Abstract
Background
The family Phyllostomidae (Chiroptera) shows wide morphological, molecular and cytogenetic variation; many disagreements regarding its phylogeny and taxonomy remains to be resolved. In this study, we use chromosome painting with whole chromosome probes from the Phyllostomidae Phyllostomus hastatus and Carollia brevicauda to determine the rearrangements among several genera of the Nullicauda group (subfamilies Gliphonycterinae, Carolliinae, Rhinophyllinae and Stenodermatinae).
Results
These data, when compared with previously published chromosome homology maps, allow the construction of a phylogeny comparable to those previously obtained by morphological and molecular analysis. Our phylogeny is largely in agreement with that proposed with molecular data, both on relationships between the subfamilies and among genera; it confirms, for instance, that Carollia and Rhinophylla, previously considered as part of the same subfamily are, in fact, distant genera.
Conclusions
The occurrence of the karyotype considered ancestral for this family in several different branches suggests that the diversification of Phyllostomidae into many subfamilies has occurred in a short period of time. Finally, the comparison with published maps using human whole chromosome probes allows us to track some syntenic associations prior to the emergence of this family.
Similar content being viewed by others
Background
The family Phyllostomidae is the third most speciose within the order Chiroptera, with 60 genera and 200 species [1], being grouped into 11 subfamilies: Macrotinae, Micronycterinae, Desmodontinae, Lonchorhininae, Phyllostominae, Glossophaginae, Lonchophyllinae, Carolliinae, Glyphonycterinae, Rhinophyllinae, and Stenodermatinae [2]. This family shows huge morphological, molecular and cytogenetic variation, both between and within species, in addition to much controversy about its taxonomy [2,3,4,5].
Traditionally, the subfamily Carolliinae consists of genera Carollia (10 spp.) and Rhinophylla (3 spp.) [3, 4, 6, 7]. However, in the classification proposed by Baker et al. [2] these two genera are not closely related. The subfamily Carolliinae (composed by the genus Carollia) was included as sister group to Glyphonycterinae, a new subfamily that includes Glyphonycteris and Trinycteris that were previously subgenera within Micronycteris [8,9,10], and later were raised to the genus level [3, 11]. Rhinophylla was included in its own subfamily (Rhinophyllinae) that was seen as closely related to the subfamily Stenodermatinae.
In the Baker et al. [2] classification, the group of Carolliinae + Glyphonycterinae + Rhinophyllinae + Stenodermatinae was designated Nullicauda, an unranked taxon, initially proposed by Wetterer et al. [3] for the group consisting of subfamilies Carolliinae and Stenodermatinae.
Cytogenetic studies in species of the Nullicauda group show extensive interspecific and intraspecific chromosome variation [12]. To further investigate the controversial phylogenetic hypotheses based on morphological and molecular data in Nullicauda, we analyzed chromosome evolution in this group through multidirectional chromosome painting using whole chromosome paint probes from two phyllostomid bats, Phyllostomus hastatus (PHA) and Carollia brevicauda (CBR) [13], in the species Rhinophylla pumilio, Rhinophylla aff. fischerae, Trinycteris nicefori and Glossophaga soricina. The results obtained here were compared with those previously published with the same probes in other species, in order to construct a phylogeny based on chromosomal homology data. We also integrated the comparative chromosome maps obtained with these probes with maps generated using human probes in Glossophaga soricina [14, 15].
Methods
Specimens examined
Chromosome banding and painting were used to analyze the karyotypes of bats from three phyllostomid subfamilies, Glossophaginae, Glyphonycterinae and Rhinophyllinae (Table 1), with the last two being members of the Nullicauda group. The specimens were captured with mist nets. Chromosome preparations and tissue biopsies were sent to the Cytogenetic Laboratory of the Federal University of Pará. Specimens were deposited in the Mastozoology collection of the Emilio Goeldi Museum and Zoology Museum of the Western Pará University.
Chromosome preparations, cell culture and chromosome banding
Metaphase chromosomes were obtained from direct bone marrow extraction [16], and through primary culture of fibroblasts [17]. G-banding patterns were obtained from incubation in enzymatic trypsin solution [18], and subsequent incubation in saline solution (0.5X SSC) and Wright dye staining diluted in phosphate buffer at a ratio of 1: 3 v / v. Staining time was 5 min. Karyotypes were organized following the pattern in the literature for each species.
Fluorescence in situ hybridization (FISH)
Multidirectional chromosome painting was performed using whole chromosome probes from CBR and PHA (Phyllostomidae), generated by flow cytometry [13]. The probes were amplified and labeled by DOP-PCR [19, 20] and hybridized following procedures previously described [13, 20]. Briefly, the slides were incubated in a pepsin enzyme solution, washed in 2× SSC and dehydrated in an alcohol series (70%, 90% and 100%). Subsequently the slides were oven aged at 65 °C for 1 hour, denatured for 1 min in formamide solution (70% formamide/ 2× SSC) and incubated in hybridization solution (14 μl of solution containing: 50% formamide, 2× SSC, 10% dextran sulfate, 5 μg of salmon sperm DNA, 2 μg mouse Cot-1 DNA and 1 μl of labelled PCR product) for 3 days. After post-hybridization stringency washing, biotin-labeled probes were detected with avidin-Cy3 or avidin-FITC (1 μg/ml; Amersham). For dual color-FISH we used both biotin-labeled and direct-labeled probes. Digital images were captured using Axiovision 3.0 software via an Axiocam mRM CCD camera, coupled to a Zeiss Axioplan 2 microscope or with the Nis-Elements software on a Nikon H550S microscope. Chromosomes were identified by chromosomal morphology and by staining patterns with the fluorochrome DAPI (4 ‘, 6-diamidino-2-phenylindole).
Literature data
The results obtained here were compared with those using PHA and CBR probes [13] previously published for Chiroderma villosum, Mesophylla macconnelli, Vampyressa thyone, Vampyriscus bidens and Vampyriscus brocki from subtribe Vampyressina [21]; Artibeus obscurus, Uroderma bilobatum and Uroderma magnirostrum from the subfamily Stenodermatinae [22], thus allowing phylogenetic analysis (see below) of the Nullicauda group. Finally, the mapping data performed here from Glossophaga soricina (GSO) were compared with those of [14, 15], allowing the establishment of chromosomal correspondence between PHA and CBR bats and human.
Phylogenetic analysis using chromosomal characters
A binary matrix was constructed showing the presence and absence of discrete characters through chromosome homologies among the species analyzed in this paper and those from literature (see above) and is available as a Additional file 1. The results generated were used in a parsimony cladistic analysis. We used the PHA species as outgroup because 1) it is the most basal among the species analyzed here; 2) its karyotype is similar to Macrotus californicus (MCA), a species of the most basal genus of Phyllostomidae according to Baker et al. [2]; and 3) its karyotype is similar to the ancestral karyotype [22]. The analysis of Maximum Parsimony was made using PAUP 4.0b10 (Phylogenetic Analysis Using Parsimony) [23]. A heuristic search to find the most parsimonious tree(s) was performed using Tree Bisection Reconnection (TBR) branch-swapping; the posterior bootstrap probability was obtained with one thousand replicates.
Results
Rhinophylla pumilio
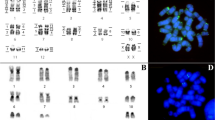
Rhinophylla pumilio (RPU) shows a karyotype with 2n = 34, and FN = 62 (Fig. 1a), as well as G-, C-banding and Ag-NOR staining, FISH with telomeric and ribosomal DNA probes (not shown) similar to those from literature [24].
Hybridization of PHA whole chromosome probes onto the genome of RPU revealed 17 homologous segments (Fig. 1a). Fifteen PHA paints (PHA 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15 and X) are fully preserved in RPU 1, 2, 4, 3, 5, 6, 7, 8, 9, 10, 11, 12, 14, 16 and X chromosomes, respectively. Only PHA13 showed two hybridization signals, in RPU 13 and RPU 15q.
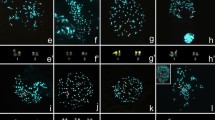
Hybridization of CBR whole chromosome probes onto the genome of RPU revealed 26 homologous segments (Fig. 1a). Eleven chromosome pairs of RPU hybridized with only one probe of CBR: RPU 4 (CBR 1), RPU 5 (CBR 1), RPU 10 (CBR 3), RPU 15q (CBR 5), RPU 12 (CBR 6), RPU 11 (CBR 7), RPU 13 (CBR 8), RPU 14 (CBR 9), RPU 16 (CBR 6), RPU X (CBR X) and RPU Y (CBR Y1). CBR probes 7, 8, 9 and X were fully conserved in RPU. Six chromosome pairs of RPU hybridized with two different probes of CBR: RPU 1 (CBR 3/4), RPU 3 (CBR 2/5), RPU 6 (CBR 1/Y2), RPU 7 (CBR 3/1), RPU 8 (CBR 4/2) and RPU 9 (CBR 2/1). One chromosome pair of RPU (RPU 2) hybridized with two probes of CBR, but with four signals: CBR 2/Y2/2/Y2. Examples of painting with CBR and PHA probes can be seen in Fig. 2a.
Rhynophylla aff. fischerae
Rhinophylla aff. fischerae (RFI) has a karyotype with 2n = 38 and FN = 68 (Fig. 1b). The results on G-, C-banding and Ag-NOR staining, FISH with telomeric and ribosomal DNA probes (not shown) are similar to those from literature [25].
Chromosome painting with whole chromosome probes of PHA revealed 24 homologous segments in RFI (Fig. 1b). Fourteen chromosome pairs of RFI hybridized with only one probe of PHA each: RFI 5 (PHA 6), RFI 6 (PHA 1), RFI 7 (PHA 1), RFI 8 (PHA 3), RFI 9 (PHA 9), RFI 10 (PHA 11), RFI 11 (PHA 12), RFI 12 (PHA 14), RFI 14 (PHA 4), RFI 15 (PHA 2), RFI 16 (PHA 15), RFI 17 (PHA 7), RFI 18 (PHA 13) and RFI X (PHA X). Five chromosome pairs of RFI hybridized with two probes of PHA: RFI 1 (PHA 8/5); RFI 2 (PHA 2/3); RFI 3 (PHA 6/4); RFI 4 (PHA 8/7); RFI 13 (PHA 10/13).
The RFI genome revealed 29 hybridization signals with whole chromosome probes of CBR (Fig. 1b). Eleven chromosome pairs of RFI hybridized with only one probe of CBR each: RFI 5 (CBR Y2), RFI 6 (CBR 3), RFI 7 (CBR 4), RFI 8 (CBR 1), RFI 10 (CBR 7), RFI 11 (CBR 6), RFI 12 (CBR 9), RFI 14 (CBR 2), RFI 16 (CBR 6), RFI 17 (CBR 1), RFI 18 (CBR 5) and RFI X (CBR X). Five chromosome pairs of RFI hybridized with two probes of CBR: RFI 1 (CBR 2/1), RFI 3 (CBR 1/5); RFI 4 (CBR 4/3), RFI 9 (CBR 2/1) and RFI 13 (CBR 3/8). The chromosome pairs RFI 2 and RFI 15 had four (CBR 2/Y2/2/1) and three (CBR 2/Y2/2) hybridization signals, respectively. Pair RFI 16 hybridized only in its short arm (CBR 6), while the long arm was fully heterochromatic. Examples of painting with CBR and PHA probes can be seen in Fig. 2b.
Trinycteris nicefori
Trinycteris nicefori (TNI) has a karyotype with 2n = 28 and FN = 52 (Fig. 1c), a similar result to those from literature [26]. The chromosome complement consists of 10 meta/submetacentric and three subtelocentric chromosomes. The X chromosome is submetacentric and the Y is acrocentric. The sequential G-, C-banding pattern showed constitutive heterochromatin blocks in the pericentromeric region of all chromosomes and faint blocks in the distal portion of the short arm of pairs 4 and 6. Staining with silver nitrate showed the NOR in the proximal region of the short arm of pair 6. Fluorescence in situ hybridization with telomeric probes showed signs only at chromosomal tips and 18S rDNA probes confirmed the silver nitrate staining. These data were used to confirm the karyotype analysis but are not shown here.
Chromosome painting with whole chromosome probes of CBR and PHA onto the genome of TNI revealed 24 and 21 conserved homologous segments, respectively (Fig. 1c). Eight PHA paints (PHA 1, 6, 7, 8, 9, 10, 11 and X) are fully preserved in TNI 1, 5, 6, 7, 8, 9, 10 and X chromosomes, respectively. Five TNI hybridized with two different probes of PHA: TNI 2 (PHA 4/3), TNI 3 (PHA 3/2), TNI 4 (PHA 5/2), TNI 12 (PHA 13/5) and TNI 13 (PHA 13/4). Only TNI 11 hybridized with 3 PHA probes (PHA 12/15/14).
Hybridizations with CBR probes showed that four TNI pairs hybridized with a single CBR probe each: TNI 9 (CBR 3), TNI 10 (CBR 7), TNI 13 (CBR 5) and TNI X (CBR X). The remaining pairs hybridized with two CBR probes each: TNI 1 (CBR 4/3), TNI 2 (CBR 2/1), TNI 3 (CBR 1/Y2), TNI 4 (CBR 1/2), TNI 5 (CBR 1/Y2), TNI 6 (CBR 3/1), TNI 7 (CBR 4/2), TNI 8 (CBR 2/1), TNI 11 (CBR 6/9) and TNI 12 (CBR 8/1). Examples of painting with CBR and PHA probes can be seen in Fig. 2c.
Glossophaga soricina
Glossophaga soricina (GSO) has a karyotype with 2n = 32 and FN = 60 (Fig. 1d). These data, as well as those of classical cytogenetics in this species, are similar to those from the literature [14, 15, 27]. The karyotype was organized following Sotero-Caio et al. [15], with the aim of comparing the PHA and CBR probes with human probes (see below).
Chromosome painting with whole chromosome probes of CBR and PHA onto the genome of GSO revealed 18 and 26 conserved homologous segments (Fig. 1d). Fourteen chromosome pairs of GSO hybridized with only one probe of PHA each: GSO 1 (PHA 1), GSO 2 (PHA 4), GSO 3 (PHA 2), GSO 4 (PHA 3), GSO 5 (PHA 5), GSO 6 (PHA 6), GSO 7 (PHA 7), GSO 9 (PHA 8), GSO 10 (PHA 9), GSO 11 (PHA 10), GSO 13 (PHA 13), GSO 14 (PHA 14), GSO 15 (PHA 13) and GSO X (PHA X). Two chromosome pairs of GSO hybridized with two probes of PHA: GSO 8 (PHA 11/12) and GSO 12 (PHA 15/12).
Eight chromosome pairs of GSO hybridized with only one probe of CBR each: GSO 4 (CBR 1), GSO 5 (CBR 1), GSO 11 (CBR 3), GSO 12 (CBR 6), GSO 13 (CBR 8), GSO 14 (CBR 9), GSO 15 (CBR 5) and GSO X (CBR X). Seven chromosome pairs of GSO hybridized with two probes of CBR: GSO 1 (CBR 3/4), GSO 2 (CBR 2/5), GSO 6 (CBR 1/Y2), GSO 7 (CBR 3/1), GSO 8 (CBR 7/6), GSO 9 (CBR 4/2) and GSO 10 (CBR 2/1). Pair GSO 3 hybridized with two probes of CBR, but with four signals: CBR 2/Y2/2/Y2. Examples of painting with CBR and PHA probes can be seen in Fig. 2d.
Correspondence among PHA, CBR and human whole chromosome probes
Once we mapped the GSO genome with the PHA and CBR probes, we were able to determine the correspondence of these probes with human (HSA) chromosomes, since previous works [14, 15] performed HSA mapping in GSO (Fig. 3).
Phylogenetic analyses using chromosomes as characters
Based on chromosome homologies identified by multidirectional chromosome painting, we analyzed the genera here mapped, together with the previously mapped PHA, CBR, Uroderma magnirostrum (UMA, 2n = 36, FN = 62), Uroderma bilobatum (UBI, 2n = 42, FN = 50), Artibeus obscurus (AOB, 2n = 30/31, FN = 56), and the Vampyressina Chiroderma villosum (CVI, 2n = 26, FN = 48), Mesophylla macconnelli (MMA, 2n = 21/22, FN = 18), Vampyressa thyone (VTH, 2n = 23/24 FN = 20), Vampyriscus bidens (VBI, 2n = 26, FN = 48) and Vampyriscus brocki (VBR, 2n = 24, FN = 44). We used a total of 93 discrete chromosomal characters to build a matrix of their presence or absence (Additional file 1: Table S1). The Maximum Parsimony analysis (MP) resulted in nine equally parsimonious trees (Tree length = 124, Consistence index = 0.75, retention index = 0.7578, Homoplasy index = 0.25). The main branch leads to all analyzed species except the outgroup (Fig. 4). After the split from PHA, the next branch leads to RPU (Rhinophyllinae), then GSO (Glossophaginae), TNI (Glyphonycterinae), CBR (Carolliinae), RFI (Rhinophyllinae) and the Stenodermatinae. In the last subfamily the phylogenetic relationships included Vampyressina, previously analyzed [21]. The present study confirmed the internal relationships of Stenodermatinae presented in that paper. RPU and RFI belong to the same subfamily but are in different branches because of the lack of phylogenetic signal in RPU (see Discussion).
Maximum parsimony tree obtained with PAUP software, using chromosome characters resulted from comparative mapping of the species here studied. Abbreviations are described in Table 1. Other symbols: “p” = short arm; “q” = long arm; “meta” = metacentric chromosome; “acro” = acrocentric chromosome; “/” = syntenic groups physically linked; “i” = inversion; “p + q” = part of short arm linked with part of the long arm. On the top are the subfamilies. Bold numbers (over the branches) are the bootstrap values for one thousand replicates. Ideograms: 1) into the box are the chromosomes of Phyllostomus hastatus, used as probes; each chromosome is represented by G-banding and a specific color. The numbering on all the figure are related to this species karyotype, the outgroup. 2) Below the tree is the proposed ancestral karyotype of Phyllostomidae [22]. 3) Below each branch are represented the chromosomes found on each species or group of species karyotypes which resulted from rearrangements and are relevant to the present analysis
Mapping of chromosome changes in the phylogeny obtained
Comparing the data matrix with the phylogeny of Fig. 4 it was possible to list the chromosomal rearrangements that served as chromosome signatures for each branch.
Discussion
Whole chromosomes probes from CBR and PHA, previously described [13], were hybridized onto the genomes of RPU, RFI, TNI and GSO in order to establish chromosomal homologies between these four species. Our results were integrated with those present in the literature for both these probes and also for human probes.
Intra-specific and intrageneric chromosome variation in Rhinophylla
Multidirectional chromosome painting in RFI compared to RPU show many rearrangements between the two species. When comparing these results with Colombian RFI [28], the karyotype of Colombian RFI is similar to the RPU analyzed here, which emphasizes the differences between RFI of the two regions (Colombia and Brazil). These differences support the hypothesis that postulate the RFI from Brazil as a new species [25], Rhinophylla aff. fischerae, based on its reproductive isolation from the RFI of Colombia (due to multiple chromosomal rearrangements).
Karyotypes and phylogeny
When comparing the nine equally parsimonious phylogenies obtained, we found that they were similar, with some differences due to polytomy. Considering the molecular phylogenies already published [1, 2] we opted for the phylogeny presented in Fig. 4. Analyzing the phylogeny, it is necessary to take into account that RPU, GSO and TNI show practically identical karyotypes to PHA, which in turn is similar to the MCA karyotype and to the ancestral karyotype of Phyllostomidae [22]. Thus the branching that places RPU near PHA is artificial and reflects the absence of phylogenetically informative signal in the RPU karyotype. This occurs more or less similarly with GSO and TNI, although some phylogenetic signal is shared with CBR, placing them in an intermediate position (see below). Thus, except for the RPU position, the phylogeny is similar to [1]. The recurrence of the karyotype similar to the hypothetical ancestral karyotype of Phyllostomidae throughout this phylogeny confirms its origin before the diversification of most subfamilies, implying that each subfamily developed its karyotypes independently from the ancestral karyotype. This would occur, for example, if the diversification of the branches that formed the subfamilies occurred in a short period of time. Evidence that this actually occurred was obtained by concatenating mitochondrial genomes with nuclear genes [29]. This fact explains the considerable difficulty in constructing Phyllostomidae phylogenies from chromosome data (even with precise tools such as chromosome painting) due to the few synapomorphies between subfamilies, which can be seen in the data matrix (Additional file 1).
The ancestral karyotype of Phyllostomidae and chromosome signatures
Placing the chromosomal rearrangements in the phylogeny (Fig. 4) shows that almost all nodes are characterized by chromosomal rearrangements. We highlight the rearrangement PHA 7q10, which is present from CBR to MMA, uniting the subfamilies Carolliinae, Rhinophyllinae and Stenodermatinae. PHA 12/15, PHA 13/4 and the inversion in PHA 2p2q groups TNI and CBR species. Thus, although bootstrap values are eventually low, the occurrence of these rearrangements generates considerable confidence in this phylogeny.
Shared rearrangements with other families
The correspondence among the human whole chromosome probes with PHA and CBR (Fig. 3) allows the comparison of the Phyllostomidae karyotypes with representatives of other families where the human probes were mapped. A comparison of the human syntenic associations present in the karyotypes of several families was previously made [30]. For Phyllostomidae they used the mapping of GSO [14]. Yangochiroptera (superfamilies Emballonuroidea, Noctilionoidea and Vespertilionoidea) the association HSA 5/7/16 was shared. With the exception of Emballonuridae, the other families shared HSA 4/10/14/15/14/15, HSA 7a/7b and HSA 18/20. HSA 13/8/4 is an association even older, shared between Yangochiroptera and Megadermatidae (Yinpterochiroptera). Thus, all these associations already existed before the appearance of Phyllostomidae and therefore were part of the ancestral karyotype of the family. HSA 5/7/16 corresponds to PHA 3q; HSA 4/10/14/15/14/15 to PHA 1; HSA 7a/7b to PHA 10; HSA 18/20 to PHA 9 and HSA 13/8/4 to PHA 11 (Fig. 3). The PHA chromosomes mentioned above are conserved in most of the analyzed species of Phyllostomidae and are also present in the family ancestral karyotype [22], with the exception of PHA 1, which is separated as PHA 1p (HSA 4 / 10) and PHA 1q (HSA 14/15/14/15). The same occurs in MCA [15], where HSA 4/10 corresponds to MCA 5 and HSA 14/15/14/15 to MCA 10. One point raised is that Robertsonian rearrangements are prone to homoplasy [30]. Thus, although PHA, GSO, RPU and TNI share the association found in Molossidae, Phyllostomidae and Vespertilionidae, the question remains whether the ancestral karyotype of Phyllostomidae had the chromosome arms corresponding to PHA 1 associated or separated.
Finally, the association HSA 4/10 is reshuffled via an inversion, which resulted in the HSA 4/10/4 arrangement in all species of Phyllostomidae so far studied. However, this inversion is absent from the other families of Chiroptera, representing a potential chromosomal signature exclusive for Phyllostomidae.
Conclusions
In conclusion, although the phylogenetic power of chromosome painting data has somehow been affected by the rapid radiation of subfamily lineages, the data here presented show the chromosome signatures that support the validity of the Nullicauda by demonstrating phylogenetic associations between Rhinophyllinae-Stenodermatinae and Carolliinae-Glyphonycterinae.
Abbreviations
- 2n:
-
Diploid number
- Ag-NOR:
-
Silver nitrate staining
- AOB:
-
Artibeus obscurus
- CBR:
-
Carollia brevicauda
- CVI:
-
Chiroderma villosum
- DAPI:
-
4′, 6-diamidino-2-phenylindole
- DOP-PCR:
-
Degenerate Oligonucleotide-Primed-Polymerase Chain Reaction
- FISH:
-
Fluorescence In Situ Hybridization
- FN:
-
Fundamental Number
- GSO:
-
Glossophaga soricina
- HSA:
-
Homo sapiens
- MCA:
-
Macrotus californicus
- MMA:
-
Mesophylla macconnelli
- MP:
-
Maximum parsimony
- NOR:
-
Nucleolar Organizer Region
- PAUP:
-
Phylogenetic Analysis Using Parsimony
- PHA:
-
Phyllostomus hastatus
- RFI:
-
Rhinophylla aff. fischerae
- RPU:
-
Rhinophylla pumilio
- SSC:
-
Saline Sodium Citrate
- TBR:
-
Tree Bisection Reconnection
- TNI:
-
Trinycteris nicefori
- UBI:
-
Uroderma bilobatum
- UMA:
-
Uroderma magnirostrum
- VBI:
-
Vampyriscus bidens
- VBR:
-
Vampyriscus brocki
- VTH:
-
Vampyressa thyone
References
Baker RJ, Solari S, Cirranello A, Simmons NB. Higher level classification of phyllostomid bats with a summary of DNA synapomorphies. Acta Chiropterologica. 2016;18(1):1–38.
Baker RJ, Hoofer SR, Porter CA, Van Den Bussche RA. Diversification among new world leaf-nosed bats: an evolutionary hypothesis and classification inferred from digenomic congruence of DNA sequence. Occas Pap Mus Texas Tech Univ. 2003a;230:1–32.
Wetterer AL, Rockman MV, Simmons NB. Phylogeny of phyllostomid bats (Mammalia: Chiroptera): data from diverse morphological systems, sex chromosomes and restriction sites. Bull Am Mus Nat Hist. 2000;248:1–200.
Jones JK, Purvis A, Maclarnon A, Bininda-Emonds ORP, Simmons NB. A phylogenetic supertree of the bats (Mammalia: Chiroptera). Biol Rev. 2002;77:223–59.
Baker RJ, Porter CA, Patton JC, Van Den Bussche RA. Systematics of bats of the family Phyllostomidae based on RAG2 DNA sequences. Occas Pap Mus Texas Tech Univ. 2000;202:1–16.
Simmons NB. Order Chiroptera. In: Wilson DE, Reeder DM, editors. Mammal species of the world: taxonomic and geographic reference. 3rd ed. Baltimore: Johns Hopkins University Press; 2005. p. 312–529.
Solari S, Baker RJ. Mitochondrial DNA sequence, karyotypic, and morphological variation in the Carollia castanea species complex (Chiroptera: Phyllostomidae) with description of the new species. Occas Pap Mus Texas Tech Univ. 2006;254:1–16.
Sanborn CC. Bat of the genus Micronycteris and its subgenera. Chicago Nat Hist Mus. 1949;31:215–33.
Koopman KF. Chiroptera: Systematics. Handb. Der Zool. 1994;7:60. Mammalia: 1–217
Simmons NB. A new species of Micronycteris (Chiroptera: Phyllostomidae) from northeastern Brazil, with comments on phylogenetic relationships. Am Mus Novit. 1996;3158:1–34.
Simmons NB, Voss RS. The mammals of Paracou, French Guiana: a Neotropical lowland rainforest fauna. Part. 1. Bats. Bull Am Mus Nat Hist. 1998;237:1–219.
Moratelli R, Morielle-Versute E. Métodos e aplicações da citogenética na taxonomia de morcegos brasileiros. In: Reis NR, Peracchi AL, Pedro WA, Lima IP, editors. Morcegos Brasileiros. Londrina: Universidade Estadual de Londrina; 2007. p. 197–218.
Pieczarka JC, Nagamachi CY, O’Brien PCM, Yang F, Ren W, Barros RMS, Noronha RCR, Rissino J, Oliveira EHC, Ferguson-Smith MA. Reciprocal chromosome painting between two south American bats: Carollia brevicauda and Phyllostomus hastatus (Phyllostomidae, Chiroptera). Chromosom Res. 2005;13:349–7.
Volleth M, Klett C, Kollak A, Dixkens C, Winter Y, Just W, Vogel W, Hameister H. ZOO-FISH analysis in a species of the order Chiroptera: Glossophaga soricina (Phyllostomidae). Chromosom Res. 1999;7:57–64.
Sotero-Caio CG, Volleth M, Gollahon LS, et al. Chromosomal evolution among leaf-nosed nectarivorous bats – evidence from cross-species chromosome painting (Phyllostomidae, Chiroptera). BMC Evol Biol. 2013;13:276.
Baker RJ, Hamilton M, Parish DA. Preparations of mammalian karyotypes under field conditions. Occas Pap Mus Texas Tech Univ. 2003b;228:1–7.
Moratelli R, Andrade CM, Armada JLA. A technique to obtain fibroblast cells from skin biopsies of living bats (Chiroptera) for cytogenetic studies. Genet Mol Res. 2002;2:128–30.
Seabright M. A rapid banding technique for human chromosome. Lancet. 1971;2:971–2.
Telenius H, Pelmear A, Tunnacliffe A, et al. Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer. 1992;4:257–63.
Yang F, Carter NP, Shi L, Ferguson-Smith MA. A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma. 1995;103:642–52.
Gomes AJB, Nagamachi CY, Rodrigues LRR, Benathar TCM, Ribas TFA, O’Brien PCM, Yang F, Ferguson-Smith MA, Pieczarka JC. Chromosomal phylogeny of Vampyressine bats (Chiroptera, Phyllostomidae) with description of two new sex chromosome systems. BMC Evol Biol. 2016;16:119.
Pieczarka JC, Gomes AJB, Nagamachi CY, Rocha DCC, Rissino JD, et al. A phylogenetic analysis using multidirectional chromosome painting of three species (Uroderma magnirostrum, U. bilobatum and Artibeus obscurus) of subfamily Stenodermatinae (Chiroptera-Phyllostomidae). Chromosom Res. 2013;21:383–92.
Swoford DL. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods). Massachusetts: Version 4. Sinauer Associates, Sunderland; 2002.
Gomes AJB, Nagamachi CY, Rodrigues LRR, Farias SG, Rissino JD, Pieczarka JC. Karyotypic variation in Rhinophylla pumilio Peters, 1865 and comparative analysis with representatives of two subfamilies of Phyllostomidae (Chiroptera). Comparative Cytogenetics. 2012;6:213–25.
Gomes AJB, Rodrigues LRR, Rissino JD, Nagamachi CY, Pieczarka JC. Biogeographical karyotypic variation of Rhinophylla fischerae (Chiroptera: Phyllostomidae) suggests the occurrence of cryptic species. Comparative Cytogenetics. 2010;4:79–85.
Gardner AF. Chromosomal variation in Vampyressa and a review of chromosomal evolution in the Phyllostomidae (Chiroptera). Syst Zool. 1977;26(3):300–18.
Ribeiro NAB, Nagamachi CY, Pieczarka JC, Rissino JD, Neves ACB, Gonçalves ACO, Marques-Aguiar S, Assis MFL, Barros RMS. Cytogenetic analysis in species of the subfamily Glossophaginae (Phyllostomidae-Chiroptera) supports a polyphyletic origin. Caryologia. 2003;56(1):85–95.
Baker RJ, Bleier WJ. Karyotypes of bats of the subfamily Carolliinae (Mammalia; Phyllostomatidae) and their evolutionary implications. Experientia. 1971;27:220–2.
Botero-Castro F, M-k T, Justy F, Catzeflis F, Delsuc F, Douzery EJP. Next-generation sequencing and phylogenetic signal of complete mitochondrial genomes for resolving the evolutionary history of leaf-nosed bats (Phyllostomidae). Mol Phylogenet Evol. 2013;69:728–39.
Mao X, et al. Karyotype evolution in Rhinolophus bats (Rhinolophidae, Chiroptera) illuminated by cross-species chromosome painting and G-banded comparison. Chromosom Res. 2007;15:2–14.
Acknowledgments
We thank Manoel Rodrigues (Juris Ambientes Consultores), Flávio Eduardo Pimenta (Aotus Consultoria) for helping us in the field expedition to the area of Alcoa’s Juruti Bauxite mine. We thank Biodinamica-Rio, Aotus Consultoria, Eloiza Soarez, Fabio Sarmento, Fabio Augusto and Ramon Araujo for helping us with the expedition from Oriximina (Pará state) to Cariri (Amazonas state). This study is part of the Doctoral thesis of AJBG who was a recipient of a CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) Scholarship in Genetics and Molecular Biology, UFPA.
Funding
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação Amazônia Paraense (FAPESPA) on projects coordinated by CYN (Edital Universal, TO 064/2008; Edital BIONORTE-CNPq, Proc 552032/2010–7; Edital BIONORTE-FAPESPA, ICAAF 007/2011); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), on a Edital Pró-Amazônia project coordinated by CYN; the FAPESPA (Edital Vale – ICAAF 064/2011) and Banco Nacional de Desenvolvimento Econômico e Social – BNDES (Operação 2.318.697.0001) on a project coordinated by JCP; CYN (308428/2013–7) and JCP (308401/2013–1) are granted as CNPq researchers.
Availability of data and materials
All the data supporting findings are contained within the manuscript.
Author information
Authors and Affiliations
Contributions
AJBG collected the samples, collaborated in all of the cytogenetic data and phylogenetic analyze, undertook the bibliographic review. CYN collaborated in all cytogenetic procedures and analyzes. LRRR collected part of the samples, collaborated with cytogenetic data and reviewed the paper. MAFS helped on the development of the probes, participated in the cytogenetic analyses, and reviewed the manuscript. PCMOB and FY helped on writing the paper and discussed the data. JCP helped conceive the study and participated in developing the laboratory techniques, and coordinated the writing of this paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The necessary euthanasia occurred in accordance with animal welfare guidelines established by the Animal Ethics Committee (Comitê de Ética Animal) from Universidade Federal do Pará (Permit 68–2015). JCP has a permanent field permit, number 13248 from “Instituto Chico Mendes de Conservação da Biodiversidade”. The Cytogenetics Laboratory from UFPa has a special permit number 19/2003 from the Ministry of Environment for samples transport and 52/2003 for using the samples for research.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Table S1. Basic data matrix on chromosome rearrangements. The numbers of chromosomes are the ones from PHA, the outgroup. The meaning of the abbreviations can be found at Table 1. Other symbols: “/” = syntenic groups physically linked; “i” = inversion; “p” = short arm; “q” = long arm; “p + q” = part of short arm linked with part of the long arm; “-”= part of the arm is missing. (DOCX 158 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gomes, A.J.B., Nagamachi, C.Y., Rodrigues, L.R.R. et al. Chromosomal evolution and phylogeny in the Nullicauda group (Chiroptera, Phyllostomidae): evidence from multidirectional chromosome painting. BMC Evol Biol 18, 62 (2018). https://doi.org/10.1186/s12862-018-1176-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-018-1176-3