Abstract
Rice kernel smut (RKS), caused by the fungus Tilletia horrida, has become a major disease in rice-growing areas worldwide, especially since the widespread cultivation of high-yielding hybrid rice varieties. The disease causes a significant yield loss during the production of rice male sterile lines by producing masses of dark powdery teliospores. This review mainly summarizes the pathogenic differentiation, disease cycle, and infection process of the T. horrida, as well as the decoding of the T. horrida genome, functional genomics, and effector identification. We highlight the identification and characterization of virulence-related pathways and effectors of T. horrida, which could foster a better understanding of the rice–T. horrida interaction and help to elucidate its pathogenicity molecular mechanisms. The multiple effective disease control methods for RKS are also discussed, included chemical fungicides, the mining of resistant rice germplasms/genes, and the monitoring and early warning signs of this disease in field settings.
Similar content being viewed by others
Background
Tilletia horrida Takahashi is a pathogenic basidiomycete fungus that causes rice kernel smut (RKS), a devastating grain disease in the production of rice male sterile lines in most hybrid rice-growing regions of the world. First reported in Japan in 1896 (Takahashi 1896), by the onset of the twentieth century RKS had already been found in India, Java, Siam, and China, and categorized then as a minor disease with sporadic occurrence in these rice-growing areas (Gade 2000; Giri et al. 2000; Akhtar and Sarwar 1987; Ayado et al. 1993; Iguchi et al. 1987; Chahal 2001; Biswas 2003; Carris et al. 2006). However, in order to ensure the higher seed production of rice male sterile lines, their exserted stigma has become increasingly common, resulting a greater incidence and impact of RKS (Webster and Gunnell 1992). RKS is now recognized as among the most crucial diseases globally affecting the majority of production areas cultivating hybrid rice varieties (Uppala et al. 2017; Chen et al. 2016). In China, the hybrid rice- growing area covers ca. 1.6 million acres, where the annual prevalence of RKS is 40% to 60% of these hybrid rice fields, with the disease causing 5% to 20% yield loss (Chen et al. 2016; Wang et al. 2015). In Pakistan, the disease incidence was reportedly as high as 87% in hybrid rice fields (Akhtar and Sarwar 1987). Currently, RKS poses a mounting threat to hybrid seed production across Asia, Oceania, Europe, America, and Africa (Brooks et al. 2009; Mohamed et al. 2014; Sharma 1999; Ribeiro et al. 1973; Gade 2000; Giri et al. 2000; Sharif et al. 2014; Ayado et al. 1993; Iguchi et al. 1987). In addition, there are several kinds of viewpoints about the classify of RKS during the past few decades, once lots of scholar regard RKS as Neovossia horrida (Huang et al. 2003, 1995); however, with the completion of genome sequencing together with the systemic identified of pathogenicity characters (Wang et al. 2015, 2018a, 2018b), RKS has been belonged to T. horrida.
T. horrida infects rice flowers and colonizes their inner organs with mycelia, which eventually produce masses of dark powdery teliospores (Fig. 1). Teliospores are the main form of T. horrida during the overwintering period, and they can survive for more than 1 year in the soil, and at least 3 years on the surface of host seeds (Webster and Gunnell 1992). The early infective stage of T. horrida is asymptomatic (Zhu et al. 1998), and grain filling after pollination is blocked by the masses of dark powdery spores in the grains, thus causing a considerable reduction in the rice grain yield (Liu 2008).
Field control of RKS relies heavily on chemical fungicides and cultivation practices, and the breeding of rice male sterile lines resistant to this disease remains unsuccessful due to the paucity of robust resistance resources in the currently available rice germplasms. Moreover, environmental factors can affect the accuracy of RKS resistance phenotyping, and the ability of the pathogen to survive form one crop season to the next by forming teliospores makes it harder to control the fungal pathogen (Webster and Gunnell 1992; Teng et al. 2002; Dai et al. 2011). In light of this, here we also reviewed the control strategies, namely chemical and biological applications as well as the breeding for resistance traits in rice varieties.
In this current review, we summarize the pathogenic differentiation, disease cycle, infection process, functional genomics, and effector identification of the T. horrida. We also provide a thorough and critical discussion on rice defense responses to T. horrida infection and effective disease control strategies.
Overview of Pathogens and Diseases
Variability in Pathogenicity
Many plant pathogens have produced multiple physiological races whose virulence clearly differs during long the coevolution of pathogen–host interactions (Choi et al. 2013; Hamer et al. 1989; Kato et al. 2000). For example, Magnaporthe grisea that causes rice blast has dozens of physiological races. However, studies of the physiological races and pathogenic differentiation of T. horrida are quite scarce. Shao et al. (1997) inoculated two rice male sterile lines (Zhenshan 97A and V20A), each with four T. horrida strains separately, finding that the respective pathogenicity of these four strains was similar. Over the next 20 or so years, the number of rice male sterile lines bred has increased substantially, this providing stronger host conditions for both the pathogenic and population differentiation of T. horrida. Recently, Wang et al. (2018a) inoculated a susceptible rice male sterile lines (9311A) with seven T. horrida strains respectively isolated from different areas in China; their results showed these strains featured significantly pathogenic differentiation. Recently, the genetic diversity of the 63 T. horrida strains that isolated from different rice-growing areas in USA were investigated using multi-locus sequence analysis, which revealed the existence of five different groups of the T. horrida populations (Khanal et al. 2022). Collectively, these findings indicate that the T. horrida presently harbors significantly pathogenic and population differentiation.
Disease Cycles
The smuts fungi are multicellular organisms, whose teliospores are widespread in soil and the seeds of host plants. To date, 80 smut genera (4200 species) with distinct morphological characters have been reported and all these infect higher plants, including many economically important crops, such as Zea mays, Hordeum vulgare, Triticum aestivum, Oryza sativa, Saccharum officinarum, and Zizania latifolia (Rogerson 1988; Stirnberg and Djamei 2016; Grewal et al. 2004; Imbaby and Eldaoudi 2002; Nasiru and Ifenkwe 2004). Those smut fungi parasitic on key crops have multiple phylogenetically separate lines, and include Ustilago, Sporisorium, and Tilletia spp. (Roux et al. 1998).
T. horrida of the Tilletia genus belongs to the basidiomycota Tilletiaceae family (Wang et al. 2015). As a biotrophic fungus, T. horrida can grow on artificial media and relies on a living host to reproduce, both sexually and asexually (Wang et al. 2018a). Under natural conditions, T. horrida is known to infect Oryza sativa. Through artificial inoculation tests of 32 species of grasses with T. horrida, it was found that T. horrida could infect Oryza sativa as well as Aegilops sharonensis (Royer and Rytter 1988). Furthermore, Singh et al. (1999) found that the secondary sporidia of T. horrida were capable of germinating on multiple non-host plants, such as Cyperus rotundus, Echinochloa crusgalli, Zea mexicana, Sorghum vulgare, and Z. mays. We provide an overview of the current understanding of asexual and sexual cycles of T. horrida (Fig. 2). After successfully infecting rice flowers, T. horrida produces many mycelia on the stigma and then infects other floral organs to form dark, powdery teliospores (Templeton 1961; Tao et al. 1998). These teliospores are round to elliptical in shape and possess colorless warty on their surface (Wang et al. 2018a, b, c, d, e). The diameters of teliospores are ca. 25–30 × 23–30 μm (Wang et al. 2018a). Thick-walled teliospores may sticks to ripe host seeds or to soil, and are disseminated by airflow and wind-blown rain in fields, where they survive over the winter. Abundant teliospores will germinate after overwintering under suitable temperature and humidity conditions, producing a promycelium that displays distal verticillated digitations (Wang et al. 2018a). Microspores capable of infecting rice flowers at the late booting stage grow in these verticillated digitations and the shape of these microspores is linear or curved (Wang et al. 2018a). Therefore, these secondary microspores serve as primary infection agents in the disease cycle, and they may exhibit epiphytic budding growth on the surface of not only host but also weed plants from the vegetative stage to flowering stage (Huang et al. 2003). Unfortunately, it is difficult to detect the germination process of overwintered teliospores in actual fields conditions. The function of overwintering teliospores in the disease cycle of T. horrida needs further in-depth investigation.
The disease cycle of rice kernel smut. a Tilletia horrida produces dark, powdery teliospores in the rice panicles. b Mature dark powdery teliospores. c Germinating teliospores. d Distal verticillated digitations. e Leaves or seeds of rice and paddy field weeds may exhibit epiphytic budding by T. horrida, which may contribute to the initial infection of rice flowers. f The secondary microspores of T. horrida. g The exserted stigma of rice male sterile lines. h Rice stigma infected by T. horrida. After removing to ovaries (i). j Primary teliospores were produced in ovaries. Shown are the paths for the sexual (solid lines) and asexual (dashed line) growth in the disease cycles. Figure adapted from Wang et al. (2018a) and Tao et al. (1998)
For T. horrida, like most basidiomycete fungi, the asexual reproduction stage in their life cycles is unimportant or absent entirely, instead reproducing sexually under field conditions. Early work indicated that meiosis of T. horrida occurs in the stage of basidium formation (Singh and Pavgi 1972). Additionally, T. horrida is heterothallic, as clearly proven by inoculation experiments using single and paired monospore lines of T. horrida, in that single basidiospore lines of T. horrida are avirulent (Singh 1998). After teliospores’ germination, the mononucleosis promycelium will form secondary hyphae through hyphal or basidium mating. Mitotic divisions of single haploid nuclei and subsequent formation of septa in the basidiospores after the formation of a basal septum delineating the basidiospore, results in two- to four-celled basidiospores (Carris et al. 2006). Each haploid cell of the basidiospore can produce hyphae to directly infect rice flowers.
Infection Processes
Flower-infecting pathogens have evolved multiple strategies to infect their host plants. The pathogen responsible for rice false smut disease, Ustilaginoidea virens, specifically attacks the stamen filaments between the lodicules and ovaries; this results in significantly reduced growth of stamen filaments which precluded the formation of mature pollen, leaving the process of fertilization suppressed (Sun et al. 2020). Accordingly, U. virens infection induces the expression of many grains filling–related genes, such as grain starch biosynthetic genes and seed storage protein-encoding genes through imitate the process of ovules fertilization (Chao et al. 2014; Chen et al. 2010; Fan et al. 2015). This further activates the grain filling signaling pathway of rice, providing nutrients for the pathogen’s population growth. The study by Song et al. (2016) also showed that U. virens could hijacks the rice nutrients supply through blocking and imitating the fertilization of rice ovary. Botrytis cinerea infects alfalfa pollen grains mostly via pollen germ pores and goes on to obtains nutrients from the pollen exudates (Huang et al. 1999). Other phytopathogens which attack other parts of host flowers, such as their sepals, petals, and nectaries, have been summarized by Ngugi and Scherm (2006).
The infection processes of T. horrida was discerned through cytological observations of inoculation (Tao et al. 1998; Wang and Ouyang 1989; Zhu et al. 1998). Wang and Ouyang (1989) revealed that T. horrida hyphae spread over the ovaries of flowers through the stigmas and can reach nucellar tissue, and ultimately forming dark, powdery teliospores in aleurone cells or in the intercellular space between them. Tao and colleagues (1998) inoculated four rice male sterile lines (Zhenshan 97A, D90A, G46A, and K17A) with T. horrida, finding that the hyphae directly infected stigmas and further extended into nucellar tissue. Their research also demonstrated that T. horrida hyphae occur in ovaries at 8 h post inoculation (hpi), that primary teliospores form in the seed coat and intercellular space of aleurone cells at 7 days post inoculation (dpi), with teliospores maturating at 9 dpi. Interestingly, the process by which T. horrida infected the four rice male sterile lines was similar, not requiring pollination for infection (Tao et al. 1998). However, unlike U. virens, the teliospores formed only in ovaries that endosperm could normally fertilization (Zhu et al. 1998).
In rice, the floral organs’ tissue is not destroyed at the initial stage of T. horrida infection and lack obvious disease symptoms; the characteristics dark, powdery teliospores only appear at yellow maturity stage of rice (Zhu et al. 1998). This poses formidable challenge to early disease monitoring and further timely intervention of disease-controlling methods. Zhu et al. (1998) reported that T. horrida hyphae do not invade the cells and embryo sacs, such that the stigma cells and ovaries undergo no obvious visible changes and retain the normal ability of plasmolysis at 12 hpi. Collectively, these findings indicate that the infection strategy of T. horrida is unique and distinctive from other flower-infecting fungi.
Pathogenic Mechanism of T. horrida
T. horrida Genome
Draft genome sequences of the T. horrida strain QB-1 were completed in 2015, by using Illumina Solexa GAII sequencing technology, resulting in an assembled genome size ca. 20 Mb in size, encoding 9038 predicted proteins (Wang et al. 2015). Later, the high-quality genome sequences of T. horrida strain JY-521 were obtained using the PacBio RS II sequencing strategy combined with three single-molecule real-time sequencing (Wang et al. 2018b). The assembled genome size of JY-521 is ca. 23.2 Mb and encodes 7729 predicted proteins, of which 6973 were supported by the RNA-seq data (Wang et al. 2018b).
Genome annotation and comparative genomics provide much crucial information pertinent to the pathogenic mechanisms of T. horrida. As a biotrophic pathogens, T. horrida has a genome with fewer carbohydrate-active enzymes (CAZymes) than some hemi-biotrophic and necrotrophic fungi (Wang et al. 2018b). Among these CAZymes are glycoside hydrolases, which break down cellulose and xylan in the plant cell wall, and pectin lyases that degrade pectin. The reduced polysaccharide degradation machinery indicates a diminished cell wall–degrading ability. This finding explains why T. horrida infection does not destroy the cells of stigmas and embryo sacs at early stage (Zhu et al. 1998).
In fact, biotrophic pathogens usually minimize CAZymes in their arsenal, to avoid the destruction of plant cell wall. The lysis of cell walls is often recognized by the host plant as a danger-associated molecular patterns, so ensuring an intact cell wall could avoid triggering plant immunity (Kämpe et al. 2006; Kemen et al. 2011; Zhang et al. 2014). This strategy is in accordance with the biotrophic lifestyle of T. horrida (Wang et al. 2018b). Although the T. horrida genome contains fewer genes related to cell wall degradation, it carries 1697 genes involved in pathogen-host interactions (PHI), which account for 21.96% of the total protein-coding genes (Wang et al. 2018b). Furthermore, T. horrida contains 4410 genes that have sequence similarity with those of four other smut fungi. Nevertheless, 2472 predicted genes are unique to T. horrida (Wang et al. 2018b). These findings could explain the differential hosts of smut fungi or diseases induced by these fungi, including T. horrida, Ustilago hordei, U. maydis, Sporisorium reilianum, and S. scitamineum. Importantly, the assembled genome of T. horrida and its annotation is especially valuable for the functional identification of pathogenicity genes and virulence effectors.
Pathogenicity Pathway and Genes in T. horrida
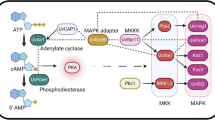
Despite the genome of T. horrida having been sequenced and annotated, our knowledge of the molecular mechanisms underlying T. horrida virulence and pathogenicity is very limited. Functional genomics can contribute to the clarifying of pathogenic pathways and genes in T. horrida. Transcriptome analysis of the T. horrida virulent strain JY-521 at different times since its inoculation (8, 12, 24, 48, and 72 hpi) of susceptible rice accessions revealed 500 differentially expressed genes (DEGs) of which most were induced at 8 hpi (Wang et al. 2020a). This shows the primary host colonization by T. horrida occurred at the early stage of infection. Furthermore, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis of these DEGs indicated that autophagy processes and lipid degradation were pivotal pathways involved in T. horrida pathogenicity (Wang et al. 2020a).
For successful colonization of plants, fungal pathogens generally secrete multiple kinds of CAZymes to break down the physical barrier of the host immune system (Cantarel et al. 2009). Thus, despite having fewer CAZymes in its genome, evidently these are critical for enhancing T. horrida’s virulence. Transcriptome analyses detected a carbohydrate esterases (CEs) family protein (smut_2980) involved in chitin deacetylase that was up-regulated at 12 hpi (Wang et al. 2020a). As previously reported, chitin deacetylase can prevent pathogenic fungi from being recognized by the plant immune system during the infection process (Gong et al. 2020; Sanchez-Vallet et al. 2020; Shimizu et al. 2010). Two glycosyltransferases (GTs) family proteins, smut_1230 and smut_1222 were also up-regulated at 8 hpi and may also play an important role during early infection of hosts. Furthermore, 64 genes related to PHI in T. horrida, such as smut_1409, smut_3510, smut_6708, and smut_2974 were greatly induced during infection, suggesting these genes are likely involved in the infection of rice by T. horrida (Wang et al. 2020a). The conserved mitogen-activated protein kinases (MAPKs) have been widely identified as being pathogenicity factors in multiple phytopathogenic fungi (Dean et al. 2005; Soanes et al. 2007; Jiang et al. 2018). For example, Ubc3, a key gene involved in the MAPKs pathway of Ustilago maydis, is related to its hyphal growth and regulates its pathogenicity by interacting with Prf1 (Mayorga and Gold 2010). Smut_0057 is the homeotic gene of Ubc3 and may be associated with the pathogenicity of T. horrida. G protein-coupled receptors (GPCRs) and G-proteins are important proteins that involved in the MAPKs pathway (Dean et al. 2005; Soanes et al. 2007). Two nonsynonymous SNPs were found in the GPCR proteins smut_1863 and smut_4953 of the three weakly pathogenic strains vis-à-vis a strongly pathogenic strain, with smut_4953 found up-regulated by T. horrida inoculation (Wang et al. 2020a). In addition, several putative secondary metabolites-related genes are up-regulated during host infection by T. horrida such as smut_2974 that is associated with sterigmatocystin biosynthesis (Wang et al. 2020a). Hence, we speculated that sterigmatocystin is necessary for the pathogenicity of T. horrida. Besides, the cytochrome P450s-encoding genes associated with secondary metabolites possess many nonsynonymous SNPs when weakly versus strongly pathogenic strains are compared, implying that these genes contribute to T. horrida pathogenicity (Wang et al. 2018b; 2020a). These findings emphasize the essentiality of secondary metabolites for conferring pathogenicity to T. horrida.
Secreted Proteins and Effectors
Effectors are crucial pathogenicity factors that suppress plant immunity and promote the successful infection of pathogens. Through comparative genomics and secretome analyses, 597 secreted proteins were predicted for the T. horrida genome (Wang et al. 2018b). Of the 597, 367 are small (< 400aa) cysteine-rich (SCR) secreted proteins; among the latter, the up-regulation of 131 SCRs was induced by T. horrida infection and these are considered candidate effectors (Wang et al. 2018b). The highly conserved effector gsr1 in T. horrida, which contains four conserved RNase active sites, is known to trigger cell death and an immune response in Nicotiana benthamiana, the former requiring the RNase active site (Wang et al. 2019). The effector uan2, which is unique to T. horrida—its homologous protein has not been found in other fungi—also triggers non-host cell death and an immune response when transiently expressed in N. benthamiana, and the predicted signal peptide of uan2 is essential for its cell death-inducing ability (Wang et al. 2019). Both effectors undergo up-regulation when T. horrida infects a host plant (Wang et al. 2019). Thus, according to transcriptome data, 26 putative effectors that presented the same expression pattern as gsr1 and uan2 were subsequently detected (Wang et al. 2019). The LysM effectors are widely distributed in plant pathogens and probably figure prominently in pathogenesis, with some identified in many fungi (Dölfors et al. 2019; Bolton et al. 2008; de Jonge et al. 2010; Gruber et al. 2011). In the T. horrida genome, seven putative LysM effectors (smut_1650, smut_2305, smut_4126, smut_4802, smut_5680, smut_7273, and smut_7501) have been annotated (Wang et al. 2018b). Recently, the putative LysM effectors smut_1650 was shown to induce cell death in N. benthamiana (Shu et al. 2021). Moreover, some fungal glycoside hydrolases and chitinases were recently confirmed to function as effectors (Zheng et al. 2013; Han et al. 2019; Yang et al. 2019). For example, MpChi in Moniliophthora perniciosa encodes a secreted enzymatically-inactive chitinase consisting of 438 aa residues, to sequester chitin-triggered immunity. In T. horrida, a glycoside hydrolase family effector containing 268 aa residues with four cysteines has been identified to trigger non-host cell death and an immune response (Shu and Wang unpublished data). Interestingly, like uan2, multiple predict effectors that induce non-host cell death and an immune response unique to T. horrida have been predicted to exist (Jiang and Wang unpublished data). To sum up, the effectors in T. horrida fulfill important roles in its pathogenicity. The knock-out of effector proteins-encoded gene in U. virens also confirmed its functioning in conferring virulence and pathogenicity to this biotrophic pathogens (Sun et al. 2020). Next, it is imperative we identify the pathogenicity functions of effectors in T. horrida by using gene-knockout techniques, and more importantly, to identify the host targets of these T. horrida effectors, which could be utilized as elegant probes for the identification of potentially resistant proteins in plants.
Management and Control Measures
As the incidence of RKS increases, measures for the effective control of this disease are urgently needed. In this respect, researchers have tried many approaches (Biswas 2001; Wang et al. 2018a, b, c, d, e; Yang et al. 2022; Jiang et al. 2021; Nataraja 2002; Brinck and Gärdenfors 2007), including the mining of resistance traits in rice cultivars or genes, chemical fungicides control, different cultivation practices, and better monitoring protocols and early warning systems. Yet research into breeding resistance in rice to RKS is proceeding slowly.
Searching for Resistant Rice Cultivars or Resistance Genes
Detecting the RKS-resistant rice cultivars or genes and then breeding disease-resistant lines are considered the most efficient, economical, and environmentally friendly methods for disease control. But it is difficult to assess RKS resistance in field settings due to immature inoculation methods and the instability of environmental conditions. Natural infection in the field is one of the most commonly used methods to evaluate the disease resistance of a crop (Wang et al. 2018a, b, c, d, e; Deng et al. 1997). To evaluate RKS, the rice plants are grown in a field under standard field management conditions without any fungicide application. Then, at crop maturity, the incidence of RKS disease is determined by counting the number of single-ear affected grains and deriving an infected seed rate, or by using evaluation criteria as described by Deng et al. (1997). It should be noted that such natural infection experiments should be performed across years and sites because the RKS incidence varies under different environmental conditions. However, the identification of RKS-resistant rice plants through artificial inoculation under controlled conditions is more efficient and reproducible than pursuing it under natural infection (Cartwright et al. 1996). The methods of artificial inoculation mainly include spore suspension spraying and single floret injection. Concerning the former, inocula were produced with ∼5–7 days, through mycelia of T. horrida grown on potato sucrose fluid medium, and spore concentration is adjusted to 1 × 106 conidia/mL with sterilized potato sucrose fluid medium. At the blooming stage of a rice plant, the spore suspension is sprayed onto the stigma of a floret with a pocket-sized sprayer, but the glume must be cut open and the stigma was exserted. For floret injection, the spore suspension is injected into a rice floret with a syringe at the late booting stage. After inoculation, rice male sterile lines need to be pollinated and are maintained at 28 °C and at least 95% relative humidity. Several rice male sterile lines featuring resistance to RKS in the field have been detected and these may be used in mining resistance genes and disease-resistance breeding through artificial inoculation experiments (Wang et al. 2018a, b, c, d, e; Zong et al. 2004; Akhtar and Sarwar 1988). The RKS resistance of 16 rice cultivars was determined at the anthesis stage using an artificial inoculation method: the disease incidence ranged from 6.25% to 97.25% at Sheikhupura and from 5.25% to 95.25% at Gujranwala, with the C-622 cultivar presenting the highest resistant level (Akhtar and Sarwar 1988). Earlier, the rice line IR-579 was also found to harbor resistance to T. horrida (Singh and Pavgi 1970). Yu and colleagues (2004) examined the RKS resistance of 10 photo-thermo-sensitive genic male sterile rice lines, found that their resistance levels differed markedly, with line S25 not infected by kernel smut disease. Recently, Wang and colleagues (2018c) evaluated the RKS resistance of 78 rice male sterile lines in the field over 3 years; their results also indicated that the resistance levels of different rice lines varied significantly, among which four lines that present more than medium resistance 4766A, Jiangcheng3A, Jufeng2A, and TianfengA were obtained. However, genes or quantitative trait loci (QTLs) involved in RKS resistance are rarely discussed (Dai et al. 2011; Liu 2008). Using the F2 population from a cross between the resistant line Jiangcheng 3B and the highly susceptible cultivar 9311B, resistance QTLs were mapped to chromosome 6 and 11 by bulk segregate analysis, and this further identified a resistant gene located in chromosome 11 when combined with transcriptome and transgenesis techniques (Wang and Zheng unpublished data). Recently, five QTLs for RKS resistance were mapped using a recombinant inbred lines population constructed by the resistant cultivar W9593S and the susceptible cultivar Aipei64S, using a composite interval mapping method (Yang et al. 2022). Among these five QTLs, qRRKS-11 and qRRKS-8 respectively explained 13.64% and 12.07% of the phenotypic variation (Yang et al. 2022).
The disparate transcriptome responses between resistant and susceptible rice male sterile lines against T. horrida infection indicate that encoding calmodulin-like proteins genes OsCML7 and OsCML14, involved in reactive oxygen species burst genes Os09g0467200, Os01g0369700, and Os01g0949800, the NADPH oxidase encoded gene Osrboh9, and the salicylic acid signaling pathway-related gene OsNPR1 may all contribute to RKS resistance (Wang et al. 2020b; 2018d). Interestingly, two flowering time regulation genes in rice, OsCTR2 and OsRR1, were exclusively up-regulated in the resistance line after T. horrida inoculation, and the overexpression these two genes resulted in a late-flowering phenotype (Wang et al. 2013; Cho et al. 2016). And flowering time regulation genes OsABF1, which RNA interference lines present late-flowering phenotype (Zhang et al. 2016a, b), was down-regulated in resistance line, but not induced expression in susceptible line. Field observations suggest that rice cultivars with a short-duration of flowering often have a greater kernel smut disease incidence than do long-duration flowering cultivars. Muthusamy and Ahmed (1977) detected the responses of 10 early-maturing and 9 later-maturing rice varieties to infection with T. horrida, finding that those with a shorter growing season were more likely to get infected than those with a longer growing season. Therefore, the delaying of flowering or start of the growing season should be considered in the control of RKS in rice production.
Chemical Fungicides Control
In fact, RKS disease control largely relies on fungicide applications during the typical production of hybrid rice (Tang et al. 2010; Feng and Lu 2009; Grewal et al. 1996; Hakro et al. 1988). Among many of tested fungicides, trifloxystrobin tebuconazole WG, benazoxystrobin SC, acetobacter fluconazole, diniconazole carbendazim WP, Keheijing, and Kanghei 95 are all highly effective against T. horrida infection and have been utilized in controlling kernel smut disease (Ding et al. 2020; Ye et al. 2005; Zhang et al. 1998). The ensuing control effect is often higher for mixed application of multiple fungicides than a single one. For example, Kanghei 95, a mixture of Jinggangmeisu, carbendazim, triadimefon, KH2PO3, and sodium borate, exerted a preventive effect against RKS of more than 90%, which exceeded that obtained by the individual application of Jinggangmeisu and carbendazim (Ye et al. 2005). In particular, the timing of fungicide application is pivotal for high efficacy in the control of RKS. The best practices for fungicide spraying in RKS management is to apply once from the end of boot stage to the initial heading stage, and once again at the early flowering stage, and one more time at the peak flowering stage, respectively (Honkura and Osada 1993; Yang et al. 2001; Shu et al. 1993). Additionally, using a biotic pesticide is an environmentally friendly methods to control plant diseases. It is well-known that many Bacillus subtilis and Trichoderma strains are used to successfully manage a suite of plant diseases (Andargie et al. 2017; El-Naggar et al. 2015). For example, several Trichoderma spp., such as T. viride, T. harzianum, and T. koningii have high rice false smut control efficiencies ranging from 46.8% to 100% (Andargie et al. 2017; El-Naggar et al. 2015; Liang et al. 2014). For T. horrida, El-Kazzaz et al. (2015) found that a Bacillus pumilus strain markedly presented promising characteristics for controlling the RKS. However, biocontrol microorganisms that control RKS have not been described; hence, it is necessary to strengthen this aspect in future studies.
Cultivation Practices
Excessive applications of nitrogen fertilizer and a high humidity environment will promote the incidence of RKS (Slaton et al. 2004). Thus, curbing nitrogen fertilization and modified planting density, as well as establishing a field microenvironment that is detrimental to the pathogen’s growth and development, could be effective measures to control RKS in rice production. Moreover, the teliospores of T. horrida can overwinter epiphytically on surviving rice plants, and act as the primary source of infection the next year. Accordingly, eliminating plants’ survival in field and irrigation canals could reduce the primary infection reservoir of RKS. The paddy (rice)-upland rotation (with a drought crop, such as maize), along with furrow irrigation, rational fertilization, appropriate early sowing, and cultivating strong cuttings are also effective at suppressing the occurrence rate of RKS (Slaton et al. 2007; Brooks et al. 2009; Kalboush et al. 2018).
Monitoring and Early Warning
Because obvious disease symptoms are absent during the initial stage of T. horrida infection, implementing control measures when the symptoms appear is too late to avoid yield losses. Thus, monitoring protocols and early warning systems are both very important for managing kernel smut in situ. Chen et al. (2016) reported on specific internal transcribed spacer (ITS) primers, which can be used to detect the presence of T. horrida in the floret of rice at the early inoculation stage. This finding could be very useful for devising rational and effective control measures. In fact, integrated multiple management methods are recommended for those rice-growing areas incurring a high kernel smut incidence.
Study of Tilletia Species
The genus Tilletia is a grass disease fungus infecting cereal crop either locally or systemically (Carris et al. 2006). The cereal-infecting Tilletia species that forms teliospores in the ovaries of their hosts are defined as bunt fungi (Bishnoi et al. 2020; Muhammad et al. 2013). Among these Tilletia species, there are five pathogens capable of infecting economically important crops; except T. horrida, the other four can also infect wheat plants. Although T. caries and T. laevis are responsible for common bunt of spring and winter wheat crops, the teliospore wall structure of these two fungi is not the same: T. laevis has a smooth surface and T. caries has a reticulated surface (Zhang et al. 2002). T. controversa is a quarantine-listed pest in many countries and causes dwarf bunt of autumn-planted wheat, yet it has never been found on spring-planted wheat (Xu et al. 2021). The symptoms caused by T. controversa in wheat are similar to T. caries, but these two smut pathogens differ in the composition of their teliospore structures (Zhang et al. 2002). The teliospores of T. controversa have a conspicuous hyaline gelatinous sheath whose thickness is 1.5–5.5 μm. The paramount smut fungus infecting wheat is Karnal bunt, caused by T. indica, whose teliospores also are covered by a conspicuous hyaline gelatinous sheath, and densely echinulate or finely cerebriform distributed on their surface (Pady et al. 1961; Zhang et al. 2002). Moreover, like T. horrida, these four fungi also survive on the seed surface of host seeds and in soil, and diseased seeds are the most important potent source of infection. However, these four pathogens infect wheat hosts at the seedling stage, in stark contrast to T. horrida (Zhang et al. 2002).
The high-quality genome sequences of four Tilletia species mentioned are now available. The assembled genome size of T. controversa, T. caries, T. laevis, and T. indica is ca. 49.87, 35.8, 28.78, and 37.46 Mb, these encoding 10459, 10043, 9799, and 9664 predicted proteins, respectively (Kumar et al. 2017; Gurjar et al. 2019; https://ncbi.nlm.nih.gov/). The genetic diversity of the 20 T. indica strains that isolated from different locations in Indian were also detected using seven multilocus sequence fragments, the results revealed that the population of T. indica was highly diverse (Gurjar et al. 2021). However, functional research of the virulence or effector proteins in these smut fungi very limited. For the control of these diseases, like that caused by T. horrida, much progress has been made by researchers (Kumar et al. 2016; Tan and Murray 2006; Gupta et al. 2019; Singh et al. 2020; Gurjar et al. 2022). For example, many wheat varieties, including Paroli, Gluten, NGB-9015, Maribos, PG3540, and Kranich, are described as being resistant to T. caries, and several major genes in them have been are identified as able to control this disease (Gupta et al. 2019). Synthetic hexaploid wheats, derived from Triticum turgidum × T. tauschii, is resistant to T. indica (Villareal et al. 2010). With the rapid development of high-throughput technologies, more genome-wide association studies (GWAS), bulk segregant analysis (BSA) and transcriptomic analyses, have been carried out to to detect the genes or locus that are resistant to these smut diseases in wheat (Singh et al. 2020; Gupta et al. 2019). Furthermore, a loop-mediated isothermal DNA amplification tool that can used be to quickly detect T. caries, T. laevis, and T. controversa in wheat grain was also reported on (Pieczul et al. 2018). This provides a crucial method to manage these smut diseases through their in situ monitoring in fields as part of an early warning system.
Conclusions
RKS has emerged as globally important disease, causing serious yield losses to hybrid rice seed production in recent years. Developing disease-resistant varieties using resistant genes is often viewed as the most economic and effective strategy to control this worsening disease threat. However, no RKS resistance gene has yet been identified and there are only few reports of mapping of RKS resistance QTLs, leaving the major QTLs with high phenotypic variance unfound. Consequently, we lack reports addressing RKS resistance breeding that would utilize those QTLs. The principal reason for this phenomenon is less innate resistance in rice germplasms and mapping populations currently available for RKS study. But now, with the rapid development of sequencing technology, it is easier to explore natural variation that associated with RKS resistance in hidden defense-related QTLs or genes among rice male sterile lines. Therefore, upcoming studies should focus on detecting effects of QTLs or genes that are involved in the RKS resistance trait, and further pyramiding several favorable consistent QTL alleles into a single cultivar to enhance the overall RKS resistance of rice male sterile lines is crucial.
Researching the function of effectors or virulence factor genes from T. horrida’s genome sequence has significantly contributed towards a better understanding of its pathogenesis and for subsequently developing sound management strategies. Although the candidate effectors of T. horrida have been predicted based on its available genome sequence (Wang et al. 2018a, b, c, d, e), their transgene study has not been performed to characterize its pathogenicity. The CRISPR-mediated genome editing technologies have enormous potential to validated the pathogenic functioning of effectors in fungi (Molla and Yang 2019; Pickar-Oliver and Gersbach 2019). To accomplish this target, a standardized genetic transformation system of T. horrida needs to be established.
Furthermore, to enhance the disease resistance of rice germplasms, impairing expression of pathogenicity-related genes in pathogen by using their small interference RNAs (siRNAs) sequences inserted into rice plants is a promising strategy. For example, the UvAspE, UvCom1, and UvPro1 are three virulence-related proteins of U. virens, for which the insertion of their siRNAs’ fragments into the rice cultivar Nipponbare significantly augmented its resistance to U. virens (Chen et al. 2021). The role of effectors siRNAs of T. horrida in RKS resistance are still poorly explored, however. Thus, analyzing the role of T. horrida’s effectors siRNAs in conferring host resistance would significantly advance our understanding of how to better manage this disease.
Detecting resistant proteins that recognize pathogen effectors using modern molecular biology experimental techniques, such as yeast-two-hybrid system, bimolecular fluorescence complementation, and co-immunoprecipitation (Wang et al. 2018a, b, c, d, e) will also help to control RKS and improve our understanding of T. horrida–rice interactions. Yet such study is hindered by the lack of virulence gene resources. Thus, we should enhance the study of antifungal proteins, which recognize pathogen effectors, as this could provide insight for clarifying the molecular mechanism underpinning the interaction of T. horrida and rice. Despite all this, any possible methods that reliably control RKS disease should not be ignored in the near future.
Like RKS, the diseases caused by T. controversa, T. caries, T. laevis, and T. indica are problematic, causing economical losses by extensively impairing wheat crop health and quality worldwide. For these Tilletia fungi, mechanism study of the molecular interaction between the pathogen and host are also limited. As mentioned above, the future studies should focus on distinguishing the effects of resistant QTLs or genes, the function of effectors based on the published genomic data of the involved Tilletia fungus, and further identify potential resistant proteins in host plants. More important is applying resistant genes of host and siRNAs of pathogen effectors to resistance breeding effectively, this being a necessary measure to settle the problem to food security posed by these diseases. In this way, not only may we gain the effective preventive and control of these smut diseases but also better understand the molecular mechanisms of their interactions between host and pathogen.
Availability of data and materials
Not applicable.
References
Akhtar MA, Sarwar M (1987) Incidence of rice kernel smut (KSm) in Pakistan. Int Rice Res Newsletter 12:4–15
Akhtar MA, Sarwar M (1988) Rice cultivars resistance to kernel smut. Pak J Agric Res 9(2):266–267
Andargie M, Congyi Z, Yun Y, Li J (2017) Identification and evaluation of potential bio-control fungal endophytes against Ustilagonoidea virens on rice plants. World J Microbiol Biotechnol 33:120
Ayado T, Yano T, Yoshitake K, Komatsu T (1993) On the kernel smut (Tilletia barclayana Brefeld) of rice plant which occurred in Fukuoka Prefecture in 1992. Kyushu Plant Protect Res 39:8–10
Bishnoi SK, He XY, Phuke RM, Kashyap PL, Alakonya A, Chhokar V, Singh RP, Singh PK (2020) Karnal bunt: a re-emerging old foe of wheat. Front Plant Sci 11:569057
Biswas A (2001) Field reaction of hybrid rice varieties to false smut FSm and kernel smut KSm diseases in West Bengal, India. Environ Ecol 21:336–351
Biswas A (2003) Kernel smut disease of rice: current status and future challenges. Environ Ecol 21:336–351
Bolton MD, van Esse HP, Vossen JH, de Jonge R, Stergiopoulos I, Stulemeijer IJE, van den Berg GCM, Borrás-Hidalgo O, Dekker HL, de Koster CG, de Wit PJGM, Joosten MHAJ, Thomma BPHJ (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol Microbiol 69:119–136
Brinck I, Gärdenfors P (2007) Measures for preventing and controlling kernel smut in hybrid rice seed production. Hybrid Rice 18(5):484–501
Brooks SA, Anders MM, Yeater KM (2009) Effect of cultural management practices on the severity of false smut and kernel smut of rice. Plant Dis 93(11):1202–1208
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238
Carris LM, Castlebury LA, Goates BJ (2006) Nonsystemic bunt fungi-Tilletia indica and T. horrida: a review of history, systematics, and biology. Annu Rev Phytopathol 44:113–133
Cartwright RD, Lee FN, Crippen DL, Eason R, Templeton GE (1996) Artificial inoculation of kernel smut of rice in Arkansas. Res Ser Arkansas Agric Exp Stat 453:107–110
Chahal SS (2001) Epidemiology and management of two cereal bunts. Indian Phytopathol 54:145–157
Chao J, Jin J, Wang D, Han R, Zhu RS, Zhu YG, Li SQ (2014) Cytological and transcriptional dynamics analysis of host plant revealed stage-specific biological processes related to compatible rice–Ustilaginoidea virens interaction. PLoS ONE 9:e91391
Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, Chermak D, Antony G, White FF, Somerville SC, Mudgett MB, Frommer WB (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468:527–532
Chen Y, Yang X, Yao J, Kyaw EP, Zhang AF, Li YF, Gu CY, Zang HY, Gao TC (2016) Simple and rapid detection of Tilletia horrida causing rice kernel smut in rice seeds. Sci Rep 6:33258
Chen XY, Pei ZX, Liu H, Huang JB, Chen XL, Luo CX, Hsiang T, Zheng L (2021) Host-induced gene silencing of fungal-specific genes of Ustilaginoidea virens confers effective resistance to rice false smut. Plant Biotechnol J 20(2):253–255
Cho LH, Yoon J, Pasriga R, An G (2016) Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol 170:2159–2171
Choi J, Park SY, Kim BR, Roh JH, Oh IS, Han SS, Lee YH (2013) Comparative analysis of pathogenicity and phylogenetic relationship in Magnaporthe grisea species complex. PLoS ONE 8:e57196
Dai L, Zhang DH, Yu LF, Wang WX, Ye ZH, Zhang AF, Chen Y (2011) Advance in rice kernel smut. Chin Agric Sci Bull 27(12):261–265
de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MHAJ, Thomma BPHJ (2010) Conserved fungal LysM efector Ecp6 prevents chitin-triggered immunity in plants. Science 329:953–955
Dean RA, Talbot NJ, Ebbole DJ, Farman ML, Mitchell TK, Orbach MJ, Thon M, Kulkarni R (2005) The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434:980–986
Deng GS, Yang ZH, Chen JF (1997) Study on standardized index for grain smut rice. Acta Phytophylacica Sinica 24(2):189–190
Ding SF, Chen H, Zhu XB, Wu HX, Che JY, Dian KY, Cui JH (2020) The control effect experiment of several fungicides for rice kernel smut. Anhui Agri Sci Bull 26(24):116–117
Dölfors F, Holmquist L, Dixelius C, Tzelepis G (2019) A LysM efector protein from the basidiomycete Rhizoctonia solani contributes to virulence through suppression of chitin-triggered immunity. Mol Genet Genomics 294:1211–1218
El-Kazzaz MK, Salem EA, Ghoneim KE, Elsharkawy MM, Kalboush ZA (2015) Biocontrol of Tilletia barclayana, the causal agent of kernel smut disease in rice. Egypti J Biol Pest Control 25(3):535–544
El-Naggar MM, Elsharkawy MM, Almalla RA, El-Kot GAN, Alwakil AM, Badr MM (2015) Control of Ustilaginoidea virens, the causal agent of rice false smut disease in Egypt. Egypt J Pest Control 25:555–564
Fan J, Guo XY, Li L, Huang F, Sun WX, Li Y, Huang YY, Xu YJ, Shi J, Lei Y, Zheng AP, Wang WM (2015) Infection of Ustilaginoidea virens intercepts rice seed formation but activates grain-filling-related genes. J Integr Plant Biol 57:577–590
Feng FB, Lu Q (2009) The occurrence reasons and control measures of rice kernel smut (Neovossia horrida) in Zhong 9A seed production. J Guangxi Agric 24(3):25–26
Gade RM (2000) Occurrence of rice kernel smut in Vidarbha region of Maharashtra. GK Giri RM Gade CU Patil J Soils Crops 10(2):309–310
Giri GK, Gade RM, Patil CU (2000) Occurrence of rice kernel smut in Vidarbha region of Maharashtra. J Soils Crops 10(2):309–310
Gong BQ, Wang FZ, Li JF (2020) Hide-and-seek: chitin-triggered plant immunity and fungal counterstrategies. Trends Plant Sci 25:805–816
Grewal RK, Chahal SS, Aulakh KS (1996) Effectiveness of triazole fungicides against kernel smut of rice. Indian Phytopathol 49(4):404–405
Grewal TS, Rossnagel BG, Scoles GJ (2004) Mapping of a covered smut resistance gene in barley (Hordeum vulgare). Can J Plant Path 26(2):156–166
Gruber S, Vaaje-Kolstad G, Matarese F, Lopez-Mondejar R, Kubicek CP, Seidl-Seiboth V (2011) Analysis of subgroup C of fungal chitinases containing chitin-binding and LysM modules in the mycoparasite Trichoderma atroviride. Glycobiology 21:122–133
Gupta V, He X, Kumar N, Fuentes-Davila G, Sharma RK, Dreisigacker S, Juliana P, Ataei N, Singh PK (2019) Genome wide association study of karnal bunt resistance in a wheat germplasm collection from Afghanistan. Int J Mol Sci 20:3124
Gurjar MS, Aggarwal R, Jogawat A, Kulshreshtha D, Sharma S, Solanke AU, Dubey H, Jain RK (2019) De novo genome sequencing and secretome analysis of Tilletia indica inciting Karnal bunt of wheat provides pathogenesis-related genes. 3 Biotech 9(6):219
Gurjar MS, Aggarwal R, Jain S, Sharma S, Singh J, Gupta S, Agarwal S, Saharan MS (2021) Multilocus sequence typing and single nucleotide polymorphism analysis in Tilletia indica isolates inciting karnal bunt of wheat. J Fungi 7:103
Gurjar MS, Jain S, Aggarwal R, Saharan MS, Kumar TPJ, Kharbikar L (2022) Transcriptome analysis of wheat–Tilletia indica interaction provides defense and pathogenesis-related genes. Plants 11:3061
Hakro AA, Akhtar MA, Sarwar M (1988) Chemical control of kernel smut of rice. Pak J Agric Res 3:422–423
Hamer JE, Farrall L, Orbach MJ, Valent B, Chumley FG (1989) Host species-specific conservation of a family of repeated DNA-sequences in the genome of a fungal plant pathogen. Proc Natl Acad Sci USA 86:9981–9985
Han YJ, Song LL, Peng CL, Liu X, Liu LH, Zhang YH, Wang WZ, Zhou J, Wang SH, Ebbole DJ, Wang ZH, Lu GD (2019) A Magnaporthe chitinase interacts with a rice jacalin-related lectin to promote host colonization. Plant Physiol 179:1416–1430
Honkura R, Osada S (1993) The time of chemicals application for control of kernel smut of rice plant. Annu Rep Soc Agric Chem North Jpn 44:10
Huang F, Pan XX, Wang YH, Cheng KL, Zhang CW (1995) Effects of host factors on the occurrence of rice kernel smut. Southwest China J Agric Sci 8(1):53–58
Huang HC, Kokko EG, Erickson RS (1999) Infection of alfalfa pollen by Botrytis cinerea. Bot Bull Acad Sin 40:101–106
Huang F, Pan X, Cheng K, Liu X (2003) Studies on the epiphytic budding of secondary sporidia of Neovossia horrida causing rice kernel smut on the plant surfaces. Southwest China J Agric Sci 16(2):59–62
Iguchi K, Fukuda H, Izuki A, Murakami K, Nakagawa M (1987) Occurrence of kernel smut of rice caused by Tilletia horrida in Chiba Prefecture. Annu Rep Kanto-Tosan Plant Protect Soc 34:13–14
Imbaby IA, Eldaoudi YH (2002) Sources of resistance for wheat, Triticum aestivum loose smut, caused by, Ustilago tritici, Pers. Assiut. J Agric Sci 33(2):209–220
Jiang C, Zhang X, Liu HQ, Xu JR (2018) Mitogen-activated protein kinase signaling in plant pathogenic fungi. PLOS Pathog 14:e1006875
Jiang YQ, Shu XY, Zheng AP, Wang AJ (2021) Recent progress in molecular mechanism of interaction between rice and Tilletia horrida. Biotechnol Bull 37(9):248–254
Kalboush Z, Ghoneim A, Hassan A (2018) Influence of mineral and organic fertilizers on rice kernel smut disease. J Plant Protect Pathol 9(7):431–440
Kämpe J, Kahmann R, Bölker M, Ma LJ, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Muller O (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97–101
Kato H, Yamamoto M, Yamaguchi-Ozaki T, Kadouchi H, Iwamoto Y, Nakayashiki H, Tosa Y, Mayama S, Mori N (2000) Pathogenicity, mating ability and DNA restriction fragment length polymorphisms of Pyricularia populations isolated from Gramineae, Bambusideae and Zingiberaceae plants. J Plant Pathol 66:30–47
Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, Bailey K, Holub EB, Studholme DJ, MacLean D, Jones JDG (2011) Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLOS Biol 9:e1001094
Khanal S, Antony-Babu S, Gaire SP, Zhou XG (2022) Multi-locus sequence analysis reveals diversity of the rice kernel smut populations in the United States. Front Microbiol 13:874120
Kumar S, Mishra CN, Gupta V, Singh R, Sharma I (2016) Molecular characterization and yield evaluation of near isogenic line (NIL) of wheat cultivar PBW 343 developed for Karnal bunt resistance. Indian Phytopath 69:119–123
Kumar A, Pandey V, Singh M, Pandey D, Saharan MS, Marla SS (2017) Draft genome sequence of Karnal bunt pathogen (Tilletia indica) of wheat provides insights into the pathogenic mechanisms of quarantined fungus. PLoS ONE 12(2):e0171323
Liang Y, Zhang X, Li D, Huang F, Hu P, Peng Y (2014) Integrated approach to control false smut in hybrid rice in Sichuan Province. China Rice Sci 21:354–360
Liu H (2008) Research advance on Neovossia horrida in rice. Jiangxi Plant Prot 31(1):3–6
Mayorga ME, Gold SE (2010) A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol Microbiol 34(3):485–497
Mohamed EK, Gabr EK, Kamal EG, Mohsen ME, Zeinab AEK, Mitsuro H (2014) Incidence of kernel smut caused by Tilletia barclayana in Egyptian rice cultivars. Afr J Microbiol Res 8(32):3052–3063
Molla KA, Yang Y (2019) CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol 37:1121–1142
Muhammad A, Muhammad R, Muhammad S, Aftab B, Muhammad I (2013) Response of some commercial cultivars and advanced lines of wheat against Karnal bunt of wheat and its management through chemicals. Int J Plant Res 3:47–51
Muthusamy M, Ahmed NJ (1977) Varietal susceptibility of rice to kernel smut [India]. Madras Agric J 64(10):692
Nasiru I, Ifenkwe OP (2004) Screening of bi-parental and mutant clones of sugarcane Saccharum officinarum L. for resistance to smut disease. Tropicultura 22(4):173–175
Nataraja S (2002) Biology and control of seed borne diseases of rice with special reference to kernel smut and stack burn diseases. Doctor’s Thesis
Ngugi HK, Scherm H (2006) Biology of flower-infecting fungi. Annu Rev Phytopathol 44:261–282
Pady SM, Duran R, Fischer GW (1961) The genus Tilletia. Mycologia 53(2):213
Pickar-Oliver A, Gersbach CA (2019) The next generation of CRISPR–Cas technologies and applications. Nat Rev Mol Cell Biol 20:490–507
Pieczul K, Perek A, Kubiak K (2018) Detection of Tilletia caries, Tilletia laevis and Tilletia controversa wheat grain contamination using loop-mediated isothermal DNA amplification (LAMP). J Microbiol Methods 154:141–146
Ribeiro M, Costa W, Drummond OA (1973) Occurrence of kernel smut or bunt of rice (Oryza sativa L.) in the state of Rio de Janeiro. Arquivos Da Universidade Federal Rural Do Rio De Janeiro 1–6.
Rogerson CT (1988) Illustrated genera of smut fungi. Brittonia 40:107
Roux C, Almaraz T, Durrieu G (1998) Phylogeny of some smuts fungi based on ITS [International transcribed spacer] sequence analysis. Comptes rendus de I Académie des Sciences. Series 3. Sciences De La Vie 321:603–609
Royer MH, Rytter J (1988) Comparison of host ranges of Tilletia indica and T. barclayana. Phytopathology 72:133–136
Sanchez-Vallet A, Tian H, Rodriguez-Moreno L, Valkenburg DJ, Saleem-Batcha R, Wawra S, Kombrink A, Verhage L, Jonge R, Esse HP, Zuccaro A, Croll D, Mesters JR, Thomma BPHJ (2020) A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization. PLoS Pathog 16:e1008652
Shao JY, Zhang XX, Lin SS (1997) Study on biological characteristics of Neovossia horrida. Acta Agric Jiangxi 9(4):27–32
Sharif MK, Butt MS, Anjum FM, Khan SH (2014) Rice bran: a novel functional ingredient. Crit Rev Food Sci Nutr 54:807–816
Sharma RC (1999) Kernel smut—A major constraint in hybrid seed production of rice and its remedial measures. Seed Res 27:82–90
Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, Shibuya N (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64:204–214
Shu ZY, Wang ZK, Ouyang Z (1993) The epiphytic stage of rice kernel smut caused by Neovossia horrida in producing rice hybrid seeds for fields planting. Acta Phytopathologica Sinica 23(3):209–210
Shu XY, Jiang YQ, Jiang B, Wang AJ, Zheng AP (2021) Cloning, expression and sequence analysis of carbohydrate gene 1650a of rice kernel smut pathogen Tilletia horrida. In: Proceedings of the Annual Meeting of Chinese Society for Plant Pathology page 172.
Singh R (1998) Kernel Smut of rice: present status. Int J Trop Plant Dis 16(2):149–167
Singh RA, Pavgi MS (1970) Varietal reaction and resistance to kernel bunt of rice. Indian Phytopathol 23:51–53
Singh R, Pavgi M (1972) Cytology of teliospore germination and development of Neovossia horrida. Riso 21:259–268
Singh CS, Pps P, Singh G (1999) Multiplication and infectivity of sporidia of Tilletia barclayana, the causal organism of kernel smut of rice. Indian Phytopathol 52(1):35–38
Singh S, Sehgal D, Kumar S, Arif M, Vikram P, Sansaloni CP, Fuentes-Dávila G, Ortiz C (2020) GWAS revealed a novel resistance locus on chromosome 4D for the quarantine disease Karnal bunt in diverse wheat pre-breeding germplasm. Sci Rep 10:5999
Slaton NA, Gbur EE, Cartwright RD, Delong RE, Brye KR (2004) Grain yield and kernel smut of rice as affected by preflood and midseason nitrogen fertilization in Arkansa. Agron J 96(1):91–99
Slaton NA, Delong RE, Norman RJ, Cartwright RD, Wilson CE (2007) Cultivar and seeding date effects on kernel smut of rice. Agron J 99(2):521–529
Soanes DM, Richards TA, Talbot NJ (2007) Insights from sequencing fungal and oomycete genomes: what can we learn about plant disease and the evolution of pathogenicity? Plant Cell 19:3318–3326
Song JH, Wei W, Lv B, Lin Y, Yin WX, Peng YL, Schnabel G, Huang JB, Jiang DH, Luo CX (2016) Rice false smut fungus hijacks the rice nutrients supply by blocking and mimicking the fertilization of rice ovary. Environ Microbiol 18:3840–3849
Stirnberg A, Djamei A (2016) Characterization of ApB73, a virulence factor important for colonization of Zea mays by the smut Ustilago maydis. Mol Plant Pathol 17(9):1467–1479
Sun WX, Fan J, Fang AF, Li YJ, Tariqjaveed M, Li DY, Hu DW, Wang WM (2020) Ustilaginoidea virens: insights into an emerging rice pathogen. Annu Rev Phytopathol 9:37
Takahashi Y (1896) On Ustilago virens Cooke and a new species of Tilletia parasitic on rice plant. Tokyo Bot Mag 10:16–20
Tan M-K, Murray GM (2006) A molecular protocol using quenched FRET probes for the quarantine surveillance of Tilletia indica, the causal agent of Karnal bunt of wheat. Mycol Res 110:203–210
Tang YQ, Li Y, Li KK (2010) Occurrence characteristics and control strategy of rice kernel smut in hybrid rice seed production. Hybrid Rice 25(2):19–21
Tao JF, Zhou KD, Zhu JQ (1998) Infection process of Neovossia horrida in male sterile rice. Southwest China J Agric Sci 11(2):68–77
Templeton GE (1961) Local infection of Rice florets by the Rice kernel smut organism, Tilletia Horrida. Phytopathology 51(2):131–132
Teng B, Li GY, Teng JS, Teng SX (2002) Analysis of the causes and control measures of rice kernel smut in combination with Peiai 64S series. Seed 21(6):81–83
Uppala SS, Liu G, Liu B, Wu M, Zhou XG (2017) Texas Rice Kernel Smut Research P22.
Villareal RL, Mujeeb KA, Fuentes DG, Rajaram S, Toro E, Del TE (2010) Resistance to karnal bunt (Tilletia indica Mitra) in synthetic hexaploid wheats derived from Triticum turgidum×T. tauschii. Plant Breeding 112(1):63–69
Wang ZK, Ouyang Z (1989) Studies on the pathogenic biology of the kernel smut of rice. Southwest China J Agric Sci 11(4):331–335
Wang Q, Zhang W, Yin ZM, Ma B, Wen CK (2013) Rice constitutive triple-response2 is involved in the ethylene-receptor signalling and regulation of various aspects of rice growth and development. J Exp Bot 64:4863–4875
Wang N, Ai P, Tang YF, Zhang J, Dai XJ, Li P, Zheng AP (2015) Draft genome sequence of the rice kernel smut Tilletia horrida Strain QB-1. Genome Announc 3:e00621-e715
Wang AJ, Jiang B, Fu R, Wang N, Yin DS, Li P, Zhang J, Zheng AP (2018a) Identification and pathogenicity of pathogen causing kernel smut on rice. Acta Phytopathol Sin 48(3):297–304
Wang AJ, Pan LX, Wang N, Ai P, Yin DS, Li SC, Deng QM, Zhu J, Liang YY, Zhu JQ, Li P, Zheng AP (2018b) The pathogenic mechanisms of Tilletia horrida as revealed by comparative and functional genomics. Sci Rep 8:15413
Wang AJ, Yin DS, Fu R, Pan LX, Gu SS, Jiang B, Zheng AP (2018c) Evaluation of resistance to rice kernel smut in seventy-eight species of rice sterile lines. Acta Phytopathol Sin 48(2):207–212
Wang AJ, Shu XY, Niu XY, Zhao WJ, Ai P, Li P, Zheng AP (2018d) Comparison of gene co-networks analysis provide a systems view of rice (Oryza sativa L.) response to Tilletia horrida infection. PLoS ONE 13(10):e0202309
Wang Y, Xu YP, Sun YJ, Wang HB, Qi JM, Wan BW, Ye WW, Lin YC, Shao YY, Dong SM, Tyler BM, Wang YC (2018e) Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat Commun 9:594
Wang AJ, Pan LX, Niu XY, Shu XY, Yi XQ, Yamamoto N, Li SC, Deng QM, Zhu J, Liang YY, Wang LX, Li P, Zheng AP (2019) Comparative secretome analysis of different smut fungi and identification of plant cell death-inducing secreted proteins from Tilletia horrida. BMC Plant Biol 19(1):360
Wang AJ, Shu XY, Niu XY, Yi XQ, Zheng AP (2020a) Transcriptome analysis and whole genome re-sequencing provide insights on rice kernel smut (Tilletia horrida) pathogenicity. J Plant Pathol 102:155–167
Wang AJ, Zha ZP, Yin DS, Shu XY, Ma L, Wang LX, Li P, Zheng AP (2020b) Comparative transcriptome analysis of Tilletia horrida infection in resistant and susceptible rice (Oryza sativa L.) male sterile lines reveals potential candidate genes and resistance mechanisms. Genomics 112(6):5214–5226
Webster RK, Gunnell PS (1992) Compendium of rice diseases. Mycologia 84:953
Xu TS, Qin DD, Din GMU, Liu TG, Chen WQ, Gao L (2021) Characterization of histological changes at the tillering stage (Z21) in resistant and susceptible wheat plants infected by Tilletia controversa Kühn. BMC Plant Biol 21:49
Yang YF, Yang AP, Zhang YJ, Wang JK, Jiang CY (2001) Chemical control of kernel smut in hybrid seed production of Pei’ai 64S. Hybrid Rice 16(1):19
Yang C, Yu YQ, Huang JK, Meng FW, Pang JH, Zhao QQ, Islam MA, Xu N, Tian Y, Liu J (2019) Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell 31:172–188
Yang YS, Sun PY, Yu ML (2022) QTL mapping for resistance to rice kernel smut of male sterile line. Biotechnol Bull 38(3):16–21
Ye XY, Wang LH, Li SJ, Liu Z, Zhu JQ (2005) Effects of Kanghei95 on controlling rice kernel smut in hybrid rice seed production. Hybrid Rice 20(6):35–36
Yu SZ, Lv CG, Yao KM, Zou JS (2004) The kernel smut resistance of ten new photo-thermo sensitive rice male sterile. Jiangsu Agric Sci 3:18–19
Zhang L, Hu J, He ZF, Zhuo ZB (1998) Prevention and cure effect of Keheijin in hybrid rice breeding. J Anhui Agrotech Teachers Coll 12(2):24–29
Zhang GM, Qi PK, Zhang Z, Yao YJ (2002) Comparative morphology of Tilletia idica and ITS related species. Mycosystema 21(3):412–418
Zhang Y, Zhang K, Fang A, Han Y, Yang J, Xue M, Bao J, Hu D, Zhou B, Sun X (2014) Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat Commun 5:3849
Zhang HF, Zheng XB, Zhang ZG (2016a) The Magnaporthe grisea species complex and plant pathogenesis. Mol Plant Pathol 17(6):796–804
Zhang CY, Liu J, Zhao T, Gomez A, Li C, Yu CS, Li HY, Lin JZ, Yang YZ, Liu B, Lin CT (2016b) A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol 171:334–343
Zheng AP, Lin RM, Zhang DH, Qin PG, Xu LZ, Ai P, Ding L, Wang YR, Chen Y, Liu Y, Sun ZG, Feng HT, Liang XX, Fu RT, Tang CQ, Li Q, Zhang J, Xie ZL, Deng QM, Li SC, Wang S, Zhu J, Wang LX, Liu HN, Li P (2013) The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat Commun 4:1424
Zhu JQ, Zhou KD, Tao JF (1998) Cytological study on Neovossia horrida infection in male sterile lines of rice. Southwest China J Agric Sci 11(1):67–72
Acknowledgements
Not applicable.
Funding
The work is supported by National Natural Science Foundation of China (grants 32001490), Key Project of Wuhan Science and Technology Bureau (Grants 2022022202015037), and Key Project of Hubei Science and Technology Department (Grants 2022BBA001).
Author information
Authors and Affiliations
Contributions
AW proposed the concept; AW, XS, YJ and JL drafted the manuscript; DY, DX, XY, JZ, FY, CJ, AZ and PL revised and finalized the manuscript. All the authors have participated in the work sufficiently. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, A., Shu, X., Xu, D. et al. Understanding the Rice Fungal Pathogen Tilletia horrida from Multiple Perspectives. Rice 15, 64 (2022). https://doi.org/10.1186/s12284-022-00612-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-022-00612-1