Abstract
Background
TP53 gene mutations occur in more than 50% of human cancers and the vast majority of these mutations in human cancers are missense mutations, which broadly occur in DNA binding domain (DBD) (Amino acids 102–292) and mainly reside in six “hotspot” residues. TP53 G245C and R273H point mutations are two of the most frequent mutations in tumors and have been verified in several different cancers. In the previous study of the whole genome sequencing (WGS), we found some mutations of TP53 DBD in esophageal squamous cell carcinoma (ESCC) clinical samples. We focused on two high-frequent mutations TP53 p.G245C and TP53 p.R273H and investigated their oncogenic roles in ESCC cell lines, p53-defective cell lines H1299 and HCT116 p53−/−.
Results
MTS and colony formation assays showed that mutant TP53 G245C and R273H increased cell vitality and proliferation. Flow cytometry results revealed inhibition of ultraviolet radiation (UV)- and ionizing radiation (IR)- induced apoptosis and disruption of TP53-mediated cell cycle arrest after UV, IR and Nocodazole treatment. Transwell assays indicated that mutant TP53 G245C and R273H enhanced cell migration and invasion abilities. Moreover, western blot revealed that they were able to suppress the expression of TP53 downstream genes in the process of apoptosis and cell cycle arrest induced by UV, which suggests that these two mutations can influence apoptosis and growth arrest might be due, at least in part, to down-regulate the expression of P21, GADD45α and PARP.
Conclusions
These results indicate that mutant TP53 G245C and R273H can lead to more aggressive phenotypes and enhance cancer cell malignancy, which further uncover TP53 function in carcinogenesis and might be useful in clinical diagnosis and therapy of TP53 mutant cancers.
Similar content being viewed by others
Background
TP53, as an important tumor suppressor gene, plays a crucial role in the process of maintenance of normal cellular function. Wild-type (WT) TP53 can be activated to regulate many cellular programs like cell cycle arrest, DNA repair, apoptosis, autophagy, senescence, metabolic remodeling and innate immunity [1,2,3]. TP53 gene mutations occur in more than 50% of human cancers, including liver cancer, breast cancer, bladder cancer, stomach cancer, colon cancer, prostate cancer, soft tissue sarcoma, ovarian cancer, brain tumor, esophageal cancer, lung cancer and osteosarcoma [4, 5]. The vast majority of TP53 mutations in human cancers are missense mutations, which broadly occur in DBD (Amino acids 102–292) and mainly reside in six “hotspot” residues (p.R175, p.G245, p.R248, p.R249, p.R273, and p.R282) [4, 6, 7].
The majority of TP53 gene mutations in human cancers abolish its tumor-suppressive function to bind to specific DNA sequences recognized by wild-type TP53, which is called loss of function (LOF) [4, 8]. Some TP53 mutations reduce the reaction with wild-type TP53 downstream genes, resulting in the inactivation of wild-type TP53 or its response elements, which lead to gain of oncogenic function (GOF) [9,10,11,12]. Moreover, the mutant P53 proteins frequently exhibit a dominant negative effect on the wild-type TP53 allele by interacting with wild-type TP53 and reducing cellular concentration of functional wild-type TP53, which can form wild-type TP53 tetramer structure but lose the activity of wild-type TP53 [1, 3, 4, 13].
As previously reported, TP53 G245C and R273H point mutations are two of the most frequent mutations in tumors and have been verified in several different cancers [7]. It has been reported that R273H can enhance invasion of lung cancer cells [14] and promote invasion and migration in endometrial cells [8]. G245C has been confirmed to result in changes in the conformation of the TP53 DNA-binding domain, compared with wild-type TP53 [15]. However, the properties of such mutations are not well characterized and there is little information on G245C and R273H mutations in ESCC and p53-defective cancer cells. From the previous results of WGS in ESCC patients’ samples [16], we focused on these two mutations and verified their tumorigenicity in ESCC cell lines, p53-defective cell lines H1299 and HCT116 p53−/−. We applied to determine the influence of G245C and R273H mutations of TP53 on cell proliferation, apoptosis and cell cycle arrest induced by UV, IR and Nocodazole in human cancer cells.
The current study aims to explore the function and impact of TP53 G245C and R273H mutations on cancer cell proliferation, migration, invasion, apoptosis and cell cycle arrest after UV, IR and Nocodazole treatments, which might serve as a potential diagnostic and therapeutic target in TP53 mutant cancers.
Results
TP53 G245C and R273H mutations analysis in ESCC patients’ samples and cell lines
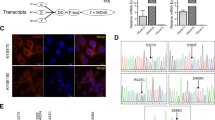
According to the previous results of whole genome sequencing (WGS) in ESCC patients’ samples [16], we found that TP53 somatic mutations were present in sequenced tumors. TP53 at codons G245 and R273 were identified in respectively as shown in Tables 1 and 2. Total genomic DNA of ten ESCC cell lines were extracted and the mutation sites in 11 TP53 exons were validated by Sanger Sequencing (Table 3) and their expression were assessed by real time PCR assays (Fig. 1a). Short interfering RNAs (siRNAs) were applied to knock down P53 expression levels in KYSE150 and COLO680 cells, which express wild-type and relatively higher levels of P53. The knockdown efficiency of P53 by siRNAs were verified by Western blot (Fig. 1b). We designed gray scale scanning in KYSE150 and COLO680 to examine the efficiency of silencing of p53, and the result indicated that p53 was knockdown by siRNAs obviously (Fig. 1c). Additionally, siRNAs and TP53 plasmids were transiently cotransfected in KYSE150 and COLO680 cells, and we validated the effects of TP53 G245C and R273H mutant transfectants on cellular malignant phenotypes including cell growth, colony formation, migration and invasion.
TP53 expression levels in ESCC cell lines. a qRT-PCR analysis of the relative TP53 mRNA levels in nine ESCC cell lines. TP53 mRNA levels in KYSE150 and COLO680 were considerate high. b Western blot assay was applied to verify the expression of P53 in KYSE150 and COLO680 after siRNAs treatment, which express relatively higher levels of P53 and contain wild-type P53. c Gray scale scanning analysis was employed to examine the efficiency of silencing of p53 in KYSE150 and COLO680
TP53 G245C and R273H mutants positively modulate tumor cell proliferation activities
To determine the role of mutant TP53 on tumor cell malignancy, MTS assays were used to test the impact of mutant TP53 on cell proliferation, the different characteristics of the various TP53 transiently cotransfected cell lines were analyzed and shown in Fig. 2a. The effects of the TP53 mutations on proliferation were reflected in colony formation assays (Fig. 2b), more colonies were presented in cells expressing mutant TP53 than that seen in wild-type TP53 cells. Additionally, the mutant TP53 led to a significantly greater promotion of cell growth than wild-type TP53 transfectants. Compared with control cells, wild-type TP53 suppressed cell proliferation. Taken together, these results illustrated that the TP53 G245C and R273H mutant cells strengthened cell proliferation abilities.
TP53 G245C and R273H mutants accelerated the proliferation activities of tumor cells. a Cell proliferation activity was examined by MTS assay and each cell clusters were counted at 0 h, 24 h, 48 h, 72 h and 96 h. Data was shown as the average from at least three independent experiments. b The long-term cell growth activity was tested by colony formation assay in above four tumor cell lines. Cell numbers were counted after 14 days incubation. (*P < 0.05, **P < 0.01, ***P < 0.001)
TP53 G245C and R273H mutations positively regulate tumor invasion and migration activities
We next determined the influence of TP53 G245C and R273H on invasion and migration activities. Matrigel™ invasion chamber system was employed to test the properties of invasion and migration, the mutant transfectants showed higher levels of membrane penetration abilities. Invasion and migration were significantly higher in cells expressing TP53 G245C and R273H than vector controls in KYSE150 (Fig. 3a) and COLO680 (Fig. 3b), p53-defective cell lines H1299 (Fig. 3c) and HCT116 p53−/− (Fig. 3d) transfected with relative plasmids. Based on the proliferation and invasion assays, we concluded that TP53 G245C and R273H mutations are gain of oncogenic function in ESCC cell lines and p53-defective cell lines H1299, HCT116 p53−/− cells.
TP53 G245C and R273H mutants promoted tumor cells invasion and migration. a Invasion and migration activities were examined by Matrigel™ invasion chamber in KYSE150 cell line. b Invasion and migration activities were assessed in COLO680 cell line. c Invasion and migration abilities were verified in H1299 cell line. d Invasion and migration activities were tested in HCT116 p53−/− cell line. Data was shown as mean ± SE. (*P < 0.05, **P < 0.01, ***P < 0.001)
The influence of TP53 G245C and R273H mutants on tumor cell apoptosis and cell cycle arrest
We also examined effects of TP53 G245C and R273H on cell apoptosis and cell cycle arrest following UV, IR and Nocodazole treatments. We assessed UV- and IR-induced apoptosis in KYSE150 and COLO680 cotransfected with control, wild-type TP53, TP53 G245C, TP53 R273H plasmids and siRNAs as well as H1299, HCT116 p53−/− cells. As shown in Fig. 4a, the percentage of cells undergoing apoptosis after UV 30 J for 12 h treatment was less in G245C and R273H than WT and control cells, compared to UV 30 J for 0 h in ESCC cell lines. Furthermore, the ratio of apoptosis cells after IR 4Gy for 12 h in G245C and R273H was dramatically less than WT and control cells in KYSE150 and COLO680 cell lines (Additional file 1: Figure S1A), which were consistent with the data described above. In the analysis of cell cycle, we found that the abilities of cells performing G1 arrest after UV 30 J for 12 h treatments in the G245C and R273H were weaker than WT and control cells, compared to UV 30 J for 0 h (Fig. 4b). In addition, this phenomenon was also observed in KYSE150 and COLO680 cells which were treated with IR 4Gy for 12 h (Additional file 1: Figure S1B). Nocodazole is a microtubule-disrupting agent to arrest cells in mitosis by triggering the mitotic checkpoint. Furthermore, we also determined cell cycle arrest induced by Nocodazole (0.5μg/ml) for 12 h. We demonstrated that, in KYSE150 cell line, G2/M ratio was diminished in these two mutant transfected cells after adding Nocodazole for 12 h. Meanwhile, the G1 phase enhanced relative to the wild-type cells. While in COLO680 cell line, the reducing ratio of G2/M in TP53 R273H mutant transfected cells after adding Nocodazole treatment was not significantly statistic different, nevertheless, the decreasing ratio of G2/M in TP53 G245C did have significantly difference, which suggest that G245C and R273H can abrogate cell response to Nocodazole induced cell cycle arrest (Additional file 1: Figure S1C). These results indicate that TP53 G245C and R273H confer stronger resistance to UV- and IR- induced apoptosis and attenuate UV, IR as well as Nocodazole induced cell cycle arrest.
G245C and R273H conferred cells stronger resistance to UV-induced apoptosis and attenuated cell cycle arrest. a Apoptosis degree induced by UV 30 J for 0 h and 12 h in cell lines. The data were presented as the portion of cells undergoing apoptosis tested by flow cytometry, the cells were dyed by PI and AnnexinV. b Quantitation of cell cycle arrest ratio in cells transfected by WT and mutant TP53 plasmids after UV 30 J for 0 h and 12 h treatment. Data was shown as the percentage of DNA amounts dyed by PI which was detected by flow cytometry. Data was shown as mean ± SE. (*P < 0.05, **P < 0.01, ***P < 0.001)
The impacts of TP53 R273H and R273C on the expression of P21, GADD45α and PARP
Increasing evidences suggest that tumor cells express highly stabilized mutant TP53 even in the absence of cellular stress, a prominent explanation is that mutant TP53 fails to initiate the transcription of the MDM2, which consequently leads to a collapse of the negative feedback loop driven by MDM2 and the maintenance of high P53 levels in cells [17,18,19]. Additionally, the expressions of P53 were verified by Western blot in Fig. 5a. As shown in Fig. 5a, P53 expression levels were increased in TP53 G245C and R273H mutant cells than wild-type and control cells.
TP53 and its downstream proteins’ expression levels. a The expression levels of P53 in p53-defective cell lines H1299 and HCT116 p53−/− after UV 30 J for 12 h treatment. b Mutant TP53 R273H and R273C reduced the protein levels of P21, GADD45α and cleaved PARP after UV 30 J for 12 h treatment in KYSE150 and COLO680. Western blot of TP53 downstream proteins P21, GADD45α and cleaved PARP expression levels in KYSE150 (right) COLO680 (left) cell lines after UV 30 J treatment for 12 h
TP53 gene can be involved in many important cellular functions through the transcriptional regulation of its downstream genes. To further explore the mechanism of mutant TP53 responses to cell cycle arrest and apoptosis, we examined the expression levels of several TP53 downstream genes that participate in the process of cell cycle arrest and apoptosis, including P21 and GADD45α which play vital roles in cell cycle arrest and apoptosis response, acting as both mediator and effector [20, 21]. Furthermore, the impacts of G245C and R273H mutations of TP53 on the protein levels of P21 and GADD45α were determined, the two mutations were found greatly attenuated the induction of P21 and GADD45α after UV treatment (Fig. 5b). Hence, we speculate that G245C and R273H influence the process of cell cycle arrest and apoptosis might be due, at least in part, to down-regulate P21 and GADD45α protein levels. In addition, PARP is cleaved to PARP p85 rapidly during apoptosis [22]. Similarly, we have found that UV-induced cleaved PARP levels in control and WT TP53 cells were elevated, but decreased markedly in G245C and R273H cells. These results further suggest that there exist a defect in UV-induced apoptosis in mutant cells (Fig. 5b).
Discussion
TP53 plays a vital important role in preventing tumor development. Under oncogenic or genotoxic stress, wild-type TP53 is activated to inhibit cell growth through inducing cell-cycle arrest or apoptosis process [3]. It has been reported that more than 50% of human cancers carry somatic mutations of TP53 [5] and that mutant TP53 not only lose their tumor-suppressive functions [4, 8] but also acquire new transforming abilities to promote tumorigenesis, which are independent of wild-type TP53 [9,10,11,12]. Consistent with previous reports, codons R273 and G245 are two of the most frequently mutant hot spots in the IARC database [23]. These two mutations have been proved to eliminate the function of wild-type TP53 including TP53 mediated cell cycle arrest and apoptosis, as well as to contribute to tumorigenesis through regulating some downstream genes. It is also evident that missense mutations may convert TP53 from a tumor suppressor gene into an oncogene, and it is of both scientific and clinical interest to understand the gain of function activities of mutant TP53 [11]. In this study, we identified these two mutations in clinical ESCC patients’ samples and investigated their oncogenic roles in ESCC cells. We found that TP53 G245C and R273H could promote cell proliferation, colony formation, invasion and migration activities. Moreover, TP5 3G245C and R273H mutations confer stronger resistance to UV- and IR- induced apoptosis and attenuate UV, IR as well as Nocodazole induced cell cycle arrest.
Mutant P53 protein often accumulates at extremely high levels in tumors, which is partly due to the disruption of the transactivation of MDM2, which serves as a negative feedback loop down-regulating P53 protein levels in the absence of DNA damage signals and maintaining wild-type P53 at the low levels in normal tissues [17,18,19]. Wild-type P53 protein primarily functions as a transcription factor to transactivate many genes in the progress of cell cycle arrest and apoptosis, while mutant P53 protein abrogates transactivation of these target genes such as P21, GADD45α and PARP [1, 7]. Cyclin-dependent kinase (CDK) inhibitor P21 (also known as P21WAF1/Cip1), acts as an important transcriptional target of TP53 [24] to inhibit the kinase activity of CDK4,6/cyclin-D and CDK2/cyclin-E leading to G1/S and G2/M cell cycle arrest in response to TP53 activation after cellular or genomic stress [20, 25, 26]. P21 is a tumor suppressor gene, but it can also serve as an oncogene in certain cellular process of human cancers [24]. In our results, we found that both TP53 G245C and R273H mutations abolished wild-type p53 activated induction of P21 protein after UV treatment (Fig. 5b). A direct outcome of the scenario is shown by the TP53 induced P21 mediated G1/S arrest of these cancer cells, which is evacuated when the G245C and R273H mutations express high levels in the cells.
It has been repeatedly found that GADD45α is induced by DNA damage and other stress signals like UV, IR or Nocodazole, which are associated with cell cycle arrest and apoptosis [21, 27, 28]. GADD45α can displace PCNA from the cyclin D1 complex to prevent DNA replication during S phase [27], bind to CDK1 to inhibit CDK1/cyclin B1 activity in vitro [21], and behave as a bridge between P53-dependent cell cycle checkpoint and DNA repair [28]. GADD45α is necessary for normal TP53 activation by stress signals and its transcription can be stimulated by TP53. Its level rises notably in apoptotic cells, while inhibition of GADD45α expression level reduces apoptosis in response to DNA damage [28]. In our study, we have found that the induction of GADD45α expression was reduced dramatically after UV treatment in the cells expressing mutant TP53 G245C and R273H, while it is up-regulated consistently in wild-type TP53 cells (Fig. 5b), which suggests that these two TP53 mutations lose the abilities to transcriptionally activate GADD45α and such reduction of GADD45α expression further attenuate apoptosis. Hence, knowing the particular TP53 mutation of cancer may have utility for management in clinical practice since apoptosis is a major effect of chemotherapy or radiotherapy.
PARP enzyme plays an important role in DNA damage repair, differentiation, apoptosis, transcription, and DNA replication [29, 30]. PARP is converted from the 116-kDa form to fragments of 89 and 24 kDa and can be cleaved to PARP p89 early during the process of apoptosis [22, 31]. The cleaved PARP is one of biomarkers for the detection of apoptosis. In addition, PARP is a target of apoptotic caspase-3 and its two proteolytic fragments have been considered to be sign of functional caspase activation [32]. In the study, we have observed that UV treatment lead to increasing cleaved PARP levels in control and wild-type TP53 cells, but decreasing in cells expressing G245C and R273H (Fig. 5b), which suggest that there exist a defect in UV-induced apoptosis in the mutant TP53 cells.
Interestingly, we have identified TP53 mutations (G245C and R273H) are distributed only in group of smokers in ESCC (5/88) and none is found in non-smokers group (Additional file 2: Figure S2) [16], which suggest that TP53 mutations may be induced by tobacco exposure and lead to ESCC progress. As it is reported that several TP53 point mutations are related to the effect of cigarette exposure in lung cancers [33,34,35] and squamous cell carcinomas of the head and neck [36], which is similar to our finding in patients with ESCC, a group that the percentage of smokers have higher TP53 mutations. Undoubtedly, it is possible that the link between tobacco-induced TP53 mutations and ESCC is influenced by other factors like tumor type and gender, age and ethnic of the patients [33], so realizing the relationship between tobacco-induced TP53 mutations and ESCC will be a powerful tool that connect disease to its specific cause.
Conclusions
In summary, this study has demonstrated that two malignant TP53 mutations (G245C and R273H) revealed strong oncogenic properties in ESCC cells, indicating that such TP53 mutations may play important roles in carcinogenesis and development of malignancy of ESCC. Given the important roles of mutant TP53 G245C and R273H in tumorigenesis and enhancing cancer cell malignancies, it might have contributions to future diagnosis and therapy in TP53 mutant cancers.
Methods
Plasmids construction
P53-SN3 was constructed from 1.8 KB-length of TP53 cDNA sequence which was cloned into the BamHI sites of the expression vector. Two mutant TP53 plasmids G245C and R273H were generated following the manuscript of QuikChange® Site-Directed Mutagenesis Kit (Agilent, USA).
Cell culture and transiently transfection
The human ESCC cell lines (KYSE2, KYSE30, KYSE70, KYSE180, KYSE410, KYSE450, KYSE140, KYSE510 and COLO680) were provided by Professor Y. Shimada of Kyoto University. The HCT116 p53−/− cell line was brought from University of Pittsburgh by Professor Qimin Zhan. The H1299 cell line was purchased from ATCC and the catalogue is CRL-5803™. The ESCC cell lines KYSE2, KYSE30, KYSE70, KYSE140, KYSE180, KYSE410, KYSE450, KYSE510, COLO680 and the human lung cancer cell line, H1299 were cultured in 90% RPMI1640 medium, with 10% fetal bovine serum and antibiotics, at 37 °C, 5% CO2; KYSE150 was cultured in the medium of 1:1 mixture of Ham’s F12 and RPMI-1640 containing 2% FBS and antibiotics at humidified atmosphere with 37 °C, 5% CO2, the human colorectal cancer cell line, HCT116 p53−/− was grown in 90% DMEM medium with 10% fetal bovine serum, at 37 °C, 5% CO2. When cells were grown to 70–80% confluence, they were transiently cotransfected with the relative plasmids and siRNAs simultaneously using Lipofectamine2000 (Invitrogen) according to the standard procedure. For siRNAs transfection and plasmids transfection, the density of cells was grown to 40–50% and 70–80% individually.
Cell proliferation assays
MTS assays were employed to analyze cell viability by a dye staining method using MTS reagent. 3000 cells per well were suspended in 96-well plates with 200 μl medium and grown for 96 h. Then, cells were incubated with MTS (0.5 mg/ml) and measured on an ELISA plate reader with the wavelength of absorbance at 490 nm and 630 nm at 0 h, 24 h, 72 h and 96 h of culture for proliferation assays, and percentage of respective basal levels (MTS concentrations of wells without cells) was calculated.
For a colony formation assay, cells were seeded in 60 mm plates at a concentration of 500 cells per well and changes culture with media every 72 h. Colonies were fix with methanol and stained with methanol and 0.5% crystal violet-acetic acid solution after 14 days, then colonies were visualized by fluorescence microscope and quantitated.
Apoptosis and cell cycle detection assays
For apoptosis studies, an Annexin V-FITC/PI apoptosis assay kit (NeoBioscience, Shenzhen, China) was employed. After 48 h transfection, cells were washed with PBS, stained with Annexin V-FITC and propidium iodide (PI) according to the manufacturer’s protocol and quantitated by using flow cytometry (Becton-Dickinson, San Jose, CA, USA) to examine the apoptotic cell numbers.
Wild-type and mutant TP53 transfected cells were seeded in six-well plate at a density of 106 cells per well. After 48 h, cells were collected by trypsinization and centrifugation and washed with PBS. The cells were fix with precool 70% ethanol (v/v) overnight at − 20 °C, then centrifugation and washed with PBS. The cells were stained with PI (Propidium, 50μg/ml) 30 min at 37 °C and analyzed by flow cytometry (Becton-Dickinson, San Jose, CA, USA). Percentage of cell cycle cells was calculated according to the number of cells in the respective phase.
Western blotting analysis
Cells were transfected with wild-type and mutant TP53 plasmids for 48 h, total cell lysates were collected using lysis buffer (1 × PBS + 4%NP-40 + 0.2%proteinase inhibitor), 2 × SDS–PAGE sample loading buffer, and boiled for 5 min. 80 μg protein was performed and fractionated by SDS–PAGE, then semi-dry transferred to polyvinylidene difluoride membrane which was probed with antibodies for relative antibodies. Secondary antibodies were incubated, then visualized and quantified using Image Quant software (GE Healthcare Biosciences, Pittsburg, PA).
Genomic DNA extraction and sequencing
Total genomic DNA was extracted in ten ESCC cell lines using TIANamp Genomic DNA kit (TIANGEN, Cat #DP304–03) according to the standard procedure. Each DNA (2 μl) was amplificated by PCR and 11 exons of TP53 were sequenced (Sino Geno Max).
exon | Forward | Reverse |
|---|---|---|
exon1 | GGAGCCTCGCAGGGGTTGATGG | CAAGTTCAGTCAGGAGCTTACC |
exon2,3 and 4 | CCTCTTGCAGCAGCCAGACTG | GCAACTGACCGTGCAAGTCA |
exon5 and 6 | GCTGCCGTGTTCCAGTTGCT | GCCACTGACAACCACCCTTA |
exon7,8 and 9 | TGCCACAGGTCTCCCCAAGG | CCCAAGACTTAGTACCTG |
exon10 and 11 | CCTCTGTTGCTGCAGATCCG | GTAGCCTGCACTGGCGTTCACC |
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using Trizol reagent (Invitrogen) according to standard procedure. A reverse transcription reaction was performed using the one-step Reverse Transcription kit (Promega). Each cDNA (2 μg) was amplified in a SYBR Green Realtime PCR Master Mix (Thermo) and loaded on the Applied Biosystems 7300 Real-time PCR Detection System (ABI, Foster City, USA). GAPDH mRNA was employed as an endogenous control for mRNA. Thermal cycling conditions were as follows: the first one step, 95 °C for 30s and the ensuing 40 cycles, 95 °C for 5 s, 65 °C for 31 s, and melt curve step: 95 °C for 15 s, 65 °C for 1 min, 95 °C for 15 s. The relative expression levels of each group were quantified using the 2-∆CT method and each mRNA level was normalized to the GAPDH mRNA. Primers used for qRT-RCR are listed as following:
Gene | Forward | Reverse |
|---|---|---|
GAPDH | TATGACAACAGCCTCAAGAT | AGTCCTTCCACGATACCA |
TP53 | CCACCATCCACTACAACTACAT | AGGACAGGCACAAACACG |
Invasion and migration assays
To prepare the invasion assay, a Matrigel™ invasion chamber (BD Biosciences, Bedford, MA) was employed to evaluate the invasive activity of TP53 mutant and WT cells. Thoroughly mixing the sufficient Matrigel™ matrix with serum-free RPMI basal medium and carefully adding 0.1 ml of the diluted Matrigel matrix solution to each insert, then using the Matrigel™ matrix solution to incubate plates for 2 h in culture incubator, 37 °C, 5% CO2. Preparing cell suspensions in serum-free RPMI basal medium containing 3 × 104 cells/ml for the 24-well chamber and subsequently filled with 600-800 μl RPMI-1640 medium with 20% FBS into each wells under the chambers. Cell invasion chambers were incubated in a humidified tissue culture incubator at 37 °C, 5% CO2. After overnight incubation, cells which invaded through the Matrigel™ membrane were fixed with methanol and stained with methanol and 0.5% crystal violet-acetic acid solution.
For the migration studies, the method as previously described and excluding the Matrigel™ matrix solution preparation was used. Invasive and migration cell numbers were visualized by fluorescence microscope and quantitated.
Statistical analysis
Data for cell proliferation, invasion, migration, cell cycle and apoptosis were obtained at least in triplicate and employing GraphPad Prism v.5 software, expressed as mean ± SD (n ≥ 3). Quantization level of western blot was by Image J 1.8.0 software. Statistical differences were analyzed by the Student’s t-test (two-tailed), while analysis of variance (ANOVA) was used to determine whether there were any significant differences between groups. A P-value of less than 0.05 was considered to reflect significant differences. For ESCC clinical data, differences between groups were analyzed by the Fisher’s exact test.
Abbreviations
- CDK:
-
Cyclin-dependent kinase
- DBD:
-
DNA binding domain
- ESCC:
-
Esophageal Squamous Cell Carcinoma
- GADD45α:
-
Growth arrest and DNA-damage-inducible protein 45 alpha
- GOF:
-
Gain of function
- IR:
-
Ionizing radiation
- LOF:
-
Loss of function
- PARP:
-
Poly ADP-ribose polymerase
- siRNAs:
-
Short interfering RNAs
- UV:
-
Ultraviolet radiation
- WGS:
-
Whole genome sequencing
- WT:
-
Wild-type
References
Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–31.
Maddocks ODK, Vousden KH. Metabolic regulation by p53. J Mol Med. 2011;89(3):237–45.
Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413–31.
Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53.
Levine AJ, Momand J, Finlay CA. Review. The p53 tumour suppressor gene. Nature. 1998;85(11):1460–7.
Levine AJ, Wu MC, Chang A, Silver A, Attiyeh EF, Lin J, Epstein CB. The spectrum of mutations at the p53 locus. Evidence for tissue-specific mutagenesis, selection of mutant alleles, and a “gain of function” phenotype. Ann N Y Acad Sci. 1995;768:111–28.
Ran B, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9(10):701–13.
Dong P, Tada M, Hamada J, Nakamura A, Moriuchi T, Sakuragi N. p53 dominant-negative mutant R273H promotes invasion and migration of human endometrial cancer HHUA cells. Clin Exp Metastasis. 2007;24(6):471–83.
Wolf D, Harris N, Rotter V. Reconstitution of p53 expression in a nonproducer Ab-MuLV-transformed cell line by transfection of a functional p53 gene. Cell. 1984;38(1):119–26.
Milner J, Medcalf AE. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell. 1991;65(5):765–74.
Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, Finlay C, Levine AJ. Gain of function mutations in p53. Nat Genet. 1993;4(1):42–6.
Willis A, Jung EJ, Wakefield T, Chen X. Mutant p53 exerts a dominant negative effect by preventing wild. Oncogene. 2004;23(13):2330–8.
Milner J, Medcalf EA, Cook AC. Tumor suppressor p53: analysis of wild-type and mutant p53 complexes. Mol Cell Biol. 1991;11(1):12–9.
Muller PAJ, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139(7):1327–41.
Pintus SS, Ivanisenko NV, Demenkov PS, Ivanisenko TV, Ramachandran S, Kolchanov NA, Ivanisenko VA. The substitutions G245C and G245D in the Zn(2+)-binding pocket of the p53 protein result in differences of conformational flexibility of the DNA-binding domain. J Biomol Struct Dyn. 2013;31(1):78–86.
Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509(7498):91–5.
Barak Y, Juven T, Haffner R, Oren M. MDM2 expression is induced by wild type p53 activity. EMBO J. 1993;12(2):461–8.
Midgley CA, Lane DP. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15(10):1179–89.
Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–9.
Charriersavournin FB, Gire V, Sedivy J, Piette J, Dulić V. p21-mediated nuclear retention of Cyclin B1-Cdk1 in response to Genotoxic stress. Mol Biol Cell. 2004;15(9):3965–76.
Wang XW. Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18(18):2892–900.
Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53(17):3976–85.
Xu J, Qian J, Hu Y, Wang J, Zhou X, Chen H, Fang JY. Heterogeneity of li-Fraumeni syndrome links to unequal gain-of-function effects of p53 mutations. Sci Rep. 2014;4(7489):4223.
Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9(6):400–14.
Bertoli C, Skotheim JM, Bruin RAMD. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14(8):518–28.
Zaldua N, Llavero F, Artaso A, Gálvez P, Lacerda HM, Parada LA, Zugaza JL. Rac1/p21 activated kinase pathway controls retinoblastoma protein phosphorylation and E2F transcription factor activation in B lymphocytes. FEBS J. 2016;
Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, Gilmer TM, Kastan MB, O'Connor PM, Jr FA. Interaction of the p53-regulated protein GADD45 with proliferating cell nuclear antigen. Science. 1994;266(5189):1376–80.
Salvador JM, Brownclay JD, Fornace AJ. Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol. 2013;793(10):1–19.
Soldani C, Lazzè MC, Bottone MG, Tognon G, Biggiogera M, Pellicciari CE, Scovassi AI. Poly(ADP-ribose) polymerase cleavage during apoptosis: when and where? Exp Cell Res. 2001;269(2):193–201.
Vizetto-Duarte C, Custodio L, Gangadhar KN, Lago JH, Dias C, Matos AM, Neng N, Nogueira JM, Barreira L, Albericio F, et al. Isololiolide, a carotenoid metabolite isolated from the brown alga Cystoseira tamariscifolia, is cytotoxic and able to induce apoptosis in hepatocarcinoma cells through caspase-3 activation, decreased Bcl-2 levels, increased p53 expression and PARP cleavage. Phytomedicine. 2016;23(5):550–7.
Duriez PJ, Shah GM. Cleavage of poly(ADP-ribose) polymerase: a sensitive parameter to study cell death. Biochem Cell Biol. 1997;75(4):337–49.
Bressenot A, Marchal S, Bezdetnaya L, Garrier J, Guillemin F, Plénat F. Assessment of apoptosis by immunohistochemistry to active caspase-3, active caspase-7, or cleaved PARP in monolayer cells and spheroid and subcutaneous xenografts of human carcinoma. J Histochem Cytochem. 2009;57(4):289–300.
Paschke T. Analysis of different versions of the IARC p53 database with respect to G→T transversion mutation frequencies and mutation hotspots in lung cancer of smokers and non-smokers. Mutagenesis. 2000;15(6):457–8.
Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–34.
Hecht SS. Tobacco smoke carcinogens and lung Cancer. J Natl Cancer Inst. 1999;91(14):1194–210.
Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA. Mutational heterogeneity in cancer and the search for new cancer genes. Nature. 2015;499(7457):214–8.
Acknowledgments
We gratefully thank Prof. Yongmei Song for the WGS data and technical advice. We also gratefully thank Mr. Tao Xu for skillful assistance with the FACS analyses and Mr. Xiao Liang for the technical services of 7300 Real-time PCR Detection System and fluorescence microscope. In addition, we thank Dr. Y.Shimada of Kyoto University for providing the ESCC cell lines.
Ethics approvals and consent to participate
Not applicable.
Funding
The design of the study is supported by the National 973 Program (2015CB553904). The collection of the data is supported by National Natural Science Foundation of China (81321091, 81672455 and 81230047) and the analysis of the data is supported by National Sci-Tech Support Plan (2013BAI01B07). The interpretation of data and the writing of the manuscript are supported by Beijing Nova Program (xx2018040) and CAMS Initiative for Innovative Medicine (2017-I2M-3-004).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Author information
Authors and Affiliations
Contributions
NK was responsible for all the experiments and drafted the manuscript, YW revised the figures and analyzed the data, SCG mainly carried out the part of cell cycle using Nocozazole by flow cytometry, YWO and JC were responsible for acquisition of data and interpretation of data, GCW contributed the supplementary S2 data, DL and QMZ was responsible for the overall design of the study and revising of the manuscript. All the authors gave final approval of the version to be published and approved the final submission.
Corresponding authors
Ethics declarations
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. G245C and R273H reduced IR-induced apoptosis and weakened cell cycle arrest. (A) The portion of KYSE150 and COLO680 cells undergoing apoptosis was tested by flow cytometry induced by IR (4Gy) for 0 h and 12 h and the cells were dyed by PI and Annexin V. (B) Quantitation of cell cycle arrest ratio in WT and mutant TP53 cells were treated with IR at 4Gy for 0 h and 12 h in KYSE150 and COLO680 cells. Results were shown as the percentage of DNA amounts dyed by PI which was detected by flow cytometry. Data was shown as mean ± SE (*P < 0.05). (C) Ratio of cell cycle arrest in WT and mutant TP53 cells were treated with Nocodazole (0.5μg/ml) for 0 h and 12 h in KYSE150 and COLO680 cells. Data was shown as the quantitation of DNA amounts dyed by PI which was detected by flow cytometry. Data was shown as mean ± SE (*P < 0.05). (TIF 2588 kb)
Additional file 2:
Figure S2. Clinical characteristics of the ESCC patients with TP53 mutations (G245C and R273H) in smokers and non-smokers (Fisher’s exact test, P = 0.107). (TIF 8844 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kang, N., Wang, Y., Guo, S. et al. Mutant TP53 G245C and R273H promote cellular malignancy in esophageal squamous cell carcinoma. BMC Cell Biol 19, 16 (2018). https://doi.org/10.1186/s12860-018-0167-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12860-018-0167-y