, that corresponds to samples of NSP-C, and proposed that the differences between the two FN (66 and 64) are due to pericentric inversions.
We also described a new cytotype with a different FN for N. paracou (2n = 56/FN = 64), distinct from those defined before (2n = 56/FN = 62, 66) [5]. The three karyotypes of N. paracou differ by the number of one-armed and bi-armed chromosomes, probably due to two pericentric inversions. Our samples of N. paracou were collected in Mazagão (Amapá state, Brazil; Fig. 1, locality 4), which is outside the known distribution for N. paracou [3], thus extending its distribution to the southeast of Amapá state.
Recently, a taxonomic revision of Neacomys spinosus (2n = 64/FN = 68) was carried out using molecular (Cytb) and morphological data [6]. Three species were recognized: N. spinosus (sensu strictu), N. amoenus, and N. vargasllosai. Among them, only N. amoenus occurs in Brazil (from eastern Ecuador and northern Peru to central western Brazil and Bolivia). Since the karyotyped samples of the N. spinosus complex were from Brazilian localities [2], the 2n = 64/FN = 68 karyotype must be assigned to N. amoenus, which means that there is no cytogenetic data for N. spinosus at the present time.
The karyotype of Neacomys sp. E (2n = 62/FN = 60) was similar to that described for N. dubosti (2n = 62) in a previous work that provided the diploid number but not showing the karyotype for this taxon [3]. Another karyotype for N. dubosti (2n = 64/FN = 68) was also described in the literature [5]. Comparative analysis of classic banding patterns revealed that the difference between N. dubosti [5] and Neacomys sp. E is probably due to fusion/fission events. Although the two are karyotypically similar our sample was collected in Santa Bárbara (Pará state, Brazil; Fig. 1, locality 5), while the known distribution for N. dubosti is Suriname, French Guyana and north of Amapá state (Brazil) [3,4,5]. Moreover, our taxonomic identification showed that NSP-E does not correspond to any other valid species described to date for Neacomys. Thus, the karyotypic similarity may reflect a convergent evolutionary process, or these taxa may share a common ancestor that diverged recently.
A better delineation of the taxonomic status, distribution and diversity will be possible only after a taxonomic review of members of the genus, including samples from eastern Amazonia.
Complex chromosomal evolution in Neacomys
The comparative chromosome painting analyses among NSP-A (2n = 54/FN = 66), NSP-B (2n = 58/FN = 68) [7], NSP-C (2n = 58/FN = 64), NSP-D (2n = 58/FN = 70), NPA (2n = 56/FN = 64), NSP-E (2n = 62/FN = 60), and NAM (2n = 64/FN = 68) karyotypes allowed us to track the distribution of homologous regions and determine the chromosomal evolution of this group. These analyses show that the chromosomal diversity found is due to 17 fusion/fission events and 1 translocation, pericentric inversions on four syntenic blocks, and six syntenic autosomal blocks with CH amplification/deletion, plus the X chromosome (Additional file 4: Table S3; Additional file 6: Figure S2).
Only four syntenic blocks were found to be stable, with no detectable rearrangement among species of Neacomys (HME 8, HME 6, HME 15, and HME 24). Of them, blocks HME 15 and HME 24 were preserved in other karyotypes from the tribes Akodontini and Oryzomyini (with the exception of AMO, ASP and NLA), while HME 8 and HME 6 were involved in distinct rearrangements in different species.
The data above corroborate the evolutionary pattern described for rodents [25], with fusion/fission events as the most common chromosomal rearrangements, followed by pericentric inversions, and these events are considered as a well-established mode of rapid speciation. The roles of different chromosomal rearrangements in the speciation process have been discussed in the literature since the comparison of rat and mouse genomes [25]. These studies revealed that there can be numerous inversions inside conserved blocks, suggesting that these events may have played a leading role in the genomic reorganization of rodents [25].
Although variation in the number of CH blocks was found in Neacomys karyotypes, which present bi-armed chromosomes with entirely heterochromatic arms (e.g., NSP-A, NSP-C and NSP-D), this is a frequent event in rodents and is the main cause of the variability of sex chromosome morphology [26, 27]. There is no evidence that such variation is involved in speciation [28], since this kind of structural change does not act as a postzygotic isolating mechanism, or generate deleterious meiotic products that could reduce fertility. Thus, our findings are consistent with the idea that the karyotypes obtained from the Marabá and Marajó Island populations (NSP-C and NSP-D, respectively) belong to the same species and exhibit regional chromosome variation [5]. This high karyotypic reorganization is characteristic of Myomorpha rodents [26] with a few exceptions [29].
We note that the rearrangements among the seven karyotypes from six species of Neacomys are not randomly distributed. In fact, Bailey et al. [30] proposed that there are hotspots for chromosomal rearrangements that usually involve repetitive sequences grouped in heterochromatin. The probes HME 5, 11, 14, 19, 23 and 26 are involved in almost half (NSP-A, NSP-C, NSP-D, NPA) or more than half (NSP-B, NSP-E, NAM) of the chromosomes of Neacomys karyotypes, in many complex rearrangements like fusion/fission, translocation, CH amplification/deletion and pericentric inversion (Additional file 4: Table S3. Additional file 6: Figure S2).
Despite the karyotypic similarity among N. minutus (2n = 35–36/FN = 40), N. musseri (2n = 34/FN = 64–68) [2], N. rosalindae (2n = 48/FN = 50) and N. macedoruizi (2n = 28/FN = 36) [9], we cannot assume that the members of this low-2n group must share chromosomal associations with each other, or with other species of Neacomys.
Our chromosome painting analysis of two species of Proechimys with comparable 2n and karyotypes composed mostly of bi-armed chromosomes (P. roberti with 2n = 30/FN = 54 and P. goeldii with 2n = 24♀25♂/FN = 42) suggest that multiple translocations largely account for the karyotypic difference, with full preservation of only three whole chromosomes [31]. Thus, an investigation of the genomes of N. minutus, N. musseri [2], N. rosalindae and N. macedoruizi [9] could help reveal whether the signatures detected in the present study (see Phylogenetic relationships and chromosomal signatures) are maintained in these taxa despite the drastic 2n decrease, or whether they are exclusive for the high-2n group (from 54 to 64). The consolidation of this data could improve our understanding of the chromosomal evolution of Neacomys and potentially facilitate the reconstruction of its hypothetical ancestral karyotype.
Moreover, given that the resolution of chromosome painting is limited and peri/paracentric inversions and reciprocal translocations are difficult to detect, the chromosome evolution in Neacomys may be more complex than currently thought, and could involve more events than those identified in the present study.
Phylogenetic relationships and chromosomal signatures
The phylogenetic analysis using chromosomal data reflects the molecular relationships of the Akodontini and Oryzomyini tribes [32] (Fig. 4). We corroborate the ancestral traits previously proposed for the Sigmodontinae subfamily (Node A, Fig. 4) [7, 23]: HME 7/(9,10), 1/12, 6/21, 11/(16,17), 5/(16,17), 15, 24 and 26. We added HME 8, since it is shared by all Akodontini, as well as HME and OCA (Oryzomyini).
Concerning the Akodontini tribe, TNI was the sister to the other species, followed by NLA, and a clade was comprised of the species of Akodon (AMO + ASP). All members of the Akodontini tribe exhibited the synapomorphies HME 3/25 and 18/25 (Node B, Fig. 4). TNI presented the HME 14/19 association as an autapomorphy, while the clade formed by NLA and AMO + ASP preserved the syntenic blocks, HME 4/11/(16,17) and 2/18, which are chromosomal signatures for this clade.
The relationships within the clade formed by the Oryzomyini species are similar to those recovered in the molecular analysis [32], and all Oryzomyini species presented the signatures previously proposed [7]: HME 8a, 8b, 18, 25 (Node C, Fig. 4). The main difference is related to the position of NPA: In the molecular analysis [7] NPA is the sister species for all Neacomys, whereas in the cytogenetics phylogeny, this position is occupied by NAM and NPA is the sister species only of the clade “NSP-A and NSP-C+NSP-D”. This difference may be consequence of the sampling, since NPA clearly exhibits high genetic variability [13]. This species occupies a large geographic distribution and varies in its haplotypes [13] and chromosomes [5]. Better sampling of NPA, on both the molecular and karyotypic levels, may allow researchers to resolve this discrepancy. Alternatively, there may have been a Long Branch Attraction (LBA) involving NPS-E and NPS-B, since they show rapid karyotypic evolution.
The chromosomal topology analysis recovered the monophyly of Neacomys, with all species included in a polytomy. We compared our chromosomal topology with the previous molecular phylogeny (Cytb and COI) [7], which had included the three undescribed species (NSP-A, NSP-B and NSP-C + NSP-D).
Although the lack of more phylogenetic signals due to the great similarity among Neacomys karyotypes, and the moderate resolution of the chromosomal tree, both topologies (molecular and chromosomal) recovered the clade “NSP-A and NSP-C+NSP-D” as the most recent divergent branch, supported by the signatures HME 19/14/23 and 26/(13,22). All Neacomys karyotypes exhibit the synapomorphic signatures (Node D, Fig. 4) proposed [7]: (HME 20/(13,22)/4, 6a/21, (9,10)/7/(9,10) and 12/(16,17)). The entirety of HME 8 is an ancestral trait in Sigmodontinae (Node A, Fig. 4), where it corresponds to MMU 9 [23]. In Neacomys, however, HME 8 is split into two blocks, 8a and 8b. This is also found in CLA, probably reflecting a homoplasy. HME 6/21 is also an ancestral trait in Sigmodontinae (Node A, Fig. 4), where it corresponds to MMU 2 [23]. In Neacomys there was a fission in the HME 6 portion, originating HME 6a/21 and HME 6b as an independent pair.
Concerning the phylogenetic position of species of Neacomys in the molecular topology [7] and the chromosomal signatures, we noted that NPA presents two Sigmodontinae plesiomorphic characters (HME 11/16 and 19/14/19); however, there are alterations in the chromosomal morphology of both associations (probably due to inversions or centromeric repositioning, which are absent in the other karyotypes of the genus. However, NSP-A and NSP-C + NSP-D exhibit a different chromosome form (HME 19/14/23). We propose that this signature (HME 19/14/19) is present in the ancestral species of the genus, maintained in NPA, derived in NSP-A, NSP-C, and NSP-D (HME 19/14/23), while it was lost in intermediary branches (NAM, NSP-B, and NSP-E).
Another ancestral trait observed is the chromosomal association HME 1/12, present as a derived form in NSP-B (HME 1/12/(16,17)), while the other karyotypes of Neacomys exhibit HME 12/(16,17). The karyotype of this group was shaped by multiple fusion/fission events that occurred during its evolution [5]. Thus, HME 1/12/(16,17) was probably present in the ancestral karyotype of Neacomys, with the HME 1 segment becoming dissociated from the syntenic block during diversification events of the genus, and being maintained only in NSP-B (2n = 54). NSP-A and NSP-C + NSP-D exhibit one ancestral trait for the Sigmodontinae (HME 26) that is changed by the fusion (HME (13,22)/26), which is a signature for these two species.
In conclusion, based on the classic banding and comparative chromosome painting results, we observe that the karyotypic divergence among species of Neacomys followed the phylogenetic relationship, as NPA retained some ancestral traits from the subfamily, while NSP-A, NSP-B and NSP-C + NSP-D shared other synapomorphic characters. This pattern was also observed in many taxonomic groups, with a few exceptions [16].
Conclusions
We describe new karyotypes and distribution areas for species of Neacomys from eastern Amazon and detect, through chromosome painting, that multiple fusion/fission events are the main cause of the karyotypic diversity in this group with high diploid numbers, followed by pericentric inversions and amplification/deletion of CH. Besides, our chromosomal phylogeny recovered the monophyly of Neacomys, and demonstrated the synapomorphies of the genus (HME 6a/21, 6b, 8a, 8b, (9,10)/7b/(9,10), 12/(16,17), 20/(13,22)/4).
Methods
Sampling
In a previous publication [5] we described the classic cytogenetic bandings for two new karyotypes from a Neacomys taxon that we called “Neacomys sp.”. In the present work we made chromosome painting in this taxon. However, as the two karyotypes are different (see Results and Discussion), here we are calling them “Neacomys sp. C” and “Neacomys sp. D”. We studied the karyotypes of five Neacomys sp. C (three males and two females) from Marabá, Pará state, Brazil (Fig. 1, locality 1); 10 Neacomys sp. D, five of which (three males and two females) from Chaves town, Marajo island, Para state, Brazil (Fig. 1, locality 3), and five (three males and two females) from Afuá town (Fig. 1, locality 2), a different locality in Marajo Island that had not sampled before. Also we studied three specimens of N. paracou (two males and one female) from Mazagão municipality (Fig. 1, locality 4), Amapá state, Brazil; one specimen (male) of Neacomys sp. E from Santa Bárbara municipality (Fig. 1, locality 5), Pará state, Brazil; and six specimens of N. amoenus (two males and four females) from Alta Floresta municipality (Fig. 1, locality 6), Mato Grosso state, Brazil.
The samples collections were made using pitfall traps [33]. Pitfall trap lines were distributed in three plots of each locality (Table 1, Fig. 1). In each plot, two traps lines were set up using six 60-l buckets set at intervals of 10 m. The traps were checked early in the morning for eight consecutive days. The specimens were deposited at the zoological collections of Museu de Zoologia da Universidade Federal do Pará (UFPA), Belém, Pará; Museu Paraense Emílio Goeldi (MPEG), Belém, Pará; Laboratório de Mamíferos, Instituto de Pesquisas Científicas e Tecnológicas do Estado do Amapá (IEPA), Macapá, Amapá; and Universidade Federal de Mato Grosso (UFMT), Cuiabá, Mato Grosso. All institutions are located in Brazil.
Cytogenetics
The metaphasic chromosomal samples were acquired from bone marrow extraction [34] and by fibroblast cell culture made from skin biopsy, established at Centro de Estudos Avançados da Biodiversidade, Laboratório de Citogenética (ICB, UFPa, Brazil). G-banding was made following Sumner et al. [35], and C-banding was performed following Sumner [36]. C-banding was performed on G-banded metaphases for the correct chromosomal assignment. Whole chromosome probes of HME [11] were used for FISH studies, with three of the 24 HME probes corresponding to two chromosome pairs each (HME (9,10), (13,22), and (16,17)). We used sequential DAPI staining and converted the DAPI staining in G-band pattern for the correct assignment of the hybridized chromosomes. Chromosome classification was established according to Levan et al. [37]. Digital images were obtained by Nis-Elements software and Nikon H550S microscopy. The final edition of images was made on Adobe Photoshop CS6.
Phylogenetic analysis
The Maximum Parsimony (MP) phylogenetic analysis was made using PAUP program (Phylogenetic Analysis Using Parsimony) version 4.0a159 [38] and T.N.T. (“Tree analyses Using New Technologies”) version 1.1 [39]. Branch support values were calculated with 1000 bootstrap replicates for both analyses. The heuristic search was made using Tree Bisection Reconnection (TBR) in PAUP. An exhaustive search was made using T.N.T.; the Fuse algorithmic was used to examine a total of 95,636,532 rearrangements. A list of characters and character states was created and converted into a non-additive (unordered) multi-state character matrix on Mesquite program version 3.10 [40]. The chosen non-additive multi-state characters were chromosomal morphology, number and syntenic blocks based on HME probes hybridized to metaphases of Neacomys sp. C (NSP-C), Neacomys sp. D (NSP-D), Neacomys sp. E (NSP-E), N. paracou (NPA), N. amoenus (NAM) (present study), and comparing with species from previous studies: Neacomys sp. A (NSP-A), Neacomys sp. B (NSP-B) [7], Cerradomys langguthi (CLA) [11], Thaptomys nigrita (TNI), Akodon montensis (AMO) [22], Akodon sp. (ASP), Necromys lasiurus (NLA) [23], Oecomys catherinae-Pará (OCA-PA), and O. catherinae-Rio de Janeiro (OCA-RJ) [24].
We did not put weight on the characters since we cannot precisely determine the probability of each rearrangement. Also, the characters are used as qualitative traits and not quantitative, as they are rare events with strong phylogenetic signals [41]. The karyotypes of TNI, AMO [22], ASP and NLA [23] were used as outgroups since they belong to the Akodontini tribe, a sister taxon to the Oryzomyini tribe, where Neacomys and the other genera belong. Both tribes belong to the Sigmodontinae subfamily. The tree was displayed and edited in Figtree program version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Availability of data and materials
All the data supporting findings are contained within the manuscript and in the supplemental files. If any additional information is necessary, please contact the corresponding author.
Abbreviations
- 2n:
-
Diploid number
- AMO:
-
Akodon montensis
- ASP:
-
Akodon sp.
- CH:
-
Constitutive heterochromatin
- CLA:
-
Cerradomys langguthi
- COI:
-
Cytochrome C Oxidase – subunit I
- Cytb:
-
Cytochrome B
- FISH:
-
Fluorescent In-Situ Hybridization
- FN:
-
Fundamental number
- HME:
-
Hylaeamys megacephalus
- IEPA:
-
Laboratório de Mamíferos, Instituto de Pesquisas Científicas e Tecnológicas do Estado do Amapá
- IRBP:
-
Interphotoreceptor Retinoid Binding Protein
- MP:
-
Maximum Parsimony
- MPEG:
-
Museu Paraense Emílio Goeldi
- NAM:
-
Neacomys amoenus
- NLA:
-
Necromys lasiurus
- NPA:
-
Neacomys paracou
- NSP-A:
-
Neacomys sp. A
- NSP-B:
-
Neacomys sp. B
- NSP-C:
-
Neacomys sp. C
- NSP-D:
-
Neacomys sp. D
- NSP-E:
-
Neacomys sp. E
- OCA-PA:
-
Oecomys catherinae-Pará
- OCA-RJ:
-
Oecomys catherinae-Rio de Janeiro
- PAUP:
-
Phylogenetic Analysis Using Parsimony
- rDNA:
-
Ribosomal DNA
- T.N.T.:
-
Tree Analyses Using New Technologies
- TBR:
-
Tree Bisection Reconnection
- TNI:
-
Thaptomys nigrita
- UFMT:
-
Universidade Federal de Mato Grosso
- UFPA:
-
Museu de Zoologia da Universidade Federal do Pará
References
Baker RJ, Koop BF, Haiduk MW. Resolving systematic relationships with G-bands. A study of five genera of South American Cricetine rodents. Syst Zool. 1983;32:403–16.
Patton JL, Silva MN, Malcolm JR. Mammals of the Rio Juruá and the evolutionary and ecological diversification of Amazonia. Bul Am Mus Nat Hist. 2000;244:202–92.
Voss RS, Lunde DP, Simmons NB. The mammals of Paracou, French Guiana: a neotropical lowland rainforest fauna. Part 2. Nonvolant species. Bull Am Mus Nat Hist. 2000;263:236.
Redi CA, Zacharias H, Merani S, Oliveira-Miranda M, Aguilera M, et al. Genome sizes in afrotheria, xenarthra, euarchontoglires, and laurasiatheria. J Heredity. 2005. https://doi.org/10.1093/jhered/esi080.
da Silva WO, Pieczarka JC, Rossi RV, Schneider H, Sampaio I, Miranda CL, et al. Diversity and karyotypic evolution in the genus Neacomys (Rodentia, Sigmodontinae). Cytogenet Genome Res. 2015. https://doi.org/10.1159/000441173.
Hurtado N, Pacheco V. Revision of Neacomys spinosus (Thomas, 1882) (Rodentia: Cricetidae) with emphasis on Peruvian populations and the description of a new species. Zootaxa. 2017. https://doi.org/10.11646/zootaxa.4242.3.1.
Oliveira da Silva W, Pieczarka JC, Ferguson-Smith MA, O'Brien PCM, Mendes-Oliveira AC, Sampaio I, et al. Chromosomal diversity and molecular divergence among three undescribed species of Neacomys (Rodentia, Sigmodontinae) separated by Amazonian rivers. Plos One. 2017. https://doi.org/10.1371/journal.pone.0182218.
Di-Nizo CB, Banci KRS, Sato-Kuwabara Y, Silva MJJ. Advances in cytogenetics of Brazilian rodents: citotaxonomy, chromosome evolution and new karyotypic data. Comp Cytogenet. 2017. https://doi.org/10.3897/CompCytogen.v11i4.19925.
Sánchez-Vendizú P, Pacheco V, Vivas-Ruiz D. An introduction to the systematics of small-bodied Neacomys (Rodentia: Cricetidae) from Peru with descriptions of two new species. Am Mus Novit. 2018:1–38 https://doi.org/10.1206/3913.1.
Hice CL, Velazco PM. The non-volant mammals of the Reserva Nacional Allpahuayo Mishana, Iquitos, Peru. Special Publications. Mus Texas Tech Univ. 2012;60:1–135.
Nagamachi CY, Pieczarka JC, O’Brien PCM, Pinto JA, Malcher SM, Pereira AL, et al. FISH with whole chromosome and telomeric probes demonstrates huge karyotypic reorganization with ITS between two species of Oryzomyini (Sigmodontinae, Rodentia): Hylaeamys megacephalus probes on Cerradomys langguthi karyotype. Chromosome Res. 2013. https://doi.org/10.1007/s10577-013-9341-4.
Weksler M, Percequillo AR, Voss RS. Ten new genera of Oryzomyine rodents (Cricetidae, Sigmodontinae). Am Mus Novitates. 2006;3537:29.
Catzeflis F, Tilak M. Molecular systematic of neotropical spiny mice (Neacomys: Sigmodontinae, Rodentia) from the Guiana region. Mammalia. 2009. https://doi.org/10.1515/MAMM.2009.037.
Percequillo AR, Weksler M, Costa LP. A new genus and species of rodent from the Brazilian Atlantic Forest (Rodentia: Cricetidae: Sigmodontinae: Oryzomyini), with comments on oryzomyine biogeography. Zool J Linn Soc Lond. 2011;161:357–90.
Ventura K, O’Brien PC, Yonenaga-Yassuda Y, Ferguson-Smith MA. Chromosome homologies of the highly rearranged karyotypes of four Akodon species (Rodentia, Cricetidae) resolved by reciprocal chromosome painting: the evolution of the lowest diploid number in rodents. Chromosom Res. 2009;17(8):1063–78. https://doi.org/10.1007/s10577-009-9083-5.
Graphodatsky A, Ferguson-Smith MA, Stanyon R. A short introduction to cytogenetic studies in mammals with reference to the present volume. Cytogenet Genome Res. 2012. https://doi.org/10.1159/000341502.
Ao L, Mao X, Nie W, Gu X, Feng Q, Wang J, et al. Karyotypic evolution and phylogenetic relationships in the order Chiroptera as revealed by G-banding comparison and chromosome painting. Chromosom Res. 2007. https://doi.org/10.1007/s10577-007-1120-7.
Pieczarka JC, Gomes AJB, Nagamachi CY, Rocha DCC, Rissino JD, O’Brien PCM, et al. A phylogenetic analysis using multidirectional chromosome painting of three species (Uroderma magnirostrum, U. bilobatum and Artibeus obscurus) of subfamily Stenodermatinae (Chiroptera-Phyllostomidae). Chromosome Res. 2013. https://doi.org/10.1007/s10577-013-9365-9.
Ribas TFA, Rodrigues LRR, Nagamachi CY, Gomes AJB, Rissino JD, O'Brien PCM, et al. Phylogenetic reconstruction by cross-species chromosome painting and G-banding in four species of Phyllostomini tribe (Chiroptera, Phyllostomidae) in the Brazilian Amazon: an independent evidence for monophyly. PLoS One. 2015. https://doi.org/10.1371/journal.pone.0122845.
Gomes AJB, Nagamachi CY, Rodrigues LRR, Benathar TCM, Ribas TFA, O’Brien PCM, et al. Chromosomal phylogeny of Vampyressine bats (Chiroptera, Phyllostomidae) with description of two new sex chromosome systems. BMC Evol Biol. 2016. https://doi.org/10.1186/s12862-016-0689-x.
Hass I, Sbalqueiro IJ, Müller S. Chromosomal phylogeny of four Akodontini species (Rodentia, Cricetidae) from southern Brazil established by ZOO-FISH using Mus musculus (Muridae) painting probes. Chromosome Res. 2008. https://doi.org/10.1007/s10577-007-1211-5.
Suárez P, Nagamachi CY, Lanzone C, Malleret MM, O’Brien PCM, Ferguson-Smith MA, et al. Clues on syntenic relationship among some species of Oryzomyini and Akodontini tribes (Rodentia: Sigmodontinae). PLoS One. 2015. https://doi.org/10.1371/journal.pone.0143482.
Pereira AL, Malcher SM, Nagamachi CY, O’Brien PCM, Ferguson-Smith MA, Mendes-Oliveira AC, et al. Extensive chromosomal reorganization among species of New World muroid rodents (Cricetidae, Sigmodontinae): searching for phylogenetic ancestral traits. PLoS One. 2016. https://doi.org/10.1371/journal.pone.0146179.
Malcher SM, Pieczarka JC, Geise L, Rossi RV, Pereira AL, O'Brien PCM, et al. Oecomys catherinae (Sigmodontinae, Cricetidae): evidence for chromosomal speciation? PLoS One. 2017. https://doi.org/10.1371/journal.pone.0181434.
Romanenko SA, Volobouev V. Non-Sciuromorph rodent karyotypes in evolution. Cytogenet Genome Res. 2012. https://doi.org/10.1159/000339294.
Romanenko SA, Perelman PL, Trifonov VA, Graphodatsky AS. Chromosomal evolution in Rodentia. Heredity. 2011. https://doi.org/10.1038/hdy.2011.110.
Kasahara S. Introdução à Pesquisa em Citogenética de Vertebrados. 1st ed: Ribeirão Preto, SP, Brasil. Sociedade Brasileira de Genética; 2009.
King M. Chromosomal rearrangements, speciation and the theoretical approach. Heredity. 1987;59:1–6.
Swier VJ, Bradley RD, Rens W, Elder FFB, Baker RJ. Patterns of chromosomal evolution in Sigmodon, evidence from whole chromosome paints. Cytogenet Genome Res. 2009. https://doi.org/10.1159/000218747.
Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. https://doi.org/10.1186/gb-2004-5-4-r23.
Da Silva WO, Rodrigues da Costa MJ, Pieczarka JC, Rissino J, Pereira JC, Ferguson-Smith MA, Nagamachi CY. Identification of two independent X-autosome translocations in closely related mammalian (Proechimys) species. Sci Rep. 2019. https://doi.org/10.1038/s41598-019-40593-8.
Pardiñas UFJ, Teta P, Salazar-Bravo J. A new tribe of Sigmodontinae rodents (Cricetidae). Mastozool Neotrop. 2015;22(1):171–86.
Corn PS. Straight-line drift fences and pitfall traps. In: Heyer WR, Donnelly MA, McDiarmid RW, Hayek LC, Foster MS, editors. Measuring and monitoring biological standard methods for amphibians. Washington, DC: Smithsonian Institution Press; 1994. p. 109–17.
Ford CE, Hamerton JL. A colchicine, hypotonic – citrate, squash sequence for mammalian chromosomes. Stain Technol. 1956;31:247–51.
Sumner AT, Evans HJ, Buckland RA. New technique for distinguishing between human chromosomes. Nature (Lond) New Biol. 1971;31:282.
Sumner AT. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972;75:304–6 PMID: 4117921.
Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–20.
Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland: Sinauer Associates; 2002.
Goloboff PA, Farris JS, Nixon KC. T.N.T., a free program for phylogenetic analysis. Cladistics. 2008;24:774–86.
Maddison WP, and Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 3.40. 2018, http://mesquiteproject.org.
Rokas A, Holland PWH. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;15(11):454–9.
Acknowledgements
The authors thank Claudia Silva (IEPA), for helping in the field expedition to Amapá state, and Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) and Secretaria de Estado de Meio Ambiente do Pará (SEMA-PA) for the authorization of sample collections. This study is part of the Doctoral thesis of WOS in Genetic and Molecular Biology, under a CAPES Doctoral Scholarship included in a project Pró-Amazônia (Proc. 047/2012) coordinated by CYN. MJRC is a Doctoral student in Bionorte and Biodiversity, under a CAPES Doctoral Scholarship included in a project Pró-Amazônia (Proc. 047/2012) coordinated by CYN. CYN (305880/2017-9) and JCP (305876/2017-1) are grateful to CNPq for Productivity Grants.
Funding
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Fundação Amazônia Paraense de Amparo à Pesquisa (FAPESPA) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) on projects coordinated by CY Nagamachi (Edital BIONORTE-CNPq, Proc 552032/2010–7; Edital BIONORTE-FAPESPA, ICAAF 007/2011; Edital Pró-Amazônia Proc 047/2012); the FAPESPA (Edital Vale – Proc 2010/110447) and Banco Nacional de Desenvolvimento Econômico e Social – BNDES (Operação 2.318.697.0001) on a project coordinated by JC Pieczarka; the Fundação de Amparo à Pesquisa do Estado de Mato Grosso (FAPEMAT) on a project coordinated by RV Rossi (Proc 477017/2011). The funding body had no role in the design of the study and collection, analysis, and interpretation of data and in writing of the present paper.
Author information
Authors and Affiliations
Contributions
WOdS, JCP, MJRdC, MAF-S, PCMO, CYN: gave substantial contributions to the conception of the work; acquisition, analysis, and interpretation of cytogenetic data; WOdS, MJRdC, ACM-O, RVR: gave a substantial contribution to the field collection of biological samples; WOdS, JCP, MJRdC, MAF-S, PCMO, ACM-O, RVR, CYN: participated in the draft of the work or revised it critically for important intellectual content; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The animals were maintained in the lab with food and water, free from stress, until their euthanasia by IP injection of buffered and diluted barbiturates under local anesthetic (Ketamine HCl in combination with Diazepam), according to procedures recommended by the American Society of Mammalogists. The Ethics Committee (Comitê de Ética Animal da Universidade Federal do Pará) approved this research (Permit 68/2015). JCP has a permanent field permit, number 13248 from “Instituto Chico Mendes de Conservação da Biodiversidade”. The Cytogenetics Laboratory, CEABIO from UFPa has permit number 19/2003 from the Ministry of Environment for sample transport and permit 52/2003 for using the samples for research.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
C-banding in Neacomys. (A) Neacomys sp. D (NSP-D, 2n = 58/FN = 70); (B) N. paracou (NPA, 2n = 56/FN = 64); (C) Neacomys sp. E (NSP-E, 2n = 62/FN = 60); (D) N. amoenus (NAM, 2n = 64/FN = 68).
Additional file 2: Table S1.
Non-additive multi-state character list. Character descriptions are detailed in Additional file 5.
Additional file 3: Table S2.
Chromosome character data matrix of non-additive multi-state character employed in the Maximum Parsimony analysis.
Additional file 4: Table S3.
Chromosomal rearrangements among seven karyotypes of six Neacomys species, based on chromosome painting with Hylaeamys megacephalus probes (HME) [10].
Additional file 5.
Analysis and description of characters and characters states.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Oliveira da Silva, W., Pieczarka, J.C., Rodrigues da Costa, M.J. et al. Chromosomal phylogeny and comparative chromosome painting among Neacomys species (Rodentia, Sigmodontinae) from eastern Amazonia. BMC Evol Biol 19, 184 (2019). https://doi.org/10.1186/s12862-019-1515-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-019-1515-z